Abstract
Ebp1, an ErbB3 binding protein that is a member of the proliferation-associated PA2G4 family, inhibits the proliferation and induces the differentiation of human ErbB positive breast and prostate cancer cell lines. Ebp1 binds the tumor suppressor retinoblastoma protein (Rb) both in vivo and in vitro, and Rb and Ebp1 cooperate to inhibit the transcription of the E2F1-regulated cyclin E promoter. We show here that Ebp1 can inhibit the transcription of other E2F-regulated reporter genes and of several endogenous E2F-regulated genes important in cell cycle progression in both Rb positive and Rb null cells. The Ebp1-mediated transcriptional repression depended on the presence of an E2F1 consensus element in the promoters. A fusion of Ebp1 with the GAL4 DNA binding domain protein had independent transcriptional repression activity that mapped to the C-terminal region of Ebp1. This C-terminal region of Ebp1 bound functional histone deacetylase (HDAC) activity and inhibitors of HDAC significantly reduced Ebp1-mediated repression. Ebp1 bound HDAC2, but not HDAC1, in vitro. An Ebp1 mutant lacking the HDAC binding domain failed to inhibit transcription. Our results suggest that Ebp1 can repress transcription of some E2F-regulated promoters and that one mechanism of Ebp1- mediated transcriptional repression is via its ability to recruit HDAC activity.
INTRODUCTION
Ebp1, a member of the PA2G4 family of proliferation-regulated proteins, was isolated as an ErbB3 binding protein in our laboratory (1). The ectopic expression of Ebp1 inhibited the growth of human ErbB positive breast and prostate cancer cells (2) and induced cellular differentiation (3). Treatment of serum-starved human breast cancer cells with the ErbB-3/4 ligand heregulin (HRG) induced translocation of Ebp1 from the cytoplasm to the nucleus (1). The regulated nuclear accumulation of Ebp1 suggested that Ebp1 may act as a transcription factor or transcriptional coregulator. As the downstream effects of HRG-induced signal transduction are poorly understood, we were interested in determining if Ebp1, an HRG-regulated protein, could have effects on gene expression that ultimately result in cell growth inhibition or differentiation.
The E2F family of transcription factors are important in the control of cell cycle progression (4). The first E2F-type transcription factor (E2F-1) was identified as a cellular DNA binding protein involved in activation of the adenoviral E2a promoter (5). Subsequently, E2F binding sites were found in the promoters of a number of genes involved in DNA synthesis, such as DNA polymerase and dihydrofolate reductase (6) or in cell cycle control such as cyclin E, cyclin D, c-myb, c-myc and cdc2 (4). There are six known members of the E2F family in mammals (7). Heterodimerization of E2F proteins with the related DRTF-1-polypeptide (DP) family proteins enhances the DNA binding and transactivating activity of E2F family members (7). Binding of the retinoblastoma protein, Rb, to E2F on E2F-regulated promoters inhibits expression of many E2F-regulated genes resulting in withdrawal from the cell cycle (8).
Recently, the importance of histone deacetylases (HDACs) in inhibition of E2F-regulated promoters has been demonstrated (9–11). Class I HDACs (HDAC 1–3) cooperate with Rb family members and E2Fs in regulation of genes involved in cell cycle progression. It has been postulated that these enzymes are recruited to E2F target promoters where they deacetylate histones and modify chromatin structure (12). Recently, Rayman et al. (9) demonstrated that corepressor complexes consisting of HDAC1 and Sin3B were specifically recruited to endogenous E2F-regulated promoters, but that this repression required p107 and p130, rather than Rb.
The fact that Ebp1 inhibited cell growth (3) led us to speculate that Ebp1 might affect the transcription of E2F-regulated genes important in cell cycle progression. We previously demonstrated that Ebp1 could bind Rb and inhibit transcription from the E2F1-regulated cyclin E reporter plasmid in both Rb positve and Rb null cells (13). The purpose of the present study was to determine if ectopic expression of ebp1 could inhibit the activity of other E2F-regulated promoters and if that repression involved HDACs. We show here that ectopic expression of ebp1 inhibited the activity of E2F-regulated reporter and endogenous genes. Using GAL4 DNA binding domain assays, we demonstrated that Ebp1 had independent transcriptional repression activity which maps to the C-terminal region of the protein. In addition, we demonstrate that the C-terminal region of Ebp1 bound HDACs from nuclear extracts and that inhibitors of HDACs significantly reduced Ebp1-mediated transcriptional repression. These studies suggest that one mechanism of Ebp1-induced cell growth inhibition and differentiation may be via its ability to influence expression of E2F-regulated genes.
MATERIALS AND METHODS
Cell culture
All cell lines were obtained from the American Type Culture Collection (Manassas, VA) and maintained at 37°C in a humidified atmosphere of 5% CO2 in air. Cell lines were routinely cultured in RPMI 1640 media supplemented with 10% FBS.
Plasmids
The E2F1 reporter plasmid (14) contains a 728 bp fragment of the E2F1 promoter upstream of the luciferase reporter gene. The E2F1 Luc mutant plasmid contains the same promoter fragment in which the E2F sites have been mutated (14). The wild-type cyclin D1 reporter plasmid contains a –163 bp fragment of the cyclin D1 promoter in pA3 luciferase (15). The mutant contains this fragment with a mutated E2F site. The myc promoter (–1100 to +565) luciferase plasmid contains an E2F consensus sequence cloned upstream of the luciferase reporter gene. The pSG424 plasmid has been previously described (16). PSG424-ebp1 (full-length) encoding nucleotides 262–1240 (amino acids 1–372) of the Ebp1 cDNA (GenBank accession no. U87954) was created by inserting the cDNA (derived by PCR) between EcoRI–XbaI restriction sites in frame with the GAL4 DNA binding domain of the pSG424 plasmid. The N- (amino acids 1–181) and C-terminal (amino acids 182–372) ebp1 deletion mutants were similarly constructed. The pPG5-E1b-luc reporter plasmid contains five GAL4 DNA binding sites cloned upstream of a minimal E1b promoter element and the firefly luciferase gene. pG5-Tk-Luc contains five GAL4 DNA binding sites cloned upstream of a minimal TK promoter element and the firefly luciferase gene (17).
The bacterial expression vectors encoding full-length and truncated glutathione S-transferase (GST)-Ebp1 fusion proteins and a mammalian expression vector (pcDNA3) encoding full-length ebp1 were previously described (13). A mammalian expression vector encoding a truncated ebp1 (ebp1 Δ 45) was constructed by inserting a BamHI–EcoRI PCR fragment encoding nucleotides 262–1240 (GenBank accession no. U87954) (amino acids 1–327) into pcDNA3 (2). Expression of this construct was determined by western blot analysis of transfected cells using antibody to full-length Ebp1 (2; and data not shown). The orientation and integrity of the cDNA inserts in all constructs were confirmed by automated DNA sequencing in the core laboratory of University of Maryland School of Medicine.
Luciferase reporter assays
COS-7 or MCF-7 (1 × 105) cells were plated in 6-well plates in complete media. When cells reached 50–60% confluence, they were transfected with 1 µg of individual reporter plasmids and wild-type or truncated (Δ 45) ebp1 expression plasmids. Cell were transfected using the Fugene-6 reagent (Boehringer Mannheim, Indianopolis, IN). Thirty-six hours later, luciferase activity was determined as previously described (13) and normalized by protein concentration determined using a BCA Kit (Pierce, Rockford, IL) or by cotransfection with a β-galactosidase plasmid. All transfection experiments were carried out in triplicate wells and repeated three times.
For experiments using the pSG424 plasmids, cells were transfected with 0.4 µg of either pSG424, pSG424-ebp1 (full length) or the N- or C-terminal ebp1 constructs and 1 µg of reporter plasmid.
Creation of stably transfected cell lines
To establish ebp1 stable transfectants, subconfluent human prostate cancer LNCaP cells growing in 100-mm tissue culture dishes were transfected with 10 µg of pcDNA3, or pcDNA3-ebp1 expression plasmids using Lipofectamine according to the manufacturer’s protocol. Cells were selected in G418 (1000 µg/ml) for 5 weeks and mass cultures of resistant colonies expanded.
Northern blot analysis
Total RNA was extracted using STAT RNA-60 and fractionated by electrophoresis prior to blotting on Hybond-N+ filters (Amersham Pharmacia Biotech). The cDNA for E2F1 that was used as a probe was previously described (6). C-Myc was detected using a cDNA clone that encodes the third exon of the human c-myc oncogene (ATCC). A 677 bp cDNA corresponding to wild-type DHFR was a gift of Dr Arif Hussain, University of Maryland, Baltimore (18). The probes were labeled with [γ-32P]dCTP using the Prime-a-gene labeling system (Promega, Madison, WI). Hybridization was performed as previously described (2). Densitometric analysis was performed using a Molecular Dynamics Densitometer (Sunnyvale, CA).
DNA content analysis
Cells were trypsinized and washed twice with phosphate-buffered saline (PBS). Cells were resuspended, fixed in cold 70% ethanol, and stained in hypotonic propidium iodide (PI) solution (0.05 mg/ml in PBS with 1 mg/ml DNase-free RNase) for 2 h on ice. Samples were excited at 488 nm and PI fluorescence emission was measured with a 575 band pass filter in a Coulter Epics Elite ESP Flow Cytometer (Beckman-Coulter, Miami, FL). Cell cycle distribution analysis was performed using the Multicycle software package (Phoenix Flow, Sunnydale, CA).
Affinity purification of GST fusion proteins and GST pull-down assays
In vitro expression and purification of recombinant GST-Ebp1 fusion proteins were performed essentially as described (13). Briefly, the recombinant vector was introduced into BL21 cells. Overnight cultures of individual colonies of the transformants were diluted 1:10 and grown for 1 h at 37°C before induction for 4 h at 25°C with IPTG (0.1 mM). The cultures were centrifuged at 10 000 g for 15 min at 4°C and the cells resuspended in ice cold PBS. Bacterial cells were lysed by sonication on ice for 15 consecutive 15 s intervals. Following addition of Triton X-100 to a final concentration of 1% (v/v) and centrifugation, the supernatants were incubated with glutathione–agarose beads (50% slurry) for 30 min at room temperature. Beads were used directly in GST pull-down assays after extensive washing with buffer.
GST pull-down assays and western blotting were performed as previously described (13). Briefly, cells were rinsed with PBS and lysed in buffer consisting of 20 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1 µg/ml leupeptin and 1 mM PMSF. Cell lysates (1 mg of protein) were mixed with equal amounts of GST or GST-Ebp1 loaded onto glutathione–Sepharose beads and incubated overnight at 4°C with gentle rotation. An aliquot of bound GST or GST-Ebp1 fusion constructs was also analyzed by Coomassie blue staining of SDS–PAGE gels to confirm equal loading of fusion proteins. The pelleted beads were then washed in lysis buffer, mixed with SDS sample buffer, boiled, and proteins separated on SDS gels. After electrophoresis, the proteins were transferred to Immobilin-P membranes, and immunoblotted as previously described (13). The blots were probed with monoclonal antibodies diluted in PBS supplemented with 2% milk for 2 h. Blots were washed three times with PBST and proteins detected using an ECL kit (Pierce). HDAC1 antibody was obtained from UBI (Lake Placid, NY) and HDAC2 antibody was from Zymed (San Francisco, CA).
Histone deacetylase assays
HDAC assays were performed using a kit from UBI essentially as described by the manufacturer. Briefly, HeLa cells were rinsed with PBS and lysed in buffer consisting of 20 mM Tris, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1 µg/ml leupeptin and 1 mM PMSF. Cells lysates (500 µg of protein) were mixed with equal amounts (3 µg) of GST or GST-Ebp1 loaded onto glutathione–Sepharose beads as described (13). The resultant beads were incubated with 200 µl of HDAC buffer containing 20 000 c.p.m. of 3H-labeled acetylated histone H4 peptide for 2 h at 37°C. A parallel series of reactions received 250 mM sodium butyrate, an HDAC inhibitor.
Statistical analysis
Results were analyzed using a two-tailed Student’s t-test. Significance was established at P ≤ 0.05.
RESULTS
Ebp1 inhibits E2F-mediated transcription
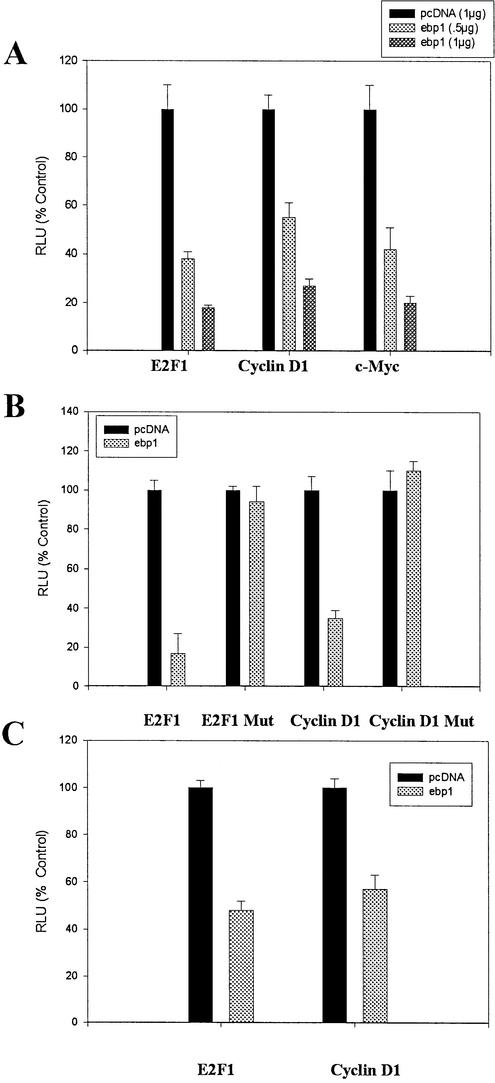
We have previously demonstrated that ectopic expression of ebp1 inhibits the activity of a cyclin E promoter luciferase reporter gene. To determine if Ebp1 would have similar effects on other E2F-regulated promoters, we transiently transfected MCF 7 cells (Rb wild-type) (19) with an E2F1 luciferase reporter plasmid in which luciferase expression is driven by a 728 bp E2F1 promoter (14). Transfected ebp1 significantly (P ≤ 0.05) inhibited luciferase activity of the E2F1 reporter construct in a concentration-dependent manner (Fig. 1A). E2F1 luciferase activity was inhibited 82% at the highest concentration of Ebp1 tested. We also examined the ability of Ebp1 to inhibit the E2F1-regulated cyclin D1 and c-myc promoters. Ebp1 also inhibited luciferase activity of these reporters in a concentration-dependent manner (Fig. 1A).
Figure 1.
Inhibition of E2F-mediated transcription by expression of Ebp1 in transfected cells. (A) Inhibition of transcription mediated by endogenous E2F. Logarithmically growing MCF-7 cells were transfected with an E2F1 luciferase reporter plasmid, a cyclin D1 reporter plasmid, or a c-myc reporter plasmid along with an ebp1 expression construct or the pcDNA vector control at the indicated concentrations. Cell extracts were assayed for relative luciferase activities (RLU) 48 h later as described in Materials and Methods. For these and other luciferase experiments the mean value ± SE derived from triplicates samples are shown. One of three representative experiments is shown. (B) Effect of E2F1 elements on the ability of Ebp1 to inhibit E2F-mediated transcription. MCF-7 cells were transfected with the E2F1 Luc (wild-type) plasmid which contains the native E2F1 promoter upstream of the luciferase gene and ebp1 (1 µg) or pcDNA3. The E2F1 Luc (mutant) plasmid, in which the E2F sites in the E2F1 promoter have been mutated, were also transfected in a similar manner. Wild-type and mutated cyclin D1 reporter constructs were similarly transfected along with either a control pcDNA vector or wild-type ebp1 (1 µg). RLU were determined 48 h later. Note that the RLU observed from cells transfected with the mutant cyclin D1 reporter and pcDNA 3 were increased 2-fold when compared to cells transfected with the wild-type cyclin D1 reporter in keeping with previous reports (15). Activity in the presence of ebp1 is compared to activity observed in the presence of pcDNA3 when using either the wild-type or mutant reporter. (C) Ebp1 inhibits E2F1 and cyclin D promoter activity in Rb negative cells. SAOS-2 cells were transfected with an E2F1 luciferase reporter plasmid, or a cyclin D1 reporter plasmid, along with 1 µg of either an ebp1 expression construct or the pcDNA vector control. Cell extracts were assayed for RLU 48 h later as described in Materials and Methods.
We were next interested in the contribution of E2F elements in the reporter constructs to the observed transcriptional repression. We first examined the ability of Ebp1 to inhibit an E2F1 promoter that contains four point mutations in the E2F recognition site (20). Ebp1 had no effect on luciferase activity of the mutant construct (Fig. 1B). We also examined the effect of Ebp1 on a cyclin D1 promoter with mutated E2F sites (15). We found that Ebp1 did not repress transcription from the cyclin D1 promoter with mutated E2F elements. Thus, the presence of E2F consensus elements in these promoters was necessary for Ebp1 to exert its effects.
Finally, we have previously demonstrated that although Ebp1 can physically associate with Rb and that Ebp1 repressed transcription more efficiently in the presence of Rb, Ebp1 could still inhibit activity of a cyclin E reporter plasmid in Rb negative SAOS-2 cells (13). We therefore examined the ability of Ebp1 to inhibit transcription of E2F1 and cyclin D reporter plasmids in Rb negative SAOS-2 cells. Ebp1 inhibited the expression of both the cyclin E2F1 and cyclin D genes (Fig. 1C). These data indicate that Ebp1 inhibited these promoters by both Rb-dependent and -independent mechanisms.
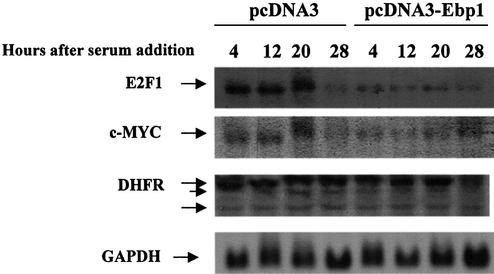
To verify that Ebp1’s effects on transcription were not caused by potential artifacts due to the use of transfected promoters, we tested whether constitutive overexpression of Ebp1 in stably transfected cell lines would also inhibit the E2F-regulated transcription of endogenous genes. For this purpose we generated stable LNCaP human prostate cancer cell lines overexpresssing Ebp1 by transfecting cells with an ebp1 expression plasmid and selecting for G418-resistant colonies. We obtained cell lines in which ebp1 was constitutively expressed 2–3-fold higher than in the control vector line (2), consistent with previous findings which indicate that high levels of Ebp1 expression are incompatible with cell growth (3). We serum starved cells for 24 h, then added serum for up to 28 h and determined steady state E2F1 message levels by northern analysis. In vector control cells, the levels of E2F1 increased 3-fold between 4 and 20 h after serum addition in keeping with previously reported data (21) (Fig. 2). No E2F1 message was observed before serum addition in either control LNCaP cells or ebp1 transfectants (data not shown). At 28 h, E2F1 steady state mRNA levels in vector controls were lower than those observed in these cells at the 4 h time point (Fig. 2). In ebp1 transfectants, E2F1 levels 4 h after serum addition were 4-fold lower than those observed in vector controls at the same time point. In contrast to vector controls, no increase in E2F1 levels were observed in ebp1 transfectants at later time points after serum addition (Fig. 2). Thus, the serum-induced increase in E2F1 levels observed in control cells were decreased in ebp1 stable transfectants.
Figure 2.
Constitutive overexpression of Ebp1 in LNCaP cells affects induction of cell cycle regulated genes in response to serum. LNCaP cells stably transfected with either pcDNA3 or ebp1 were serum starved for 24 h and then serum stimulated for up to 28 h as indicated before RNA harvesting. Northern blots were prepared using 30 µg of total RNA from each cell line. The blots were probed with 32P-labeled cDNA for the indicated genes and were visualized by autoradiography.
We next compared mRNA levels of two other E2F-regulated genes, c-myc and DHFR, in control and ebp1-transfected cells. Although both c-myc and DHFR promoters contain E2F binding sites, E2F affects these promoters differently. For example, mutation of E2F-binding sites in the DHFR promoter indicates that E2F binding is required to activate DHFR expression (22), while mutation of E2F binding sites in the myc promoter increases its activity, similar to the effect observed using the E2F1 promoter (23). Our results indicated that in vector control cells, steady state levels of c-myc mRNA increased between 4 and 20 h after serum addition similar to E2F1 (Fig. 2). The levels of c-myc mRNA then decreased at 28 h. The levels of c-myc mRNA were much lower in ebp1 transfectants 4 h after serum addition as compared to vector controls. In contrast to vector controls, addition of serum did not increase c-myc mRNA after 4 h in ebp1 transfectants. Three DHRF mRNA transcripts were observed in both vector controls and ebp1 transfectants as previously reported (24). The largest (2.1 kb) DHFR transcript was increased at 20 h as compared with 4 h in both ebp1 and control transfectants. At 28 h the levels of the 2.1 kb transcript were decreased only in ebp1 transfectants.
It was possible that the apparent lack of increase of c-myc and E2F1 mRNA in Ebp1 overexpressing cells after serum stimulation was secondary to a generalized proliferation defect in these cells. Therefore, we measured the ability of ebp1 and vector controls to progress through the cell cycle in response to serum. Cells were starved for 24 h and then fed with serum and flow cytometric analysis performed. These data revealed that both vector and ebp1-transfected cells were arrested in G1 after serum starvation and began to move out of G1 after 12 h of serum stimulation. Although Ebp1 transfected cells traversed through the cell cycle somewhat more slowly (beginning after 12 h), at 28 h equal percentages of vector and ebp1-transfected cells were in G1 and S (Table 1). However, E2F1 mRNA was not increased in ebp1 transfectants even at this time (Fig. 2). This would suggest that the decrease in E2F1 transcription observed in ebp1-transfected cells was not due to a generalized block in cell cycle traverse.
Table 1. Cell cycle distribution among Ebp1 and vector transfectants.
| Cell line | Hours after serum addition | Percentage of cells in different phases of the cell cycle | ||
|---|---|---|---|---|
| G1 | S | G2/M | ||
| Vector | 0 | 85.4 | 6.8 | 7.8 |
| Ebp1 | 0 | 77.8 | 11.9 | 10.3 |
| Vector | 4 | 87 | 5.3 | 7.7 |
| Ebp1 | 4 | 81.5 | 6.9 | 11.7 |
| Vector | 12 | 77.7 | 21 | 1.3 |
| Ebp1 | 12 | 84.5 | 7.9 | 7.6 |
| Vector | 20 | 63 | 33.5 | 3.5 |
| Ebp1 | 20 | 80 | 17 | 3 |
| Vector | 28 | 56.5 | 39.3 | 4.2 |
| Ebp1 | 28 | 69.5 | 26.5 | 4 |
LNCaP cells stably transfected with vector alone (vector) or ebp1 were serum-starved for 24 h. Cells were then refed with serum containing media and harvested at the time points indicated. Samples were fixed in ethanol, stained with propidium iodide and analyzed by flow cytometry. Average of two independent experiments, 10 000 cells per point.
Ebp1 represses transcription upon fusion to a heterologous DNA binding protein
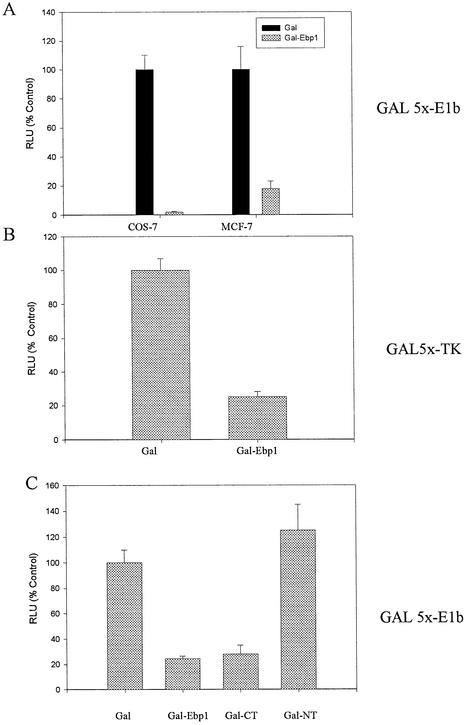
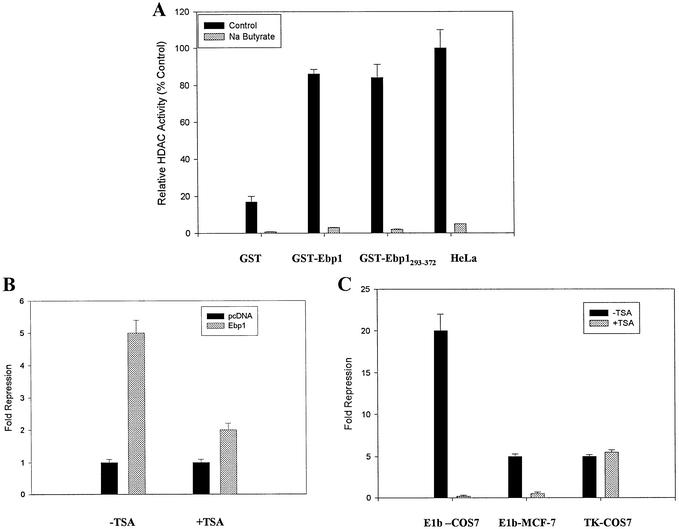
In order to analyze the mechanism through which Ebp1 regulates transcription when it is part of a DNA-bound complex, Ebp1 was targeted to a promoter by fusing Ebp1 in frame with the GAL4 DNA binding domain of the plasmid pSG424. The expression of the fusion protein was assessed by western blot analysis using antibodies to the GAL4 binding domain (data not shown). MCF-7 and COS-7 cells were transfected with a reporter plasmid containing four upstream GAL4 sites fused to an E1b minimal promoter. The ability of the GAL4 DBD-Ebp1 fusion protein (Gal-Ebp1) to modulate transcription from this minimal promoter was tested. Induction of the GAL4 Luc reporter was observed on cotransfection of the pSG424 plasmid carrying the GAL4 DNA binding domain alone, which is consistent with evidence of a previously described cryptic transcriptional activation domain between amino acids 97 and 147 of the GAL4 DBD (25). Transfection of the GAL4 DBD-Ebp1 fusion plasmid significantly (P ≤ 0.05) inhibited induction of luciferase activity by >95% in COS-7 and 82% in MCF-7 cells (Fig. 3A).
Figure 3.
Repression of GAL-4 dependent transcription by Ebp1. Ebp1 cDNA was fused to the GAL4 DNA binding domain in pSG424. Transcriptional repression was measured by determining the ability of the fused Ebp1 to reduce expression of luciferase under the control of an E1b or TK promoter as indicated. (A) COS-7 or MCF-7 cells were transfected with a GAL4 DNA binding domain vector (Gal), or the GAL4-Ebp1 construct, and a luciferase reporter plasmid containing five Gal4 binding sites upstream of the E1b promoter as indicated. RLU was determined 48 h later. (B) COS-7 cells were transfected with the GAL4 DNA binding domain parent vector, or the GAL4-Ebp1 construct, and a reporter plasmid containing five GAL4 DNA binding sites cloned upstream of a minimal TK promoter. Luciferase activity was determined 48 h later. (C) COS-7 cells were transfected with a GAL4 DNA binding domain vector (Gal), the GAL4-Ebp1 expression construct, or constructs encoding N-terminal (NT) (amino acids 1–186) or C-terminal (CT) (amino acids 187–372) Ebp1 fused to the GAL4 DNA binding domain. The E1b reporter construct was used in these experiments. Luciferase activities were measured 48 h later as described.
In order to determine the effects of the GAL-Ebp1 fusion protein on another reporter, we also examined the ability of Ebp1 to repress transcriptional activation of a reporter construct in which five GAL4 DNA binding sites are cloned upstream of a minimal TK promoter element and the firefly luciferase gene (pG5-TK-luc) (17). COS-7 cells were transfected with this reporter construct and Gal4-Ebp1. Ebp1 also repressed the activity of this reporter by 75% (Fig. 3B).
We finally investigated whether the N-terminal (1–186) or C-terminal (186–372) domains of Ebp1 were important for the transcriptional repression. Activity of the luciferase reporter plasmid was inhibited almost 80% by the C-terminal plasmid. In contrast, the N-terminal domain of Ebp1 had no effect on luciferase activity of the reporter plasmid (Fig. 3C).
Ebp1 recruits HDAC activity
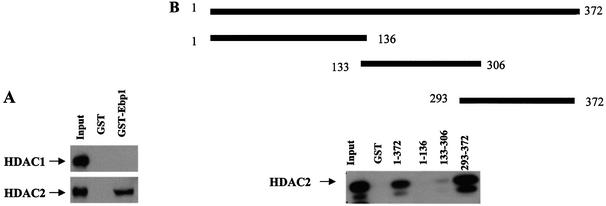
As Ebp1 contains an autonomous repression domain, we sought to begin to examine the mechanism of the transcriptional repression. Recent data indicate that many transcriptional repressors mediate repression through the ability to associate with HDACs as part of large multi-protein complexes (12). Therefore, we tested whether Ebp1 could bind endogenous functional HDAC activity. First, to determine if endogenous HDAC1 or -2 could physically associate with Ebp1, HeLa cell nuclear extracts were incubated with GST-Ebp1 and probed with antibodies to HDAC1 and -2. We found that HDAC2, but not HDAC1, was associated with Ebp1 (Fig. 4A). To determine the HDAC binding domain of Ebp1, a series of GST-Ebp1 truncated fusion proteins were prepared. Equal amounts of the fusion proteins or GST alone were incubated with nuclear lysates of HeLa cells. The results showed that the last 72 C-terminal amino acids of Ebp1 (amino acids 300–372) were sufficient to bind HDAC2 (Fig. 4B).
Figure 4.
Ebp1 binds to HDAC2 in vitro. (A) HeLa cell lysates were incubated with GST-Ebp1 or GST alone (2 µg) and associated proteins determined by western blotting using antibodies to HDAC1 and HDAC2. (B) The C-terminal end of Ebp1 is sufficient to bind HDAC2. Equal amounts of GST-Ebp1 fusion proteins as indicated or GST alone were prepared and incubated with lysates of HeLa cells. Ebp1 associated proteins were analyzed by western blotting using an anti-HDAC2 antibody. Aliquots of the cell lysates were also loaded directly onto gels and analyzed by western blotting (Input).
To determine if Ebp1 was associated with enzymatically active HDACs, GST-Ebp1 was incubated with nuclear extracts of HeLa cells and bound material assayed for HDAC enzymatic activity. Endogenous HDAC activity was readily detectable in GST-Ebp1 bound fractions as opposed to GST alone. A GST-Ebp1 fusion protein encoding amino acids 293–372 (encompassing the HDAC2 binding domain) bound HDAC enzymatic activity as well as full-length Ebp1. The HDAC inhibitor Na butyrate (26) (included in the UBI kit) inhibited activity of all samples by at least 95% (Fig. 5A).
Figure 5.
Transcriptional repression by Ebp1 involves HDACs. (A) GST-Ebp1 binds HDAC activity from Hela cells. Hela nuclear extract (500 µg) was incubated with GST, wild-type GST-Ebp1 or a mutant fusion protein (amino acids 293–372) and bound HDAC activity was determined in the presence or absence of Na butyrate as described in Materials and Methods. As a positive control for HDAC activity, HeLa nuclear extract alone (HeLa) (5 µg) was used. The c.p.m. released in the presence of this control were 4605 ± 565. (B) Ebp1-mediated repression can be relieved by TSA. MCF-7 cells were transfected with pcDNA or pcDNA-ebp1 expression vectors and the E2F1 reporter plasmid. Twenty-four hours later, TSA (160 nM) was added for 16 h and luciferase activity measured. Luciferase activity in the vector control cells (in either the presence or absence of TSA) was given a value of 1. Note however that TSA alone induced a 1.5-fold increase in the activity of the reporter plasmid. (C) COS-7 or MCF-7 cells (as indicated) were transfected with a GAL4 DNA binding domain vector (Gal), or the GAL4-Ebp1 construct, and luciferase reporter plasmids containing five GAL4 binding sites cloned upstream of either the E1b or TK promoter as indicated. TSA (160 nM) was added 24 h after transfection. RLU was determined 16 h later. Fold repression was determined by comparing RLU activity from cells transfected with the GAL-Ebp1 fusion construct versus the GAL DBD alone. TSA alone did not significantly change RLU observed in the presence of the GAL DBD alone.
To address the functional relevance of the ability of Ebp1 to recruit HDAC activity, we tested whether Ebp1-mediated transcriptional repression was sensitive to inhibitors of HDAC activity. We examined the effect of the specific and irreversible HDAC inhibitor trichostatin A (TSA) (27) on the repression mediated by Ebp1 on the E2F1 luciferase promoter construct. As shown in Figure 5B, we observed that an 18 h treatment of MCF-7 cells with 160 nM TSA inhibited Ebp1-mediated transcriptional repression 65%, suggesting that Ebp1 repression of this promoter was partially dependent upon HDAC activity.
To determine if HDAC activity is also required for repression by GAL-Ebp1 fusion proteins, we examined the effects of TSA on repression mediated by GAL-Ebp1. We found that TSA abrogated the ability of Ebp1 to affect the E1b promoter. In contrast to the effects of TSA on the E1b promoter, TSA did not prevent Ebp1-induced repression of the TK promoter (Fig. 5C).
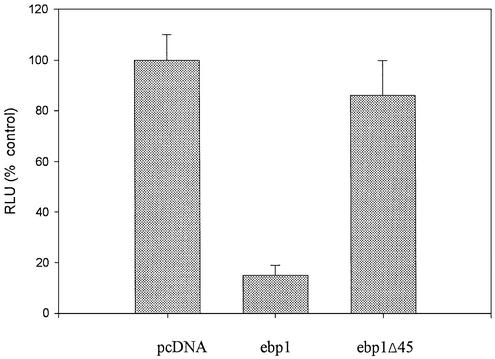
We next analyzed the ability of an Ebp1 protein (ebp1 Δ 45) (2) that lacks part of the HDAC binding domain to repress E2F1-mediated transcription. This Ebp1 mutant is stably expressed in mammalian cells (2). MCF-7 cells were transfected with either wild-type ebp1 or the deletion mutant. Deletion of the HDAC binding domain resulted in the inability of Ebp1 to repress transcription of the E2F1 promoter (Fig. 6).
Figure 6.
Ebp1 transcriptional repression relies on the presence of an HDAC binding domain. MCF-7 cells were transfected with 1 µg of pcDNA3, full-length ebp1 or an ebp1 mutant that lacks the C-terminal 45 amino acids (ebp1 Δ 45). The ability of these constructs to affect activity of an E2F1 promoter was determined as previously described in Figure 1.
DISCUSSION
Ebp1 was isolated in our laboratory as an ErbB3 binding protein that translocates to the nucleus of ErbB-3/4 expressing AU565 human breast cancer cells after treatment with the cognate ligand HRG (1). The regulated nuclear accumulation of Ebp1 in this system suggested that Ebp1 might have some activity in control of transcription. We have previously demonstrated that Ebp1 binds Rb in vitro and that ectopic expression of ebp1 inhibits transcription of the E2F1-regulated cyclin E gene in both Rb positive and Rb null cell lines (13). In this report, we demonstrate that the ectopic expression of ebp1 inhibits transcription from other E2F-regulated reporter plasmids and from endogenous E2F-regulated genes. The E2F elements in the promoters contributed to this effect. We also show, using GAL4 DNA binding domain fusion proteins, that Ebp1 contains an autonomous silencing domain within the C-terminal region which mediates transcriptional repression when tethered to a promoter sequence. This region of Ebp1 bound functional HDAC activity. The ability of Ebp1 to repress transcription appeared in part to depend on its ability to recruit HDAC activity.
We found that ectopic expression of ebp1 inhibited luciferase activity from several E2F1-regulated reporter plasmids. We have previously demonstrated that Ebp1 does not non-specifically inhibit transcription since expression of luciferase reporters driven by either the SV40 or AP1 promoter was not affected (13). In addition, the ability of Ebp1 to decrease luciferase activity of the reporter plasmids depended on the presence of E2F1 elements within the promoters. We also examined the effects of overexpression of Ebp1 on endogenous E2F1-regulated genes using stable transfectants of LNCaP cells in which the constitutive level of Ebp1 was 2–3-fold higher than in cells transfected with the control vector (2). E2F1 and c-myc mRNA were not observed in either vector control or ebp1 transfectants after serum starvation as expected (21). Serum did induce an increase in steady state levels of E2F1 and c-myc mRNA in ebp1 transfectants 4 h after serum stimulation, but this increase was 4-fold less than that observed in vector controls. In addition, E2F1 and c-myc steady state transcript levels in ebp1 transfectants did not rise at later time points after serum stimulation, in contrast to vector control cells where a peak increase was observed 20 h after serum addition. However, the expression of DHFR mRNA after serum treatment was similar in ebp1 as compared to control transfectants. It is of interest that DHFR was not affected by ebp1 overexpression in the same manner as the myc or E2F1 promoters. DeGregori et al. (6) found that transcription of DHFR was not affected by infection of REF cells with adenoviral E2F1, whereas c-myc and E2F1 were strongly induced. Thus, the DHFR promoter is not regulated by the E2F1 transcription factor in the same manner as the E2F1 or c-myc promoters. Finally, FACS analysis revealed that Ebp1 cells do enter S phase, although at a slightly later time point than vector controls. However, at 28 h similar percentages of cells were in G1 and S in vector and ebp1 transfectants. Thus, the inhibition of E2F1 mRNA levels was not due only to a generalized inhibition of cell cycle progression.
To further explore possible mechanisms of Ebp1-mediated transcriptional repression, we determined if Ebp1 could recruit HDAC activity. Recently, a large body of evidence highlights the importance of HDACs in transcriptional repression. HDACs are recruited by several transcriptional repressors such as the bHLH-zip MAD proteins, the zinc finger protein YY1 and certain unliganded nuclear receptors and, most pertinent to our work, Rb (12). Ebp1 was able to bind HDAC activity from HeLa cell nuclear extracts. The HDAC binding site of Ebp1 was mapped to amino acids 293–372. This was part of the C-terminal domain of Ebp1, which when fused to the DNA binding domain of GAL4, repressed transcription of artificial promoters containing Gal4 binding sites. Deletion of this domain abrogated the ability of Ebp1 to inhibit transcription. This domain is also critical for Ebp-1-induced growth inhibition (13), consistent with the hypothesis that the growth inhibitory effects of Ebp1 correlate with the ability to repress transcription.
In western blot analysis, we demonstrated that Ebp1 bound HDAC2 but not HDAC1. These findings suggest that the ability of Ebp1 to bind HDACs is important in its transcriptional repression effects. The fact the Ebp1 bound HDAC2 but not HDAC1 is somewhat surprising as these proteins share 84% identity. However, increasing data suggest that HDAC1 and HDAC2 may play complementary roles in transcriptional repression. For example, Humphrey et al. (28) proposed a model in which HDAC1 core complexes can recruit HDAC2 containing complexes to form higher order complexes on chromatin. It is possible that Ebp1 is part of a transiently ordered complex in vivo (28). Whether Ebp1 recruits HDAC directly or indirectly has not yet been determined. We have demonstrated that Ebp1 can bind Rb and it is well known that Rb binds HDAC activity through its interaction with the Rbp1 bridging protein (29), RbAp48 (30) or c-ski, Sno and Sin3A (31). Whether Ebp1 associates with any or all of these proteins remains to be determined. Our data do suggest that Ebp1 could be part of a transcriptional repressor complex that includes Rb or other pocket proteins, and HDAC that forms on E2F binding sites on promoters. Studies are underway in our laboratory to demonstrate the presence of Ebp1 in nuclear extracts bound to E2F1 consensus elements by EMSA and CHIP assays.
We also extended our previous work indicating that Ebp1 could inhibit transcription of E2F-regulated genes in Rb negative cells. The mechanisms of Ebp1 inhibition of reporter activity in Rb negative cells are not known at this time. However, we have found that in MCF-7 cells, Ebp1 can bind to the Rb-related p130 pocket protein, but not p107 (data not shown). The ability to bind to another member of the Rb family of proteins suggests a mechanism that could account for the ability of Ebp1 to inhibit transcription in cells such as SAOS-2 which lack functional Rb. Alternatively, Ebp1 may associate with other transcriptional repressor complexes such as Myc-Mad that regulate E2F function in an Rb-independent manner.
Our results also suggest that recruitment of HDACs may be only one mechanism of Ebp1-induced repression. For example, the TSA-mediated reversal of Ebp1’s repression of the E2F1 promoter was not complete, suggesting that Ebp1 might use other mechanisms in addition to histone deacetylation to repress transcription. Although TSA abrogated the ability of Ebp1 to affect a GAL4-E1b promoter, TSA did not affect the ability of a GAL4-Ebp1 fusion protein to inhibit the TK promoter. Differential effects of TSA on the repression ability of other proteins has similarly been found using these promoters. For example, the Rb-mediated repression of the GAL4-TK promoter was not affected by TSA, while repression of the GAL4-E1b promoter was reversed (27). As the differential effects of TSA on the repressive ability of Rb and Ebp1 are similar, it is possible that Ebp1 may, in certain contexts, function through Rb. Conversely, TSA led to relief of repression of GAL-Elk (1–206) on a TK, but not an E1b reporter (17). These findings suggest that Ebp1 has multiple mechanisms of transcription repression. Similar non-HDAC-related modes of activity have been suggested for the transcriptional repressors mSin3 and NCoR (32). Ebp1, in addition to recruiting HDACs, may also inhibit promoter activity by direct inhibition of transcription factors or by interaction with ATP-dependent chromatin remodeling complexes such as SNF/SWI.
In conclusion, the results presented here indicate that Ebp1 can inhibit transcription of cell cycle related genes containing E2F consensus sites. The transcription of both exogenous and endogenous genes was affected. The ability of Ebp1 to recruit HDAC activity contributed to its ability to repress promoter activity. The current results suggest a potential mechanism by which Ebp1 suppresses cell growth. We hypothesize that repression of E2F activation of cell cycle related genes by Ebp1 leads to inhibition of cell cycle progression and subsequent cellular differentiation. It is possible that Ebp1, via its ability to interact with ErbB3 and affect E2F1-regulated transcription, may be able to transmit HRG-induced signals from the cell membrane that ultimately result in changes in gene expression. The factors regulating the ability of Ebp1 to transmit signals from ErbB3 to E2F-regulated genes requires further study.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Richard Pestell for the cyclin D1 reporter plasmids, Dr Joseph Nevins for the E2F1 plasmids, Dr Andrew Sharrocks for the pG5-TK luc plasmid, Dr Arun Rishi for the c-myc reporter and Dr Roger Davis for the pSG424 plasmid and pG5-E1b plasmids. We thank Dr Miriam Smyth for help with flow cytometric analysis. This work was supported in part by NIH grants R01 CA76047 and R21 088882-01 (to A.W.H.).
REFERENCES
- 1.Yoo J.Y., Wang,X.W., Rishi,A.K., Lessor,T., Xia,X.M., Gustafson,T.A. and Hamburger,A.W. (2000) Interaction of the PA2G4 (EBP1) protein with ErbB-3 and regulation of this binding by heregulin. Br. J. Cancer, 82, 683–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Y., Fondell,J.D., Wang,Q., Xia,X., Cheng,A., Lu,M.L. and Hamburger,A.W. (2002) Repression of androgen receptor mediated transcription by the ErbB-3 binding protein, Ebp1. Oncogene, 21, 5609–5618. [DOI] [PubMed] [Google Scholar]
- 3.Lessor T.J., Yoo,J.Y., Xia,X., Woodford,N. and Hamburger,A.W. (2000) Ectopic expression of the ErbB-3 binding protein ebp1 inhibits growth and induces differentiation of human breast cancer cell lines. J. Cell Physiol., 183, 321–329. [DOI] [PubMed] [Google Scholar]
- 4.Lavia P. and Jansen-Durr,P. (1999) E2F target genes and cell-cycle checkpoint control. Bioessays, 21, 221–230. [DOI] [PubMed] [Google Scholar]
- 5.Yee A.S., Raychaudhuri,P., Jakoi,L. and Nevins,J.R. (1989) The adenovirus-inducible factor E2F stimulates transcription after specific DNA-binding. Mol. Cell. Biol., 9, 578–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeGregori J., Kowalik,T. and Nevins,J.R. (1995) Cellular targets for activation by the E2F1 transcription factor include DNA synthesis-regulatory and G1/S-regulatory genes. Mol. Cell. Biol., 15, 5846–5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adams P.D. and Kaelin,W.G.,Jr (1995) Transcriptional control by E2F. Semin. Cancer Biol., 6, 99–108. [DOI] [PubMed] [Google Scholar]
- 8.Nevins J.R. (1992) E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science, 258, 424–429. [DOI] [PubMed] [Google Scholar]
- 9.Rayman J.B., Takahashi,Y., Indjeian,V.B., Dannenberg,J.H., Catchpole,S., Watson,R.J., te Riele,H. and Dynlacht,B.D. (2002) E2F mediates cell cycle-dependent transcriptional repression in vivo by recruitment of an HDAC1/mSin3B corepressor complex. Genes Dev., 16, 933–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magnaghi-Jaulin L., Groisman,R., Naguibneva,I., Robin,P., Lorain,S., Le Villain,J.P., Troalen,F., Trouche,D. and Harel-Bellan,A. (1998) Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature, 391, 601–605. [DOI] [PubMed] [Google Scholar]
- 11.Brehm A., Miska,E.A., McCance,D.J., Reid,J.L., Bannister,A.J. and Kouzarides,T. (1998) Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature, 391, 597–601. [DOI] [PubMed] [Google Scholar]
- 12.Cress W.D. and Seto,E. (2000) Histone deacetylases, transcriptional control and cancer. J. Cell Physiol., 184, 1–16. [DOI] [PubMed] [Google Scholar]
- 13.Xia X., Cheng,A., Lessor,T., Zhang,Y. and Hamburger,A.W. (2001) Ebp1, an ErbB-3 binding protein, interacts with Rb and affects Rb transcriptional regulation. J. Cell Physiol., 187, 209–217. [DOI] [PubMed] [Google Scholar]
- 14.Johnson D.G., Ohtani,K. and Nevins,J.R. (1994) Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev., 8, 1514–1525. [DOI] [PubMed] [Google Scholar]
- 15.Watanabe G., Albanese,C., Lee,R.J., Reutens,A., Vairo,G., Henglein,B. and Pestell,R.G. (1998) Inhibition of cyclin D1 kinase activity is associated with E2F-mediated inhibition of cyclin D1 promoter activity through E2F and Sp1. Mol. Cell. Biol., 18, 3212–3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma J. and Ptashne,M. (1987) Deletion analysis of GAL4 defines two transcriptional activating segments. Cell, 48, 847–853. [DOI] [PubMed] [Google Scholar]
- 17.Yang S.H., Vickers,E., Brehm,A., Kouzarides,T. and Sharrocks,A.D. (2001) Temporal recruitment of the mSin3A-histone deacetylase corepressor complex to the ETS domain transcription factor Elk-1. Mol. Cell. Biol., 21, 2802–2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hussain A., Yang,H., Protzman,J. and Melera,P.W. (1994) Selection for the expression of one form of Chinese hamster dihydrofolate reductase over another during growth in methotrexate. Gene, 144, 277–282. [DOI] [PubMed] [Google Scholar]
- 19.Carroll J.S., Prall,O.W.J., Musgrove,E.A. and Sutherland,R.L. (2000) A pure estrogen antagonist inhibits cyclin E-Cdk2 activity in MCF-7 breast cancer cells and induces accumulation of p130-E2F4 complexes characteristic of quiescence. J. Biol. Chem., 275, 38221–38229. [DOI] [PubMed] [Google Scholar]
- 20.Johnson D.G., Schwarz,J.K., Cress,W.D. and Nevins,J.R. (1993) Expression of transcription factor E2F1 induces quiescent cells to enter S phase. Nature, 365, 349–352. [DOI] [PubMed] [Google Scholar]
- 21.Ohtani K., DeGregori,J. and Nevins,J.R. (1995) Regulation of the cyclin E gene by transcription factor E2F1. Proc. Natl Acad. Sci. USA, 92, 12146–12150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Means A.L., Slansky,J.E., Mcmahon,S.L., Knuth,M.W. and Farnham,P.J. (1992) The Hip1 binding-site is required for growth-regulation of the dihydrofolate-reductase gene promoter. Mol. Cell. Biol., 12, 1054–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dyson N. (1998) The regulation of E2F by pRB-family proteins. Genes Dev., 12, 2245–2262. [DOI] [PubMed] [Google Scholar]
- 24.Yang H.H. and Melera,P.W. (1994) A genetic-polymorphism within the third poly(A) signal of the DHFR gene alters the polyadenylation pattern of DHFR transcripts in CHO cells. Nucleic Acids Res., 22, 2694–2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen S.B., Nicol,R. and Stavnezer,E. (1998) A domain necessary for the transforming activity of SnoN is required for specific DNA binding, transcriptional repression and interaction with TAF(II)110. Oncogene, 17, 2505–2513. [DOI] [PubMed] [Google Scholar]
- 26.Ohno Y., Lee,J., Fusunyan,R.D., MacDermott,R.P. and Sanderson,I.R. (1997) Macrophage inflammatory protein-2: chromosomal regulation in rat small intestinal epithelial cells. Proc. Natl Acad. Sci. USA, 94, 10279–10284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo R.X., Postigo,A.A. and Dean,D.C. (1998) Rb interacts with histone deacetylase to repress transcription. Cell, 92, 463–473. [DOI] [PubMed] [Google Scholar]
- 28.Humphrey G.W., Wang,Y., Russanova,V.R., Hirai,T., Qin,J., Nakatani,Y. and Howard,B.H. (2001) Stable histone deacetylase complexes distinguished by the presence of SANT domain proteins CoREST/kiaa0071 and Mta-L1. J. Biol. Chem., 276, 6817–6824. [DOI] [PubMed] [Google Scholar]
- 29.Lai A., Lee,J.M., Yang,W.M., DeCaprio,J.A., Kaelin,W.G.,Jr, Seto,E. and Branton,P.E. (1999) RBP1 recruits both histone deacetylase-dependent and -independent repression activities to retinoblastoma family proteins. Mol. Cell. Biol., 19, 6632–6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nicolas E., Morales,V., Magnaghi-Jaulin,L., Harel-Bellan,A., Richard-Foy,H. and Trouche,D. (2000) RbAp48 belongs to the histone deacetylase complex that associates with the retinoblastoma protein. J. Biol. Chem., 275, 9797–9804. [DOI] [PubMed] [Google Scholar]
- 31.Tokitou F., Nomura,T., Khan,M.M., Kaul,S.C., Wadhwa,R., Yasukawa,T., Kohno,I. and Ishii,S. (1999) Viral ski inhibits retinoblastoma protein (Rb)-mediated transcriptional repression in a dominant negative fashion. J. Biol. Chem., 274, 4485–4488. [DOI] [PubMed] [Google Scholar]
- 32.Pazin M.J. and Kadonaga,J.T. (1997) What’s up and down with histone deacetylation and transcription? Cell, 89, 325–328. [DOI] [PubMed] [Google Scholar]