Abstract
A DNA ligase IV (LIG4)-null human pre-B cell line and human cell lines with hypomorphic mutations in LIG4 are significantly impaired in the frequency and fidelity of end joining using an in vivo plasmid assay. Analysis of the null line demonstrates the existence of an error-prone DNA ligase IV-independent rejoining mechanism in mammalian cells. Analysis of lines with hypomorphic mutations demonstrates that residual DNA ligase IV activity, which is sufficient to promote efficient end joining, nevertheless can result in decreased fidelity of rejoining. Thus, DNA ligase IV is an important factor influencing the fidelity of end joining in vivo. The LIG4-defective cell lines also showed impaired end joining in an in vitro assay using cell-free extracts. Elevated degradation of the terminal nucleotide was observed in a LIG4-defective line, and addition of the DNA ligase IV–XRCC4 complex restored end protection. End protection by DNA ligase IV was not dependent upon ligation. Finally, using purified proteins, we demonstrate that DNA ligase IV–XRCC4 is able to protect DNA ends from degradation by T7 exonuclease. Thus, the ability of DNA ligase IV–XRCC4 to protect DNA ends may contribute to the ability of DNA ligase IV to promote accurate rejoining in vivo.
INTRODUCTION
DNA non-homologous end joining (NHEJ) represents the major mechanism for the repair of DNA double-strand breaks (DSBs) in mammalian cells and also effects rearrangements at the site-specific DSBs introduced during V(D)J recombination (1). Five proteins that function in NHEJ in mammalian cells have been identified to date (2). The heterodimeric Ku protein, comprising subunits Ku70 and Ku80, binds strongly to double-stranded DNA (ds DNA) ends. When DNA bound, Ku recruits the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) and thereby activates its kinase activity. Ku and DNA-PKcs together constitute the DNA-PK complex (3,4). An additional complex of DNA ligase IV and Xrcc4 (the DNA ligase IV–XRCC4 complex) is also required for NHEJ (1,2). Recently, a further protein, Artemis, was also shown to have some involvement in NHEJ (5). Mutants lacking these components show pronounced radiosensitivity, defective ability to repair DNA DSBs and impaired V(D)J recombination. Recently, hypomorphic mutations in DNA ligase IV were identified in some human patients displaying immunodeficiency and developmental delay (6).
Although NHEJ was first identified in mammalian cells, the process is also conserved in lower eukaryotes. However, whilst NHEJ is the major mechanism for the repair of DSBs in higher organisms, its contribution to overall DSB repair in yeast may be restricted to situations in which homologous recombination (HR) cannot function (7,8). HR is an efficient and accurate DSB rejoining process since any information lost at the break site is retrieved using the undamaged template. Currently, little is known about the level of accuracy achieved by NHEJ.
The characterization of NHEJ in yeast has relied upon an in vivo plasmid end-joining assay in which the repair of DSBs generated by restriction enzymes is examined using plasmids that lack homology with chromosomal sequences. A dramatic defect in plasmid rejoining is seen in NHEJ-defective yeast strains (9–11). This assay has proved an important tool to examine both the efficiency and fidelity of the rejoining process.
A plasmid-based assay has been widely used to characterize the process of V(D)J recombination in mammalian cells. It has been shown to be dependent upon the known NHEJ components and reflects events that occur at the endogenous loci (12–14). In contrast, the use of an in vivo plasmid assay to examine DSB rejoining in rodent cell lines lacking NHEJ components failed to demonstrate any significant defect (15–17). The plasmid-based host cell end-joining assay has also been used in human cells to examine the fidelity of DSB rejoining (18–22). However, to date, it is unclear whether plasmid rejoining in human cells reflects a process controlled by the known NHEJ components (Ku, DNA-PKcs, Xrcc4 and DNA ligase IV).
In this study, we show that a LIG4-null human pre-B cell line as well as cell lines derived from LIG4 syndrome patients are markedly impaired in in vivo plasmid end joining, demonstrating that the assay monitors NHEJ. The residual end joining observed in LIG4-null cells demonstrates the presence of an error-prone DNA ligase IV-independent end-joining process in mammalian cells. Additionally, one cell line with a hypomorphic mutation in DNA ligase IV retained sufficient DNA ligase IV activity to promote substantial rejoining, but the fidelity of rejoining was markedly impaired. Thus, in human cells, DNA ligase IV is an important factor influencing the fidelity of rejoining in vivo. An examination of end joining in vitro using cell-free extracts from LIG4-defective cell lines shows that the presence of DNA ligase IV can serve to protect the ends from DNA degradation as well as to rejoin them. We also show that the presence of DNA ligase IV–XRCC4 alone or in combination with Ku contributes to the protection of DNA ends from degradation by T7 exonuclease even in the absence of ligation. The ability of DNA ligase IV–XRCC4 to restrict degradation of DNA ends may contribute towards its ability to promote accurate rejoining in vivo.
MATERIALS AND METHODS
Cell culture
The N114P2 cells and the parental cell line, Nalm-6, isolated as described previously (23), were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS), l-glutamine and penicillin/streptomcyin. 180BR primary and 180BRneo transformed fibroblasts were cultured as previously described (24). 1BR3 and 1BR3neo are a normal primary and immortalized fibroblast cell line, respectively, used as controls. Fibroblasts were cultured in minimal essential medium supplemented with 15% FCS, penicillin and streptomycin as described previously. LB2304 lymphoblastoid cells were obtained as described previously (6). AHH1, a lymphoblastoid cell line derived from a healthy donor, was used as a control (25). Lymphoblasts were grown in suspension in RPMI 1640 medium supplemented with glutamine (2 mM), gentamycin (40 µg/ml) and 10% (v/v) FCS.
N114P2 cells express normal levels of XRCC4 as visualized by immunofluorescence (data not shown). The R278H mutation in 180BR cells results in a 80–90% decrease in protein activity, but XRCC4 and DNA ligase IV are expressed normally (26). The LB2304 cell line has barely detectable levels of DNA ligase IV but normal levels of XRCC4 (6; data not shown).
Host cell end-joining assay
The recombination substrate pHRecCJ (18,27) or its modified derivatives, pHRrecCJV or pHRecCJI (see Fig. 1), were linearized by digestion with either EcoRV or EcoRI (Promega) to generate blunt-ended breaks or cohesive-ended breaks with 5′ overhangs, respectively. For the substrates pHRecCJV or pHRecCJI, linear molecules were separated from the excized fragment and any uncut molecules by agarose gel electrophoresis. Exponentially growing cells were transfected with 1.25–2 µg of linearized plasmid DNA by electroporation (lymphoblastoid cell lines) as described previously (18,27) or by Ca3(PO4)2 (fibroblast cell lines). The pre-B cells were transfected using an Easyject Plus Electroporator (Flowgen, Lichfield, UK). Parallel transfections were carried out using an equivalent quantity of circular plasmid molecules. Cells were harvested 48 h after transfection, lysed and plasmid DNA extracted using a modification of the Qiagen miniprep protocol. Since linearized plasmids have to be recircularized in order to replicate in human cells, recovered DNA samples were digested with the restriction enzyme DpnI, which recognizes and cuts only bacterially methylated GATC sequences (dam+). Thus, the DpnI digestion ensures that only plasmids repaired and replicated in human cells will be recovered. Routinely, one-half of the mini-lysis was digested with DpnI but, in experiments where the plasmid recovery was low, as much of the mini-lysis as possible was used for DpnI digestion and subsequent analysis. The DpnI-digested DNA was electroporated into bacteria, XL1-blue or DH5α, and colonies carrying the plasmids were selected on LB plates containing 100 µg/ml ampicillin. Colonies were counted following appropriate dilution, and the numbers given in Table 1 represent those estimated to be present in the 1 ml used for transformation. End-joining frequency was estimated from the number of colonies obtained after transformation with linear versus circular plasmids.
Figure 1.

The substrate pHRecCJ and modifications, pHRecCJI and pHRecCJV. All plasmids contain the prokaryotic ColE1 origin, the β-lactamase gene (AmpR), the SV40 origin of replication and large T-antigen-coding sequence. In the original substrate, pHRecCJ, the target LacZ gene is interrupted by an intervening sequence that includes the unique restriction sites EcoRI and EcoRV. The two modified pHRecCJ substrates, pHRecCJI and pHRecCJV, contain two closely located EcoRI or EcoRV restriction sites that have been generated by the insertion of a fragment at the unique EcoRI or EcoRV site of pHRecCJ.
Table 1. End-joining frequency in Lig4–/– cells.
| Cell line | Exp | EcoRV-restricted plasmid | EcoRI-restricted plasmid | ||||
|---|---|---|---|---|---|---|---|
| Coloniesd (cut/uncut) | % EJ | Lig4+/+/Lig4–/– | Coloniesd(cut/uncut) | % EJ | Lig4+/+/Lig4–/– | ||
| Nalm-6 (Lig4+/+) | 1 | 43/1411c | 3.0 | 6880/49 920c | 13.7 | ||
| 2 | 1.07 × 105/2.3 × 107c | 0.4 | 2.4 × 105/1.2 × 107b | 2.0 | |||
| 3 | 940/14 000a | 6.7 | |||||
| N114P2 (Lig4–/–) | 1 | 0/5547c | <0.018 | >167.0 | 860/1.1 × 105c | 0.8 | 17.1 |
| 2 | 3/1.3 × 106b | ∼0.0002 | ∼2000.0 | 24 186/1.5 × 107b | 0.16 | 12.5 | |
| 3 | 80/1.7 × 105a | 0.047 | 143.0 | ||||
| N114P2+pcDNA3 | 3 | 300/1.7 × 105a | 0.18 | 37.0 | |||
| N114P2+pcDNA3/Lig4 | 3 | 2540/1.7 × 105a | 1.5 | 4.5 | |||
a–cThe number of replicate transfections carried out in each experiment, with a, b and c representing 1, 2 and 3 replicated experiments, respectively.
dColonies screened represent the total colonies present in the 1 ml of transformed bacteria obtained from each transfection. In experiments with two or three replicate transfections, d represents the colonies in 2 or 3 ml, respectively.
Analysis of rejoined plasmids
To assess the accuracy of rejoining, DNA from the plasmids was digested with the restriction enzyme used to generate the DSB. Those events that had not restored the original restriction site (i.e. inaccurate repair events) were analyzed by further restriction analysis and DNA sequencing. Only DNA sequence analysis has been described here. Statistical analysis of data was performed using either the Student t-test or the χ2 test with Yates’ correction. The P-values for the test are indicated when appropriate.
Immunoblotting
Whole-cell extracts were prepared as previously described (28). The anti-DNA ligase IV antibody was raised against the whole protein.
Preparation of cellular extracts
Extracts were prepared according to the protocol of Baumann and West (29). Briefly, 1–2 × 109 cells were harvested, washed three times with phosphate-buffered saline and once with hypotonic lysis buffer [10 mM Tris–HCl pH 8.0, 1 mM EDTA, 5 mM dithiothreitol (DTT)]. Cells were resuspended in 2 vol of hypotonic buffer, incubated on ice for 20 min and then lysed by homogenization while progressively adding the protease inhibitors (phenylmethylsulfonyl fluoride 0.17 mg/ml; aprotinin, 0.01 U/ml trypsin inhibitor; and 1 µg/ml each of pepstatin, chymostatin and leupeptin). After 20 min on ice, 0.5 vol of high salt buffer (50 mM Tris–HCl pH 7.5, 1 M KCl, 2 mM EDTA, 2 mM DTT) was added and the extract was centrifuged for 3 h at 42 000 r.p.m. in a Beckman SW50.1 rotor. The supernatant was recovered and dialyzed for 3 h against E buffer [20 mM Tris–HCl pH 8.0, 0.1 M KOAc, 20% (v/v) glycerol, 0.5 mM EDTA, 1 mM DTT]. Aliquots were fast frozen and stored at –80°C.
In vitro end-joining reactions
End-joining reactions were performed with the plasmid pSJ (3.3 kb), a derivative of pHRecSJ (22). The plasmid was linearized with SalI (New England Biolabs) to create cohesive termini with 5′ overhangs. To generate a non-ligatable substrate, a single nucleotide (dTTP) was incorporated using DNA polymerase I large Klenow fragment (New England Biolabs). These plasmids were subsequently subjected to DNA ligation (T4 DNA ligase, New England Biolabs). Linear molecules, i.e those resistant to ligation due to the incorporation of a single nucleotide, were isolated by gel electrophoresis and used as a substrate for ligation. To analyze end degradation, the substrate was dephosphorylated and 5′-end labeled with [γ-32P]ATP (ICN, France) using T4 polynucleotide kinase (New England Biolabs). Reactions (15 µl) were carried out in 50 mM Tris–HCl pH 7.5, 0.5 mM Mg(OAc)2, 60 mM KOAc, 1 mM ATP, 1 mM DTT, 100 µg/ml bovine serum albumin and 20 ng of linearized plasmid. Incubation was for 2 h at 37°C. DNA products were deproteinized, separated by electrophoresis through a 0.8% agarose gels and visualized either directly by autoradiography (5′-32P-labeled DNA) or by Southern transfer and hybridization with a 0.8 kb fragment from the LacZ gene, radioactively labeled with [α-32P]dCTP (ICN, France) by random priming (Rediprime II kit, Amersham Pharmacia Biotech, France). The ligase IV–XRCC4 complex and Ku were expressed in insect cells and prepared as described previously (26). The reaction products were quantified with a Storm phosphorimager (Molecular Dynamics).
Protection of DNA ends by the DNA ligase IV–XRCC4 complex and Ku
The cohesive- and blunt-ended fragments were produced from Bluescript using AflII–PstI- or PvuII-generating fragments, respectively. A 20 ng aliquot of end-labeled fragment was pre-incubated with the indicated amounts of DNA ligase IV–XRCC4 and/or Ku 70/80 for 30 min on ice in the reaction mixture described previously (26). T7 exonuclease (5 U; New England Biolab), the minimum amount that gave nearly complete digestion, was added and the reactions were incubated at room temperature for another 30 min. After incubation, reactions were deproteinized, phenol/chloroform extracted and precipitated. Aliquotes were run on 0.8% agarose gels. Dried gels were analyzed and quantified using a STORM phosphorimager (Molecular Dynamics).
RESULTS
The frequency and fidelity of plasmid DNA end joining in vivo are dramatically impaired in a LIG4-null cell line
A human pre-B cell line, N114P2, with targeted disruptions in both LIG4 alleles, and its parental line, Nalm-6, were used to examine the requirement for DNA ligase IV in a plasmid-based host cell end-joining assay. The experimental approach is similar to that previously described (18,19,22). The plasmids used are based on pHRecCJ and are able to replicate in human cells and in Escherichia coli. For the experiments with pre-B cells, modified pHRecCJ substrates (pHRecCJV and pHRecCJI) were used, which enable uncut plasmids to be distinguished from accurately rejoined molecules (Fig. 1). The plasmids were linearized using EcoRV or EcoRI to create blunt or 5′-complementary ends, respectively, and transfected into the human cell lines. Parallel experiments were carried out using uncut circular plasmid molecules. After 48 h, plasmids were recovered and digested with DpnI, a methylation-specific restriction enzyme. Since mammalian cells do not methylate DpnI sites following replication, plasmids that have failed to be repaired and replicate within the human cells are eliminated. After transfection into bacteria, the proportion of colonies recovered using cut plasmid relative to uncut plasmid was taken to represent the end-joining frequency.
The end-joining frequency for both EcoRV- and EcoRI-generated DSBs was markedly lower in the LIG4–/– N114P2 cells relative to the parental, Nalm-6 cells (Table 1). For EcoRV-cut plasmids, the decrease was >100-fold, with a more modest decrease (∼14-fold) for EcoRI-cut plasmids. Although we observed considerable inter-experimental variation in end-joining frequency, the defect in the LIG4-null line (N114P2) was reproducible and non-overlapping with the control range.
To verify that the decreased end-joining frequency is due to the lack of DNA ligase IV, plasmid expressing LIG4 cDNA was transfected together with EcoRV-cut plasmid. Although the frequency did not reach that found in the Nalm-6 cells, significantly increased end joining was observed in the presence of LIG4 cDNA, showing that the impaired end-joining frequency in N114P2 cells is a direct consequence of the targeted disruptions in LIG4 (Table 1, experiment 3). The lack of full complementation is most likely explained by the fact that this is a transient transfection system involving co-transfection of two plasmids. It is also possible that DNA ligase IV is not fully expressed in the transfected cells.
To assess the accuracy of rejoining, recovered plasmids were digested with the restriction enzyme used for linearization (EcoRI or EcoRV). Due to the low recovery of recombinants obtained for N114P2 cells in the above experiments, several additional transfections were carried out to increase the number of recombinants available for analysis. The analysis of plasmids obtained following transfection of N114P2 cells with EcoRV-linearized substrates indicates that none of the 130 recombinants had been rejoined accurately (Table 2). However, a similar low level of accurate repair (2%) was observed with control Nalm-6 cells (Table 2). For transfections involving EcoRI-restricted plasmids, N114P2 cells displayed a signficantly lower level of accurate repair compared with the control Nalm-6 cells (4% versus 22%, P ≤0.0001) (Table 2).
Table 2. Analysis of accurate and inaccurate repair events.
| Cell line | EcoRV-restricted plasmids | EcoRI-restricted plasmids | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Accuracya | Sequence analysisb | Accuracya | Sequence analysisb | |||||||
| Nos analyzed | Mean deletion size (bp) | Insertions | MH | Nos analyzed | Mean deletion size (bp) | Insertions | MH | |||
| Nalm-6 | 2% (4/180) | 23 | 169 | 61% (14/23) | 67% (6/9) | 22% (37/171) | 27 | 42 | 85% (23/27) | 25% (1/4) |
| N114P2 | 0% (0/130) | 18 | 371 | 28% (5/18) | 92% (12/13) | 4% (7/161) | 36 | 94 | 25% (9/36) | 81% (22/27) |
| P = 0.142 | P = 0.099 | P = 0.048 | P = 0.264 | P <0.0001 | P = 0.202 | P <0.0001 | P = 0.052 | |||
| 1BR3neo | 38% (72/191) | 42 | 572 | 17% (7/42)< | 74% (26/35) | 20% (9/44) | 33 | 545 | 27% (9/33) | 75% (18/24) |
| 180BRneo | 9% (11/128) | 16 | 358 | 31% (5/16) | 73% (8/11) | 3% (1/29) | 16 | 430 | 19% (3/16) | 69% (9/13) |
| P = 0.001 | P =0.126 | P = 0.281 | P >0.999 | P = 0.044 | P = 0.384 | P = 0.726 | P = 0.716 | |||
| 1BR3 | 30% (39/130) | 17 | 520 | 29% (5/17) | 92% (11/12) | 29% (26/91) | 14 | 289 | 21% (3/14) | 55% (6/11) |
| 180BR | 0% (0/25) | 7 | 152 | 29% (2/7) | 100% (5/5) | ND | ND | ND | ND | ND |
| P <0.0001 | P = 0.053 | P >0.999 | P >0.999 | |||||||
| AHH1 | 36% (51/142) | 20 | 285 | 5% (1/20) | 68% (13/19) | 34% (56/164) | 23 | 320 | 9% (2/23) | 81% (17/21) |
| LB2304 | 0% (0/144) | 21 | 303 | 5% (1/21) | 55% (11/20) | 0% (0/95) | 14 | 202 | 14% (2/14) | 75% (9/12) |
| P <0.0001 | P = 0.964 | P >0.999 | P >0.999 | P <0.0001 | P = 0.259 | P = 0.625 | P = 0.686 | |||
MH, microhomology; ND, not determined.
aPercentage of the total number of recominants in which the original restriction site was restored.
bSequence analysis of randomly selected independent inaccurate repair events.
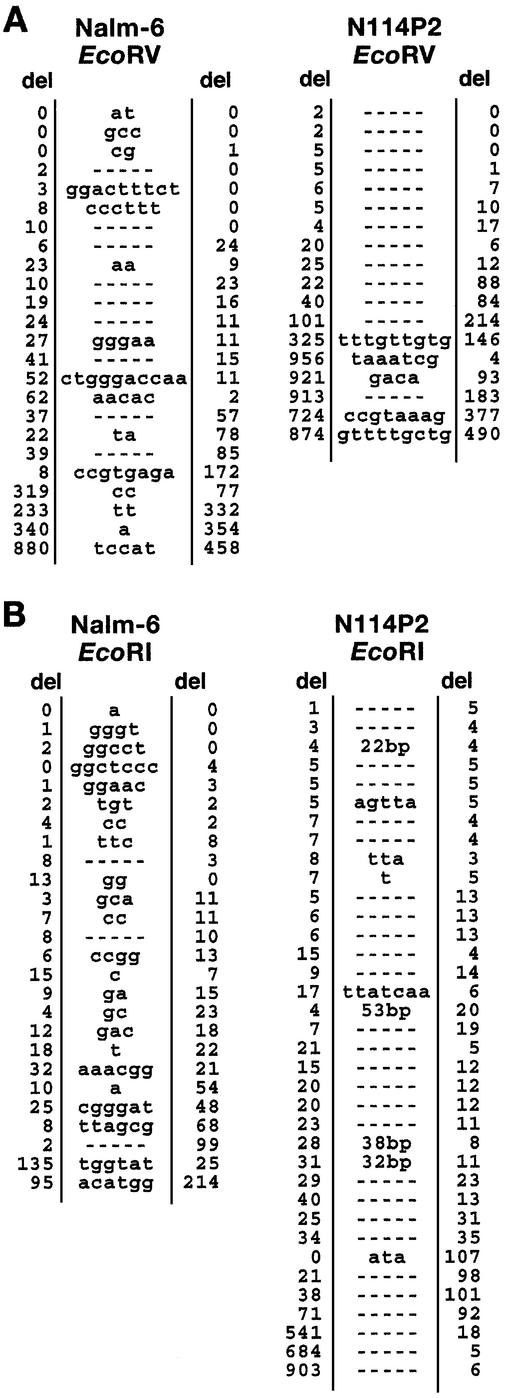
To characterize the recovered recombinants further, a sample of inaccurately rejoined plasmids was examined by sequencing (Table 2). Among the plasmids recovered from Nalm-6 and N114P2 cells, a similar range of deletion sizes was observed for both EcoRV- and EcoRI-cut substrates (1–1364 and 1–909 bp, respectively). The small increase in mean deletion size seen in N114P2 cells (Table 2) was not considered to be significant considering the wide range in deletion sizes. Sequencing also exposed further features of the junctions, namely the presence of insertions and direct repeat sequences. The presence of microhomology at the junctions can only be assessed accurately in those plasmids that did not have insertions. For both cell lines, microhomology was observed at the majority of the inaccurately rejoined junctions not associated with insertions (Table 2). A striking feature was the presence of insertions at many of the Nalm-6 junctions (61 and 85% for EcoRV and EcoRI, respectively) (Table 2). In Nalm-6 cells, these insertions, which were short (1–10 nt) and had a high GC content, strongly resemble N (non-templated) nucleotide insertions generated during V(D)J recombination by terminal deoxynucleotidyl transferase (TdT) (28). Nalm-6 cells express high levels of TdT (data not shown) and insert TdT nucleotides in vivo during V(D)J recombination (12). Our data suggest that TdT can also add nucleotides during end joining. In contrast, although N114P2 cells also express high levels of TdT (data not shown), the junctions from recombinants derived from N114P2 had significantly fewer insertions compared with those found in Nalm-6 (28 and 25% for EcoRV and EcoRI, respectively). Additionally, these insertions were frequently longer and had a lower GC content compared with those observed in Nalm-6 cells (Fig. 2). These observations provide suggestive evidence that a component of the insertions in Nalm-6 cells is both TdT and DNA ligase IV dependent.
Figure 2.
Nucleotide sequence analysis of the junctions of inaccurately rejoined plasmids from Nalm-6 and N114P2 cells. Recombinants were obtained with (A) EcoRV- or (B) EcoRI-linearized substrate. Letters in the center indicate extra nucleotides. The number of nucleotides deleted from each end is noted under del.
Taken together, these results show that DNA ligase IV is required for the majority (>90%) of end-joining events observed in these pre-B cells. Furthermore, since LIG4 is fully inactivated in the N114P2 cells, the residual rejoining observed shows that end joining can also take place via a DNA ligase IV-independent process. This alternative process(es) is more efficient in carrying out cohesive end rejoining than blunt end rejoining. Another striking feature of the processing of DSBs in the absence of DNA ligase IV is a marked decreased in the fidelity of end rejoining.
The frequency and fidelity of plasmid DNA end joining in vivo are impaired in cells harboring hypomorphic mutations in LIG4
Having established that the plasmid host cell end-joining assay monitors a DNA ligase IV-dependent process, we next examined several recently described human cell lines harboring hypomorphic mutations in LIG4. First, using the same experimental strategy as described above, we examined an SV40-transformed cell line, 180BRneo, that was derived from a leukemia patient, who proved to have a mutation in LIG4. The mutational change, a substitution of a histidine for an arginine (R278H) in the active site motif, dramatically impairs but does not abolish DNA ligase IV function (26). We observed a modest but significant decrease in end-joining frequency (4.3-fold) of EcoRV-cut plasmids in 180BRneo relative to 1BR3neo, a control SV40 immortalized line (Table 3). A modest decrease in frequency was also observed using EcoRI-cut plasmids (2.3-fold). However, due to the variation in frequency between experiments, this difference is not statistically significant.
Table 3. Frequency of end joining.
| Exp | EcoRV-restricted plasmid | EcoRI-restricted plasmid | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1BRneo Colonies (cut/uncut) | %EJ | 180BRneo Colonies (cut/uncut) | %EJ | 1BRneo Colonies (cut/uncut) | %EJ | 180BRneo Colonies (cut/uncut) | %EJ | |||
| 1 | 15 000/103 000 | 14.6 | 38/1440 | 2.6 | 2920/138 400 | 2.1 | 92/9600 | 0.96 | ||
| 2 | 1075/25 500 | 4.2 | 119/12 200 | 0.98 | 20 800/528 800 | 3.9 | 142/29 600 | 0.48 | ||
| 3 | 7900/138 400 | 5.7 | 150/9600 | 1.6 | 17 500/103 000 | 17.0 | 63/1440 | 4.4 | ||
| 4 | 9020/181 600 | 5.0 | 40/1960 | 2.0 | 1750/25 500 | 6.9 | 780/12 220 | 6.4 | ||
| 5 | 32 000/528 800 | 6.0 | 354/29 600 | 1.2 | ||||||
| Mean | 7.0 ± 3.8 | 1.6 ± 0.6 | P = 0.022 | 7.5 ± 6.0 | 3.0 ± 2.5 | P = 0.268 | ||||
| EcoRV-restricted plasmid | EcoRI-restricted plasmid | |||||||||
| 1BR3 | 180BR | 1BR3 | 180BR | |||||||
| Colonies (cut/uncut) | %EJ | Colonies (cut/uncut) | %EJ | 1BR3/180BR | Colonies (cut/uncut) | %EJ | Colonies (cut/uncut) | %EJ | 1BR3/180BR | |
| 1050/21 650 | 4.8 | 25/17 800 | 0.14 | 34.3 | 690/26 200 | 2.6 | 0/21 300 | <0.004 | >650 | |
The results shown with 1BRneo and 180BRneo cells represent the values of 4–5 experiments. The mean and SD is given. The P-values represent the significance of the difference between the two sets calculated using the t-test.
The fidelity of rejoining for both EcoRV- and EcoRI-cut plasmids was significantly lower in 180BRneo cells relative to 1BR3neo cells (Table 2). Sequencing of randomly chosen inaccurately rejoined plasmids from these two lines indicates that there was no appreciable difference in the size of deletions nor in the occurrence of junctions with insertions or microhomology.
Taken together, our results indicate that the hypomorphic LIG4 mutation in 180BR cells confers only a modest impact on the frequency of plasmid rejoining in 180BRneo cells compared with the marked decrease found in cells with inactivated LIG4. There was, however, a significant decrease in the accuracy of plasmid rejoining in 180BRneo cells. These results suggest that there is sufficient residual DNA ligase IV activity in 180BRneo cells to enable plasmids to be rejoined, albeit with impaired fidelity.
We have reported previously that the levels of NHEJ proteins (Ku70, Ku80 and DNA-PKcs) are lower in primary cells (1BR3 and 180BR) relative to transformed cells (1BRneo and 180BRneo cells), possibly reflecting the faster growth rate of transformed cells (30). We also observed lower levels of DNA ligase IV in 180BR primary cells relative to 180BRneo cells (data not shown). We therefore argued that if the considerable rejoining capacity remaining in 180BRneo cells represents residual activity due to higher levels of a mutant protein, the defect might be less marked than in 180BR primary cells. We therefore attempted to carry out the host cell end-joining assay using primary 180BR and control (1BR3) fibroblast cell lines. The primary cells have a markedly lower transformation frequency than transformed lines, making the analysis more restricted. The results in Tables 2 and 3 represent the compiled data from two experiments. Using EcoRV-cut plasmids, there was a markedly decreased (34-fold) end-joining frequency in 180BR versus 1BR3 cells. None of the 25 plasmids recovered from 180BR cells was rejoined accurately, suggesting that the fidelity of rejoining is also decreased since 30% of plasmids recovered from 1BR3 cells were rejoined accurately. Sequence analysis of a sample of these plasmids showed a small difference in deletion size between 180BR and 1BR3 cells, which was not statistically significant considering the limited number of deletions analyzed for 180BR cells and the range of deletion sizes observed. Using EcoRI-linearized plasmids, we did not recover any rejoined plasmids from 180BR cells despite a rejoining frequency of 2–3% being observed in a control primary fibroblast cell line and efficient recovery of circular plasmids. Taken together, our findings suggest that the frequency and fidelity of end joining are strongly decreased in 180BR relative to control primary cells and are more marked than the defect seen in the transformed derivative.
Recently, we described additional patients with hypomorphic mutations in LIG4 (6). These patients, in contrast to patient 180BR, have marked clinical phenotypes including immunodeficiency, microcephaly and developmental delay, and represent a new disorder called LIG4 syndrome (6). For this study, we examined LB2304, a lymphoblastoid cell line with very low levels of mutant DNA ligase IV protein (6). Due to the low numbers of colonies recovered using both cut and uncut plasmids, we were unable to estimate the frequency of rejoining. However, a sufficient number of plasmids were recovered to analyze the accuracy of rejoining (Table 2). In multiple experiments with a normal lymphoblastoid cell line (AHH1), we observed an accuracy of rejoining of 34% (EcoRI) and 36% (EcoRV) (Table 2) (27). In marked contrast, no accurately rejoined junctions were observed in three independent experiments with LB2304 cells using EcoRV-cut plasmids and two experiments using EcoRI-cut plasmids (Table 2). Sequence analysis of inaccurately rejoined plasmids derived from LB2304 cells frequently revealed the recovery of identical recombinants, a feature not observed with control cells. These products were probably derived from the same rejoining event and are indicative of a low end-joining frequency. The sequences analyzed in Table 2 represent independent recombinants. No marked or consistent changes in deletion size were observed in LB2304 cells, nor in the use of microhomology or the presence of insertions.
Thus, the LB2304 cell line, derived from a patient with severe clinical features, is, like 180BR, very dramatically impaired in the accuracy of plasmid end rejoining.
Cell extracts from LIG4 mutant cell lines are unable to promote in vitro end joining
To analyze further the ability of LIG4 mutant cell lines to carry out DNA end joining, we examined the capacity of cell-free extracts to catalyze in vitro end joining, a previously described assay that is dependent upon the known NHEJ components (29). Linearized DNA substrates with 5′-complementary (SalI) ends were incubated with cell-free extracts, and the end-joining products visualized by hybridization using a 32P-labeled probe with homology to the substrate (Fig. 3A). Extracts from control cell lines, AHH1, HSC93 and Nalm-6 (Fig. 3A, lanes 2, 3 and 6), were capable of converting 35–70% of the input substrate into multimeric products during the 2 h incubation. In contrast, extracts derived from LIG4 mutant cell lines, LB2304 (Fig. 3A, lane 4) and N114P2 (Fig. 3A, lane 7), show no detectable multimeric product formation. These results verify that the assay is dependent upon DNA ligase IV–XRCC4 and that the low level of residual activity in LB2304 cells is unable to promote DNA end joining efficiently in this assay. The addition of baculovirus-expressed wild-type DNA ligase IV–XRCC4 complex restored nearly normal levels of product formation to both LB2304 and N114P2 cell extracts (Fig. 3A, lanes 5 and 8). In the absence of extract, the rejoining catalyzed by DNA ligase IV–XRCC4 alone was negligible (Fig. 3A, lane 9). For all cell extracts, a similar low level of recircularized product is observed. Together, these results indicate that, although DNA ligase IV-independent rejoining can occur, the predominant end-joining activity in human cell extracts is dependent upon DNA ligase IV.
Figure 3.
Involvement of DNA ligase IV in plasmid end joining in vitro. (A) Protein extracts (40 µg) prepared from control lymphoblastoid cell lines, AHH1, HSC93 and Nalm-6, the LIG4 syndrome cell line LB2304 and the LIG4-null cell line N114P2 were incubated with SalI-linearized pSJ DNA (20 ng) as described in Materials and Methods. Lanes 1 and 9, no extract. Recombinant DNA ligase IV–XRCC4 (180 ng) was added where shown. The reaction products were loaded on an agarose gel, blotted and hybridized with a 32P-labeled probe that is homologous to pSJ. The percentage total radioactivity in each lane converted into product (circular, monomer, dimer, trimer, tetramer, etc.) is indicated. (B) End-joining reactions were carried out as described in (A) except that 5′-32P-end-labeled SalI-linearized pSJ (20 ng) was utilized. Recombinant DNA ligase IV–XRCC4 (180 ng) and Ku70/Ku86 (200 ng) were added where shown. Product formation was analyzed by agarose gel electrophoresis followed by autoradiography. (C) Effect of DNA ligase IV–XRCC4 protein concentration on DNA end joining. Extracts (40 µg) prepared from control lymphoblastoid cell lines, AHH1 and Nalm-6, the LIG4 syndrome cell line, LB2304 and the LIG4-null cell line, N114P2 were incubated with 5′-32P-end-labeled SalI-linearized pSJ (20 ng). Various dilutions of recombinant DNA ligase IV–XRCC4 were added as shown. Product formation was analyzed by agarose gel electrophoresis followed by autoradiography. The percentage total radioactivity in each lane converted into product (circular, monomer, dimer, trimer, tetramer, etc.) is indicated.
DNA ligase IV–XRCC4 is involved in the protection of DNA ends
In the experiments described above, the similar recovery of unrejoined plasmid and the absence of any smearing indicated that there is not any extensive degradation of the input plasmid DNA. However, since the in vitro end-joining reaction can only promote precise end-to-end joining (29; data not shown), loss of a few base pairs could potentially inhibit the reaction. To examine end degradation, we performed similar experiments using a 5′-32P-end-labeled SalI-linearized plasmid substrate (Fig. 3B). Unexpectedly, in LB2304 cell extracts, a substantial loss of signal was observed (Fig. 3B, lane 4), suggesting degradation of the terminal nucleotide(s). The use of a 3′-32P-end-labeled oligonucleotide (49mer) confirmed that the observed loss of signal was due to degradation and indicated that nucleotide loss most frequently involves between one and seven nucleotides (data not shown). Degradation, however, is not inherent to the loss of DNA ligase IV since N114P2 cell extracts do not display increased levels of degradation (Fig. 3B, lane 9). Addition of the purified Ku heterodimer to LB2304 cell extracts did not overcome the degradation (Fig. 3B, lane 5). Surprisingly, however, addition of purified DNA ligase IV–XRCC4 restored both the end-joining activity of the LB2304 extract (appearance of multimeric products) and the stability of the DNA ends (significantly increased signal from the unrejoined substrate) (Fig. 3B, lane 6). Addition of DNA ligase IV–XRCC4 also restored the end-joining activity of N114P2 cell extracts (Fig. 3B, lane 10), consistent with the results described above (Fig. 3A, lane 8). Together, these results suggest that, during the DNA end-joining process, the DNA ligase IV–XRCC4 complex plays a role in end protection as well as in ligation.
To characterize further the end joining and end protection functions of DNA ligase IV–XRCC4, we examined the impact of DNA ligase IV–XRCC4 concentration. At low concentrations of DNA ligase IV–XRCC4 (11 and 22 ng), very little, if any, effect was observed on the stability of DNA ends in LB2304 extracts (Fig. 3C, lanes 4 and 5). In contrast, these same low concentrations of DNA ligase IV–XRCC4 restored the end-joining activity (dimer formation) to N114P2 extracts to ∼40% of the level seen in Nalm-6 extracts (Fig. 3C, compare lanes 9 and 11). An increase in the stability of DNA ends and end joining in LB2304 extracts was only observed at higher levels of DNA ligase IV–XRCC4 (90 and 180 ng). These results indicate that in the LB2304 background, higher concentrations of DNA ligase IV–XRCC4 are required to prevent end degradation than are required for ligation. Moreover, they suggest that DNA degradation is not simply a consequence of lack of rejoining but rather that rejoining cannot be seen if the ends are degraded. The relative levels of ligation compared with end degradation will depend upon the endogenous level of nuclease activity, which is likely to vary between cell lines.
End protection by DNA ligase IV–XRCC4 is independent of DNA ligation
To separate further the end-joining and end-protection functions of the DNA ligase IV–XRCC4 complex, LB2304 cell extracts were incubated with a non-ligatable DNA substrate in both the presence and absence of DNA ligase IV–XRCC4 (Fig. 4). The substrate used in this experiment was generated by the incorporation of a single nucleotide at the 3′ end of a SalI-linearized plasmid (Fig. 4 and Materials and Methods). Since the in vitro end-joining assay can promote only simple ligation events, this nucleotide insertion eliminates terminal homology and prevents the majority of end-joining events in normal cell extracts (Fig. 4, lanes 2 and 5). In the presence of LB2304 cell extracts, as described previously, a significant reduction of monomer signal due to increased end degradation is observed (Fig. 4, lane 3). Addition of purified DNA ligase IV–XRCC4 complex has no effect on the formation of end-joining products, yet results in the restoration of monomer signal to levels similar to normal cells (Fig. 4, lane 4). These results indicate that the end protection observed in the presence of DNA ligase IV–XRCC4 complex is independent of DNA ligation.
Figure 4.
End protection by DNA ligase IV–XRCC4 protein can occur in the absence of DNA end joining. Protein extracts (40 µg) prepared from control lymphoblastoid cell lines, AHH1 and Nalm-6, the LIG4 syndrome cell line LB2304 and the LIG4-null cell line N114P2 were incubated with a non-ligatable 5′-32P-end-labeled substrate (20 ng). Recombinant DNA ligase IV–XRCC4 (180 ng) was added where shown. Product formation was analyzed by agarose gel electrophoresis followed by autoradiography.
DNA ligase IV–XRCC4 can protect against end degradation by T7 exonuclease
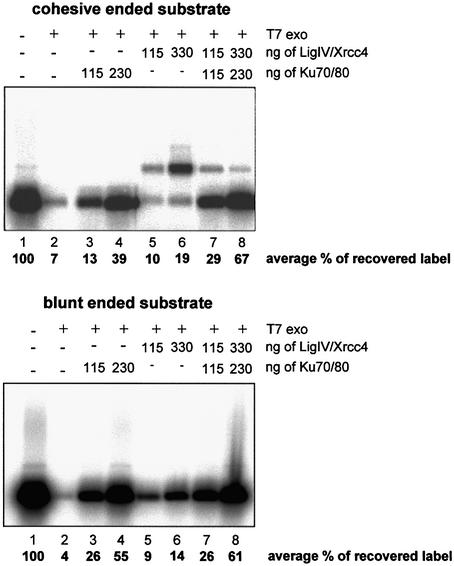
To gain further evidence for a role of DNA ligase IV–XRCC4 in protecting DNA ends from degradation, we examined the ability of in vitro expressed proteins to prevent degradation by T7 exonuclease. Addition of 5 U of T7 exonuclease resulted in significant loss (93%) of the label from a 5′-end-labeled DNA substrate with cohesive ends (Fig. 5, top, compare lanes 1 and 2). Addition of Ku prevented this degradation, partially at low concentrations and more significantly at higher concentrations. The low concentration of Ku results in 1–2 molecules bound per DNA, and the higher concentration results in multiple Ku molecules bound to the DNA (data not shown). In the presence of DNA ligase IV–XRCC4, significant ligation is observed (Fig. 5, top, lanes 5 and 6). Taking the stability of the end-labeled nucleotide to represent end protection (i.e. estimating the label present in both ligated and non-ligated substrate), DNA ligase IV–XRCC4 can also prevent end degradation. The combined presence of DNA ligase IV–XRCC4 and Ku inhibited ligation, a feature observed previously (31), but enhanced the stability of the end label to a level equivalent to an additive contribution of the separate complexes (Fig. 5, top, lanes 7 and 8). Since the buffer conditions required for ligation are similar to those required for T7 exonuclease activity, we were unable to examine end protection in the absence of ligation using a cohesive-ended substrate. Therefore, to substantiate the separation of end protection from ligation, we also employed a blunt-ended substrate, which is a poor substrate for DNA ligase IV–XRCC4 ligation (Fig. 5, bottom). Consistent with the findings above, we observed that DNA ligase IV–XRCC4 provided end protection in these conditions in which, as anticipated, no ligation was observed. Together, these results support the notion that the presence of DNA ligase IV–XRCC4 protects DNA ends from end degradation by T7 exonuclease.
Figure 5.
Ku and DNA ligase IV–XRCC4 separately and in combination can protect against T7 exonuclease digestion. T7 exonuclease (5 U) in the absence or presence of Ku and/or DNA ligase IV–XRCC4, as indicated, was incubated with an end-labeled substrate having cohesive ends (top) or blunt ends (bottom), and the products were analyzed by electrophoresis and autoradiography. The results shown are representive of three experiments undertaken, and the percentage recovery of the label represents the mean of the three experiments. Lane 1, control substrate; lane 2, substrate + 5 U of T7 exonuclease; lanes 3–8, substrate with T7 exonuclease and Ku and/or DNA ligase IV–XRCC4 as indicated.
DISCUSSION
Although NHEJ represents the major DSB repair mechanism in mammalian cells, little is known about its efficiency. Two factors, the frequency and fidelity of rejoining, are important parameters in the efficiency of repair. The frequency, representing the capacity of cells to rejoin the DSB, is likely to correlate with survival. The fidelity represents the ability to rejoin the break accurately and may influence other end points such as genomic stability and the onset of oncogenesis.
Using a LIG4-null human pre-B cell line, we show that the absence of DNA ligase IV results in decreased frequency and fidelity of end joining in an in vivo plasmid assay. The dramatic defect observed here contrasts with the extremely mild defect seen using NHEJ-defective rodent cell lines (15,16). One explanation is that rodent cells have higher levels of an alternative end-joining pathway relative to human cells, a feature also suggested from analysis of end joining using cell-free extracts (29). However, residual activity with decreased fidelity is observed in the ligase IV-null cell line, demonstrating the existence of an error-prone LIG4- independent end-joining process, a feature consistent with the residual DSB rejoining activity observed using pulsed-field gel electrophoresis (6,32). This rejoining activity appears to be more proficient in rejoining cohesive-ended relative to blunt-ended breaks. An error-prone DNA ligase IV-independent rejoining mechanism has also been described in Saccharomyces cerevisiae (7).
Analysis of lines with hypomorphic mutations in LIG4 also demonstrate that DNA ligase IV has a profound impact on the fidelity of rejoining. Indeed, no accurately rejoined junctions were recovered in our studies using 180BR primary cells or LB2304 cells. With these two lines, the frequency of rejoining was either not estimatable or very low. Thus, it is possible that the residual rejoining, like that observed in the LIG4-null line, occurs via an error-prone DNA ligase IV-independent mechanism. With 180BRneo cells, however, the frequency of rejoining is only modestly impaired. Since 180BRneo cells have higher levels of residual DNA ligase IV compared with their primary counterpart, we suggest that the residual rejoining results from residual ligase IV activity. Consistent with this notion, we have shown that 180BRneo cells have 10–20% of wild-type DNA ligase IV activity (26). However, under these conditions where the majority of rejoining occurs via a DNA ligase IV-dependent mechanism, the fidelity of rejoining is noticeably decreased. Thus, residual but impaired DNA ligase IV activity can impact upon the fidelity of rejoining. This raises the possibility that subtle alterations in ligase IV activity that may not appreciably affect the frequency of rejoining nonetheless may impair the fidelity. These findings with 180BRneo cells are consistent with results obtained using a plasmid assay for V(D)J recombination (6,30). Together, our findings show that the plasmid-based assay in human cells monitors the process of NHEJ, in contrast to findings obtained using rodent cells. Impaired DNA ligase IV activity has as great or an even greater impact on the fidelity of rejoining as it does on the end-joining frequency.
Sequence analysis of the inaccurately rejoined junctions has also provided insight into the events taking place at DNA ends. No consistent change in deletion size was seen in the DNA ligase IV-defective lines. Microhomology was observed frequently in the inaccurately rejoined junctions, demonstrating that DNA ligase IV-independent rejoining can exploit direct repeat sequences to aid rejoining. Another observation is that, in contrast to the lymphoblastoid and fibroblast cell lines examined in this study, the junctions derived from the pre-B cell line, Nalm-6, displayed a significant number of insertions (76%). The characteristics of these insertions coupled with high TdT expression suggest that they arise as TdT insertions. We also find that insertion events were slightly more prevalent with the substrate containing a 3′ extension (EcoRI, 88%) than with the blunt-ended substrate (EcoRV, 61%), consistent with the reported end preference of TdT (33). Together, these findings suggest that TdT might participate in the repair of restriction enzyme-generated DSBs as well as those generated during V(D)J recombination. Furthermore, in N114P2 cells, a significantly lower frequency of insertions (25%) is found, which do not have a marked TdT signature. Since N114P2 cells also express high levels of TdT, this suggests that TdT insertions require the presence of DNA ligase IV. This dependence may reflect a requirement for NHEJ factors in the recruitment of TdT, possibly through a direct interaction. In line with this possibility, a direct interaction between TdT and DNA ligase IV–XRCC4 has been demonstrated recently in vitro, and TdT is stably recruited to DNA only in the presence of both Ku and DNA ligase IV–XRCC4 (34).
The N114P2 cell extracts also show a dramatic defect in promoting end joining, demonstrating directly that the in vitro assay is DNA ligase IV dependent and supporting previous studies based on DNA ligase IV–XRCC4 immunodepletion (29). Surprisingly, the LB2304 cells displayed pronounced loss of the end-labeled terminal nucleotide of the substrate (Fig. 3B), which represents the loss of around 1–7 nucleotides (data not shown). This is not an inherent feature of loss of DNA ligase IV since it is not observed in N114P2 cell extracts. Increased end degradation with a wild-type lymphoblastoid cell line has been observed previously using the same assay, suggesting that it is attributable to high exonuclease activity present in certain cellular backgrounds (29). Addition of DNA ligase IV–XRCC4 complex to LB2304 extracts restored end-joining activity and the stability of DNA ends. Higher concentrations of DNA ligase IV–XRCC4 are required for end protection than for ligation, and end protection is not dependent upon ligation. Thus, end protection is not simply the result of removing the DNA ends by ligation. We also show that DNA ligase IV–XRCC4 can contribute to the protection of ends from degradation by T7 exonuclease. This effect was, again, not dependent upon ligation, demonstrated by using a non-ligatable blunt-ended substrate or a cohesive-ended substrate with high concentrations of Ku. The latter causes inhibition of ligation, a feature observed previously by ourselves and others (31). The inhibition of end degradation was observed by the presence of DNA ligase IV–XRCC4 alone or was additive with Ku (67% recovery of label in the presence of Ku and DNA ligase IV–XRCC4 versus 39% recovery in the presence of Ku alone), a finding consistent with previous reports that Ku helps to recruit DNA ligase IV–XRCC4 to DNA (31).
Together, these results support the conclusion that DNA ligase IV–XRCC4 can protect DNA ends from degradation in a manner separable from and not a direct result of ligation. Using the recombinant proteins, we have confirmed previous findings that DNA ligase IV–XRCC4 has DNA-binding activity observable by electrophoretic mobility shift assay (EMSA) using both cohesive- and blunt-ended substrates (35; data not shown). In this context, it is noteworthy that the crystal structure studies of the Chlorella virus DNA ligase provided direct evidence that the 3′ OH of the ligase-AMP directly contacts the PO4 of the nick, raising the possibility that end protection may require adenylate complex formation (36). Furthermore, XRCC4 is expressed at normal levels in LB2304 cells; thus, XRCC4 alone is not sufficient for end protection. Further work is in progress to examine the basis underlying end protection.
Our studies on plasmid rejoining in vivo demonstrate that the presence of DNA ligase IV serves to promote accurate rejoining. The observation that DNA ligase IV–XRCC4 can prevent end degradation in vitro could potentially provide one explanation for the in vivo findings. However, slower DNA ligation allowing increased time for degradation or the nature of the alternative rejoining mechanism may also contribute to the reduced fidelity observed in vivo. Intriguingly, the findings from the in vivo and in vitro studies are remarkably consistent. First, under both conditions, higher levels of DNA ligase IV appear to be required to prevent end degradation than are required for ligation. Additionally, the in vitro studies demonstrate that the end-protection function of DNA ligase IV–XRCC4 protects only a small number of nucleotides at the DNA end, which is consistent with the in vivo finding of decreased accurate rejoining in LIG4 mutants but no overt change in deletion size. Thus, although the overall level of fidelity observed in vivo may be influenced by multiple factors such as the nucleolytic capacity of the cells, the end-protection capacity of DNA ligase IV should be included as a potential determinant.
The fidelity of end joining is likely to be a major factor influencing cancer onset, and in this context it is noteworthy that the 180BR patient, whilst not displaying any overt immunodeficiency reflective of a decreased frequency of V(D)J recombination, did develop leukemia, possibly reflecting impaired fidelity. In conclusion, our findings demonstrate that DNA ligase IV is an important factor influencing the fidelity of rejoining in human cells. The newly described ability of DNA ligase IV to protect DNA ends from nucleolytic degradation may be a factor influencing the fidelity of rejoining.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to I. De Oliveira for excellent technical assistance. The laboratory of P.J. is supported by grants from the Medical Research Council, the Human Frontiers Science Programme, the Leukaemia Research Fund, the Primary Immunodeficiency Association, the Department of Health and European Union Grant Figh CT 1999. The laboratory of D.P. is supported by the Centre National pour la Recherche Scientifique, and grants from Association pour la Recherche sur le Cancer, Conseil de Radioprotection EDF and the Commission of European Community (Grant FIGH-CT 1999-00010). C.B. is the recipient of Fellowships from the Ministère de l’Education Nationale et de la Recherche.
REFERENCES
- 1.Jeggo P.A. (1998) DNA breakage and repair. Adv. Genet., 38, 185–211. [DOI] [PubMed] [Google Scholar]
- 2.Kanaar R., Hoeijmakers,J.H. and van Gent,D.C. (1998) Molecular mechanisms of DNA double strand break repair. Trends Cell Biol., 8, 483–489. [DOI] [PubMed] [Google Scholar]
- 3.Dvir A., Peterson,S.R., Knuth,M.W., Lu,H. and Dynan,W.S. (1992) Ku autoantigen is the regulatory component of a template-associated protein kinase that phosphorylates RNA polymerase II. Proc. Natl Acad. Sci. USA, 89, 11920–11924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gottlieb T.M. and Jackson,S.P. (1993) The DNA-dependent protein kinase: requirement of DNA ends and association with Ku Antigen. Cell, 72, 131–142. [DOI] [PubMed] [Google Scholar]
- 5.Moshous D., Callebaut,I., de Chasseval,R., Corneo,B., Cavazzana-Calvo,M., Le Deist,F., Tezcan,I., Sanal,O., Bertrand,Y., Philippe,N., Fischer,A. and de Villartay,J.P. (2001) Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell, 105, 177–186. [DOI] [PubMed] [Google Scholar]
- 6.O’Driscoll M., Cerosaletti,K.M., Girard,P.-M., Dai,Y., Stumm,M., Kysela,B., Hirsch,B., Gennery,A., Palmer,S.E., Seidel,J., Gatti,R.A., Varon,R., Oettinger,M.A., Sperling,K., Jeggo,P.A. and Concannon,P. (2001) DNA ligase IV mutations identified in patients exhibiting development delay and immunodeficiency. Mol. Cell, 8, 1175–1185. [DOI] [PubMed] [Google Scholar]
- 7.Boulton S.J. and Jackson,S.P. (1996) Saccharomyces cervisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J., 15, 5093–5103. [PMC free article] [PubMed] [Google Scholar]
- 8.Manolis K.G., Nimmo,E.R., Hartsuiker,E., Carr,A.M., Jeggo,P.A. and Allshire,R.C. (2001) Novel functional requirements for non-homologous DNA end joining in Schizosaccharomyces pombe. EMBO J., 20, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boulton S.J. and Jackson,S.P. (1996) Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double-strand break repair and in telomeric maintenance. Nucleic Acids Res., 24, 4639–4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Milne G.T., Jin,S., Shannon,K.B. and Weaver,D.T. (1996) Mutations in two Ku homologs define a DNA end-joining repair pathway in Saccharomyces cerevisiae. Mol. Cell. Biol., 16, 4189–4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siede W., Friedl,A.A., Dianova,I., Eckardt-Schupp,F. and Friedberg,E.C. (1996) The Saccharomyces cerevisiae Ku autoantigen homologue affects radiosensitivity only in the absence of homologous recombination. Genetics, 142, 91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gauss G.H. and Lieber,M.R. (1993) Unequal signal and coding joint formation in human V(D)J recombination. Mol. Cell. Biol., 13, 3900–3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gauss G.H. and Lieber,M.R. (1996) Mechanistic constraints on diversity in human V(D)J recombination. Mol. Cell. Biol., 16, 258–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taccioli G.E., Rathbun,G., Oltz,E., Stamato,T., Jeggo,P.A. and Alt,F.W. (1993) Impairment of V(D)J recombination in double-strand break repair mutants. Science, 260, 207–210. [DOI] [PubMed] [Google Scholar]
- 15.Kabotyanski E.B., Gomelsky,L., Han,J.O., Stamato,T.D. and Roth,D.B. (1998) Double-strand break repair in Ku86- and XRCC4-deficient cells. Nucleic Acids Res., 26, 5333–5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tzung T.Y. and Runger,T.M. (1998) Reduced joining of DNA double strand breaks with an abnormal mutation spectrum in rodent mutants of DNA-PKcs and Ku80. Int. J. Radiat. Biol., 73, 469–474. [DOI] [PubMed] [Google Scholar]
- 17.Liang F., Romanienko,P.J., Weaver,D.T., Jeggo,P.A. and Jasin,M. (1996) Chromosomal double-strand break repair in Ku80-deficient cells. Proc. Natl Acad. Sci. USA, 93, 8929–8933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Escarceller M., Buchwald,M., Jeggo,P.A., Singleton,B.K., Jackson,S.P., Moustacchi,E. and Papadopoulo,D. (1998) Fanconi anemia C gene product plays a role in the fidelity of blunt DNA end-joining. J. Mol. Biol., 279, 375–385. [DOI] [PubMed] [Google Scholar]
- 19.Escarceller M., Rousset,S., Moustacchi,E. and Papadopoulo,D. (1997) The fidelity of double strand breaks processing is impaired in complementation groups B and D of Fanconi anemia, a genetic instability syndrome. Somat. Cell Mol. Genet., 23, 401–411. [DOI] [PubMed] [Google Scholar]
- 20.Runger T.M. and Kraemer,K.H. (1989) Joining of linear plasmid DNA is reduced and error-prone in Bloom’s syndrome cells. EMBO J., 8, 1419–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Runger T.M., Poot,M. and Kraemer,K.H. (1992) Abnormal processing of transfected plasmid DNA in cells from patients with ataxia telangiectasia. Mutat. Res., 293, 47–54. [DOI] [PubMed] [Google Scholar]
- 22.Baldeyron C., Jacquemin,E., Smith,J., Jacquemont,C., De Oliveira,I., Gad,S., Feunteun,J., Stoppa-Lyonnet,D. and Papadopoulo,D. (2002) A single mutated BRCA1 allele leads to impaired fidelity of double strand break end-joining. Oncogene, 21, 1401–1410. [DOI] [PubMed] [Google Scholar]
- 23.Grawunder U., Zimmer,D., Fugmann,S., Schwarz,K. and Lieber,M.R. (1998) DNA ligase IV is essential for V(D)J recombination and DNA double-strand break repair in human precursor lymphocytes. Mol. Cell, 2, 477–484. [DOI] [PubMed] [Google Scholar]
- 24.Badie C., Iliakis,G., Foray,N., Alsbeih,G., Pantellias,G.E., Okayasu,R., Cheong,N., Russell,N.S., Begg,A.C., Arlett,C.F. and Malaise,E.P. (1995) Defective repair of DNA double-strand breaks and chromosome damage in fibroblasts from a radiosensitive leukemia patient. Cancer Res., 55, 1232–1234. [PubMed] [Google Scholar]
- 25.Crespi C.L. and Thilly,W.G. (1984) Assay for gene mutation in a human lymphoblast line, AHH-1, competent for xenobiotic metabolism. Mutat. Res., 128, 221–230. [DOI] [PubMed] [Google Scholar]
- 26.Riballo E., Doherty,A.J., Dai,Y., Stiff,T., Oettinger,M.A., Jeggo,P.A. and Kysela,B. (2001) Cellular and biochemical impact of a mutation in DNA ligase IV conferring clinical radiosensitivity. J. Biol. Chem., 276, 31124–31132. [DOI] [PubMed] [Google Scholar]
- 27.Smith J., Baldeyron,C., De Oliveira,I., Sala-Trepat,M. and Papadopoulo,D. (2001) The influence of DNA double-strand break structure on end-joining in human cells. Nucleic Acids Res., 29, 4783–4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benedict C.L., Gilfillan,S., Thai,T.H. and Kearney,J.F. (2000) Terminal deoxynucleotidyl transferase and repertoire development. Immunol. Rev., 175, 150–157. [PubMed] [Google Scholar]
- 29.Baumann P. and West,S.C. (1998) DNA end-joining catalyzed by human cell-free extracts. Proc. Natl Acad. Sci. USA, 95, 14066–14070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Badie C., Goodhardt,M., Waugh,A., Doyen,N., Foray,N., Calsou,P., Singleton,B., Gell,D., Salles,B., Jeggo,P., Arlett,C.F. and Malaise,E.-P. (1997) A DNA double-strand break defective fibroblast cell line, (180BR), derived from a radiosensitive patient represents a new mutant phenotype. Cancer Res., 57, 4600–4607. [PubMed] [Google Scholar]
- 31.Nick McElhinny S.A., Snowden,C.M., McCarville,J. and Ramsden,D.A. (2000) Ku recruits the XRCC4–ligase IV complex to DNA ends. Mol. Cell. Biol., 20, 2996–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Riballo E., Critchlow,S.E., Teo,S.H., Doherty,A.J., Priestley,A., Broughton,B., Kysela,B., Beamish,H., Plowman,N., Arlett,C.F., Lehmann,A.R., Jackson,S.P. and Jeggo,P.A. (1999) Identification of a defect in DNA ligase IV in a radiosensitive leukaemia patient. Curr. Biol., 19, 699–702. [DOI] [PubMed] [Google Scholar]
- 33.Mickelsen S., Snyder,C., Trujillo,K., Bogue,M., Roth,D.B. and Meek,K. (1999) Modulation of terminal deoxynucleotidyltransferase activity by the DNA-dependent protein kinase. J. Immunol., 163, 834–843. [PubMed] [Google Scholar]
- 34.Mahajan K.N., Nick McElhinny,S.A., Mitchell,B.S. and Ramsden,D.A. (2002) Association of DNA polymerase mu (pol mu) with Ku and ligase IV: role for pol mu in end-joining double-strand break repair. Mol. Cell. Biol., 22, 5194–5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Modesti M., Hesse,J.E. and Gellert,M. (1999) DNA binding of Xrcc4 protein is associated with V(D)J recombination but not with stimulation of DNA ligase IV activity. EMBO J., 18, 2008–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Odell M., Sriskanda,V., Shuman,S. and Nikolov,D.B. (2000) Crystal structure of eukaryotic DNA ligase-adenylate illuminates the mechanism of nick sensing and strand joining. Mol. Cell, 6, 1183–1193. [DOI] [PubMed] [Google Scholar]