Abstract
In most eukaryotic introns the 3′ splice site is defined by a surprisingly short AG consensus found a variable distance downstream of the branch site. Exactly how the spliceosome determines which AG to use, however, is not well understood. Previously we showed that when the branch site and 3′ splice site AG are supplied by separate RNA molecules, there is a strong preference for use of the 5′-most AG in the 3′ splice site-containing RNA. Here we show that this apparent 5′→3′ directionality holds even when this RNA contains four tandem repeats of a 6-nt sequence containing AG. Exactly the same pattern of 3′ splice site choice was observed when the same tandem repeats were incorporated into a full-length splicing substrate. When the 3′ splice site AG is supplied by a separate RNA, that RNA must be linear with an unobstructed 5′ end. Similarly, the branch-containing RNA must be truncated immediately 3′ to the polypyrimidine tract. A model is presented that incorporates these observations and reconciles previously proposed mechanisms for 3′ splice site selection.
Precise excision of introns is an essential step in eukaryotic gene expression. Most introns in the human genome (105–106 different sequences) are removed by a single macromolecular machine, the major spliceosome. Within this machine, intron excision occurs in two steps: cleavage at the 5′ splice site coupled with lariat formation at the branch site, followed by exon ligation at the 3′ splice site (1, 2). However, the exact mechanisms by which these sites are recognized are not entirely understood. This is particularly true for the 3′ splice site. In budding yeast, the only conserved sequence defining this key site is a YAG/trinucleotide (where Y denotes pyrimidine and “/” denotes the splice site) found a variable distance downstream of the branch site. In mammals the region between the branch site and YAG/is generally rich in pyrimidines (the polypyrimidine tract or PPT) (1, 3, 4). Notably, the natural 3′ splice site in some introns is an AAG/or GAG/(5), with the order of preference in a test system being CAG ≈ UAG>AAG>GAG (6). Thus, the sequence defining the exact site of exon ligation is apparently just AG, which could occur randomly every 16 dinucleotides. Given such odds, the spliceosome undoubtedly uses other information to distinguish the correct 3′ splice site AG from any others nearby. Exactly what that information is, however, and how it is processed by the splicing machinery remain a matter of open debate (see ref. 7 and Discussion).
To better study the mechanisms of 3′ splice site selection by the human spliceosome, we recently developed a bimolecular (trans) assay for exon ligation in HeLa nuclear extracts (8). In this system, spliceosomes assemble and undergo the first chemical step on a 5′ substrate consisting of a 5′ exon and intron truncated immediately 3′ to the PPT (Fig. 1A). Exon ligation is then initiated by addition of a separate RNA, the 3′ substrate, containing one or more AGs plus a downstream exon. Previously we found that the only sequence required for accurate 3′ splice site definition in this system is the YAG consensus sequence; there was no advantage for having a PPT covalently attached either upstream or downstream of the 3′ splice site AG in the 3′ substrate. In fact, an intronic sequence as short as 5′-GACAG/(where “/” denotes the site of exon ligation) was sufficient to designate a 3′ splice site. However, when the 3′ substrate contained more than one AG, the 5′-most invariably was used, suggesting that 3′ splice site selection in this system occurs with 5′→3′ directionality (8).
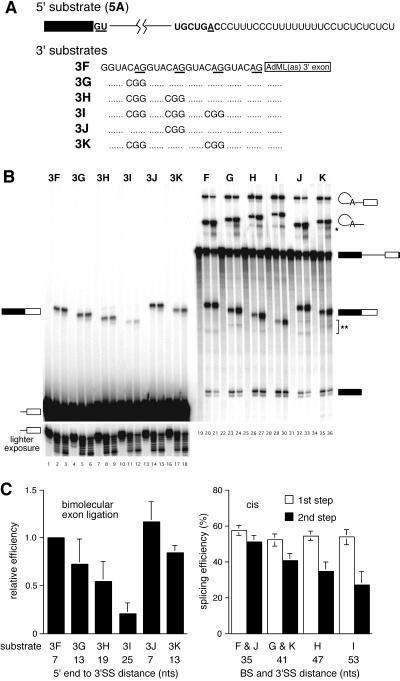
Figure 1.
Behavior of tandemly repeated 3′ splice sites in the bimolecular exon ligation assay and cis constructs. (A) 5′ and 3′ substrate sequences. Full-length RNAs corresponded to exact joining of the 5′ and 3′ substrates. (B) Denaturing polyacrylamide gel of bimolecular exon ligation (Left) and cis (Right) splicing reactions. (Upper Left) Labeled 3′ substrates were added to splicing reactions in which unlabeled RNA 5A had been preincubated for 30 min, and incubations continued for 0, 15, or 30 min. (Lower Left) Shorter exposure of the 3′ substrates to show the extent to which these RNAs were degraded during the reactions. (Right) Each cis splicing substrate was incubated under splicing conditions for 0, 30, or 60 min. Migration positions of substrates, intermediates, and products are symbolized to the left and right. Extra bands below lariat products in G, H, I, and K (*) are likely exonuclease cleavage products ending at the PPT (17). Extra bands below the ligated exons (**) have migration rates expected for debranched lariats. (C) Observed splicing efficiencies for bimolecular exon ligation (Left) and cis splicing (Right) assays. For each substrate, the distance indicated is from the 5′ end (bimolecular substrates) or branch (cis substrates) to the 3′ splice site AG used. For cis substrates, the efficiencies of RNAs J and F and RNAs K and G were combined because the branch to 3′ splice site distance was the same for each pair. In all cases, error bars indicate the SDs from at least three independent experiments, except for 3I, which was from two.
In the current study we have extended these findings by showing that this apparent 5′→3′ directionality holds even when the 3′ substrate contains four equally spaced AGs presented in identical sequences. Structural modifications of the 3′ substrate reveal that it must be linear with an unobstructed 5′ end. Taken together these results argue strongly that the splicing machinery can only engage an exogenous RNA for use as a 3′ substrate via a free end, accounting for the 5′→3′ directionality. The same pattern of 3′ splice site choice was obtained when the tandem repeats were presented to the spliceosome in a cis splicing construct, suggesting that the mechanism of 3′ splice site choice in the bimolecular system is the same as that employed on full-length introns. Because structural modifications to the 3′ end of the 5′ substrate also inhibit bimolecular exon ligation, the 3′ substrate likely gains entry to the active site for exon ligation via a space that normally is occupied by the intronic RNA downstream of the branch site in cis constructs. A model that incorporates these observations and reconciles previously proposed mechanisms for 3′ splice site selection is presented.
Materials and Methods
RNA Synthesis.
PCR products for transcription of all 5′ substrates (RNAs 5A–5D) were generated from pAdMLΔAG (8) by using primers that incorporated the appropriate 5′ and 3′ ends. All 3′ substrates contained the AdMLas exon, and templates for their transcription were generated by PCR from pAdMLpar (8). Tandem repeats containing full-length substrates (RNAs F–K) initially were produced by splinted ligation (9) of RNA 5A to the corresponding 3′ substrate (RNAs 3F–3K). Each ligated product then was reverse-transcribed, cloned, and sequenced. All full-length RNAs subsequently were transcribed from the appropriate plasmid.
All labeled RNAs were transcribed with T7 RNA polymerase (Stratagene) by using either [α-32P]UTP or [α-32P]ATP (DuPont/NEN) under standard conditions (9). All 5′ and full-length substrates were capped with G(5′)ppp(5′)G (New England Biolabs); 3′ substrates were initiated with GMP. Unlabeled RNAs were transcribed by using the T7 mMESSAGE mMACHINE kit or the T7 MEGAshortscript kit (Ambion) and quantified spectrophotometrically. All RNAs were gel-purified before use.
Splicing and Splicing Efficiency.
Full-length substrates were incubated in 40–50% HeLa nuclear extract (cells obtained from Cellex Biosciences, Minneapolis)/60–70 mM KCl/2 mM MgCl2/1 mM ATP/5 mM creatine phosphate/0.2 mg/ml tRNA/0.05 mg/ml 3′-dATP (to suppress polyadenylation) at 30°C for times indicated. For bimolecular exon ligation reactions, the 5′ substrate (35 nM input) was preincubated under splicing conditions at 30°C for 30 min to accumulate intermediate-containing spliceosomes; exon ligation then was initiated by addition of 175 nM 3′ substrate. For both assays, reactions were stopped by addition of splicing stop buffer (100 mM Tris⋅HCl, pH 7.5/10 mM EDTA/150 mM NaCl/300 mM sodium acetate/1% SDS) at the indicated time, and RNAs were extracted and separated on denaturing polyacrylamide gels. All gels were imaged and quantified with a Molecular Dynamics PhosphorImager by using the accompanying imagequant software.
The efficiency of the first step of splicing for full-length substrates was calculated as the fraction of all RNA species in the lane that had undergone lariat formation (i.e., 5′ exon, lariat intermediate, ligated exons, and lariat product). The second-step efficiency was taken to be the fraction of total RNA species that had undergone exon ligation (ligated exons and lariat product). Bimolecular exon ligation efficiencies initially were expressed as the percentage of input 5′ substrate that ended up in ligated exons. To average data from several experiments, these efficiencies were normalized to that of a standard substrate used in all experiments. For full-length substrates, second-step yields were calculated at 60-min time points—longer incubations did not significantly change the figures (data not shown). In bimolecular reactions, final product yields were calculated at 30 min, because increased degradation made quantification more difficult at longer times.
RNA Circularization.
The AdMLas 3′ substrate was annealed with a 30-nt DNA oligonucleotide (5′-CTCTTGGATCCTGTCCTGCAGGTCGACGTT-3′) whose complementarity spanned the ends of the RNA. The 3′ substrate and bridging oligo (both 0.5 μM) were annealed by heating to 90°C in 10 mM Tris⋅HCl, pH 7.5/100 mM NaCl/0.1 mM EDTA and then cooling to room temperature over the period of 1 hr. After ethanol precipitation, the RNA/DNA hybrid was incubated with T4 DNA ligase under standard conditions (9). Circular RNAs initially were identified by their anomalous migration relative to linear species on low- and high-percentage denaturing polyacrylamide gels. After gel purification, their circularity was confirmed by limited alkaline cleavage at 90°C in 250 mM NaHCO3 (pH 9.0) for 3 min followed by electrophoresis next to similarly treated linear RNAs. The accessibility of the 3′ splice site region to RNaseH cleavage was determined by incubating RNAs in nuclear extract with a 10-nt DNA oligo (5′-GATCCTGTCC-3′) whose complementarity spanned the 3′ splice site.
Results
3′ Splice Site Choice in Substrates Containing Tandemly Repeated AGs.
In our previous paper (8), we reported that the 3′ splice site chosen in the bimolecular exon ligation assay was always the AG closest to the 5′ end of the 3′ substrate. Although this was suggestive of a linear 5′→3′ search mechanism, other explanations could not be eliminated. For instance, because in those experiments the competing AGs were presented in different sequence contexts, it was possible that internal AGs always had greater propensities to be masked by intramolecular secondary structures. Furthermore, a number of proteins are known to interact with the region around the 3′ splice site AG (10–16), and it is reasonable to expect that such proteins might bind different sequences with different affinities. Therefore, for this study we constructed 3′ substrates containing four direct repeats of either GUACAG (Fig. 1A) or GUUCAG (not shown) plus various combinations of AG-to-GG mutations. These substrates were designed specifically to minimize possible sequence context effects by placing all 3′ substrate AGs within identical adjacent sequences that were neither particularly pyrimidine- nor purine-rich.
When analyzed in the bimolecular exon ligation assay, each tandem repeat-containing 3′ substrate yielded a single ligated exon product that appeared as a doublet with an intense upper band and a fainter lower band (Fig. 1B, lanes 1–18). The stepwise increase in migration of this doublet with each consecutive AG-to-GG mutation indicated that the major 3′ splice site in each 3′ substrate was the 5′-most AG. However, it was not initially clear whether the doublet reflected 3′ end heterogeneity of the spliced product or resulted from some use of the next AG downstream. Because the 3′ substrates exhibited significant degradation during the course of the reactions (Fig. 1B Lower Left), and RNA 3I exhibited this same doublet pattern even though it contained only one viable 3′ splice site AG, 3′ end heterogeneity was suspected. To test this, we constructed substrates 3J and 3K wherein the AG immediately downstream of the 5′-most AG was mutated to a nonfunctional GG. If the adjacent AG was used in RNAs 3F–3I, then the lower band in the ligated exon doublet should have disappeared or migrated more rapidly in the ligated exon products from RNAs 3J and 3K. However, because exactly the same doublets were observed with these latter constructs, the fainter band could be attributed to partial 3′ end degradation and not use of more than one AG. An additional observation with the tandem repeat-containing 3′ substrates was that the efficiency of bimolecular exon ligation decreased stepwise with each mutation that moved the 5′-most AG 6 nt further downstream such that RNA 3I spliced only 20% as efficiently as RNA 3F (Fig. 1C Left).
To determine how similar this pattern of 3′ splice site choice was to that of cis splicing reactions, we next tested the tandem repeats in full-length constructs generated by ligating the 5′ substrate to each 3′ substrate to create intact pre-mRNAs. All these RNAs underwent the first step of splicing with equal efficiency (Fig. 1C Right, open bars). Thus, neither spliceosome assembly nor lariat formation was affected by the sequence variations at the 3′ end of the intron. Notably, only one size-ligated exon product was observed for each cis construct and it was the same as that observed in the bimolecular assay (Fig. 1B, lanes 19–36). The migration of each lariat intron product also varied as expected. Moreover, as in the bimolecular assay, the efficiency of exon ligation in the cis constructs decreased stepwise as the 3′ splice site AG was moved away from the branch site and PPT in 6-nt increments (Fig. 1C Right, solid bars). For both the bimolecular and cis constructs, identical results were obtained for the GUUCAG repeat constructs (data not shown).
These experiments demonstrate that when potential effects of sequence context are minimized, the 3′ splice site chosen in the bimolecular assay is still the 5′-most AG. Our finding that this same pattern is replicated in cis constructs suggests that the mechanism of 3′ splice site selection is the same for the bimolecular and cis assays. This conclusion is supported further by the observation that the efficiency of exon ligation varied similarly in both assays as the 5′-most AG was moved further downstream.
Effects of Secondary Structures in the 3′ Substrate.
We next examined the effects of secondary structures in the 3′ substrate. If the reason the 5′-most AG always served as the site of exon ligation in the above experiments was simply because, being closer to the end, it had a lower propensity for being masked by internal secondary structures or bound proteins than downstream AGs, then placing a secondary structure upstream of it might actually allow downstream AGs to compete more effectively. On the other hand, if a linear search is involved, then hairpins placed upstream of the 5′-most AG should block bimolecular exon ligation at all downstream sites.
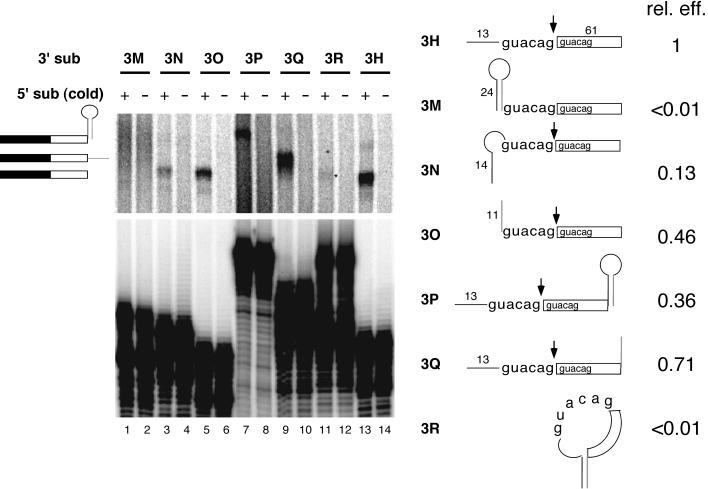
Secondary structure effects initially were assessed by appending a 24-nt hairpin upstream of the tandem AGs in RNA 3H (see Fig. 1A) to create RNA 3M. The hairpin (see Fig. 4, RNA 5B) was designed to eliminate any possible internal AG sites, and it included a CUUG tetraloop known to stabilize adjacent stem structures (18). According to Zuker's mfold program (http://www.ibc.wustl.edu/∼zuker/rna), the free energy of the hairpin is −22.6 kcal/mol at 37°C in 1 M NaCl. Therefore, at 30°C and 100 mM monovalent ions (similar to splicing conditions), this hairpin should be >99.9% formed. When tested in the bimolecular exon assay, RNA 3M yielded no detectable ligated exons (Fig. 2, lane 1). In contrast, substrates having only the first or second half of the hairpin appended to their 5′ ends (RNAs 3N and 3O, respectively), or the entire hairpin at the 3′ end (RNA 3P), gave readily detectable products (Fig. 2, lanes 3, 5, and 7). Thus, an RNA hairpin blocked bimolecular exon ligation when located at the 5′ end, upstream of the first AG, but not at the 3′ end of the 3′ substrate.
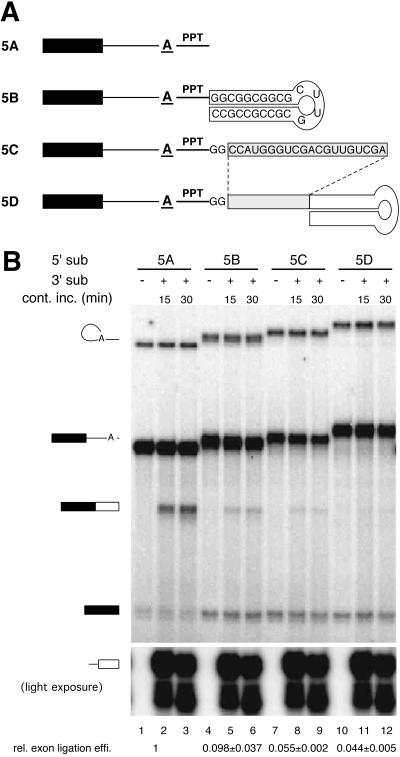
Figure 4.
Extension of the 5′ substrate interferes with bimolecular exon ligation. (A) Structures of 5′ substrates used; sequences shown were appended directly to the 3′ end of RNA 5A (Fig. 1A). (B) Bimolecular exon ligation of RNAs 5A–5D with AdMLas 3′ substrate. All RNAs were internally labeled with [α-32P]UTP; 5′ substrates had lower specific activities than the 3′ substrate. Relative bimolecular exon ligation efficiencies (data from three experiments) are indicated at the bottom.
Figure 2.
Effects of secondary structures in the 3′ substrate. (Left) Denaturing polyacrylamide gel showing splicing reactions containing preincubated, unlabeled 5′ substrate (odd lanes) or no 5′ substrate (even lanes) 30 min after addition of indicated 3′ substrates. (Right) 3′ substrate structures and relative exon ligation efficiencies. The hairpin sequence was identical to that in Fig. 4A. In RNAs 3M, 3N, 3O, and 3R, upstream sequences were attached directly to the first GUACAG repeat in RNA 3H, replacing the 5′-most 13 nt. The band in lane 11 (*) most likely resulted from splicing of RNA 3R after 3′ end degradation (which would generate RNA 3N); if full-length RNA 3R had spliced, the ligated product would have been identical to that from RNA 3Q.
To test whether an AG could serve as the 3′ splice site when it was contained in a hairpin loop, we created RNA 3R containing one-half of the above hairpin at each end of the RNA. Hybridization of the complementary sequences should generate a stem structure with a large loop containing both AGs and the entire 3′ exon within it. Like RNA 3M, RNA 3R failed to participate in exon ligation (Fig. 2, lane 11; if it had spliced, the product would have been the same size as that in lane 9), whereas RNAs 3N and 3Q, which each had only half of the stem, did give appropriately sized ligated exons. In all cases reactions containing no 5′ substrate yielded no products (Fig. 2, lanes 2, 4, 6, 8, 10, 12, and 14), confirming a requirement for both substrates. Taken together, these results indicate that a secondary structure upstream of the 5′-most AG inhibits the ability of a 3′ substrate to participate in bimolecular exon ligation.
A Circular RNA Will Not Function as a 3′ Substrate.
The above experiments suggested that a functional 3′ substrate requires a free 5′ end. However, hairpins and double-stranded regions generally are not found between the branch and 3′ splice sites in mammalian introns. Such structures could interfere with exon ligation by simple steric hindrance or by providing a target for double-strand-specific RNA-binding proteins, which, in turn, could inhibit exon ligation. To eliminate these possibilities, we next examined the ability of circular RNAs to function as 3′ substrates.
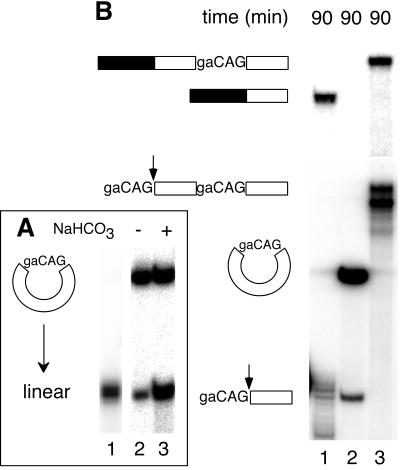
The AdMLas 3′ substrate was circularized by splinted ligation (9). This reaction yielded a number of products, including RNAs with migration patterns expected for circular monomers and linear and circular dimers. To confirm identification of the circular monomer, that RNA was subjected to partial alkaline hydrolysis (Fig. 3A). Because single nicks caused it to comigrate with the linear substrate and additional nicks generated the expected heterogeneous cleavage pattern (a smear) below the linear molecules, the isolated RNA was clearly a circle.
Figure 3.
A circular 3′ substrate does not participate in bimolecular exon ligation. (A) Partial alkaline hydrolysis of the circular monomer. A single nick (lane 3) in the circular RNA (lane 2) first generates molecules of identical migration to linear RNA (lane 1); additional nicks produce a heterogeneous cleavage pattern beginning immediately below the full-length linear species. (B) Bimolecular exon ligation reactions (90 min) containing 175 nM linear monomer (lane 1), 175 nM circular monomer (lane 2), and 52 nM linear dimer (lane 3) 3′ substrates. Substrate and product structures are symbolized to the left. If the circular 3′ substrate had spliced, the ligated exon product would have been 5 nt longer than that from linear monomer. Note that the circular 3′ substrate was more stable in nuclear extract because it resists exonuclease digestion.
The circular AdMLas 3′ substrates were tested for bimolecular exon ligation along with the linear monomer (Fig. 3B, lane 1) and linear dimer (lane 3). Ligated exons were readily detectable from both linear substrates. As expected, only the 5′-most AG was used in the linear dimer, resulting in a much longer spliced exon product. However, no product could be detected for either the circular monomer (Fig. 3B, lane 2; if this RNA had spliced, the product would have been 5 nt longer than that in lane 1) or circular dimer (data not shown).
To test the general 3′ splice site accessibility in the circular monomer compared with the linear monomer and dimer, all three RNAs were subjected to targeted RNase H cleavage. When incubated under splicing conditions with a 10-nt DNA oligo complementary to the 3′ splice site region, all three RNAs were digested similarly, whereas a nonspecific oligo did not induce cleavage (data not shown). These results indicate that all of the splice sites in all three substrates should have been equally available to the active site for exon ligation, unless access to that site is somehow limited such that only linear RNAs can gain entry. Taken together with the secondary structure studies above, these results clearly demonstrate that the 3′ substrate must have a free 5′ end to function in bimolecular exon ligation.
Extension of the 5′ Substrate Severely Decreases Bimolecular Exon Ligation Efficiency.
If a free 5′ end is required on the 3′ substrate, is an unobstructed 3′ end also needed on the 5′ substrate? To test this, the same 24-nt hairpin as above was appended to the 3′ end of RNA 5A. The hairpin was placed either immediately adjacent to the PPT (RNA 5B) or 22 nt further downstream (RNA 5D). RNA 5C had the same 22-nt extension, but no hairpin (Fig. 4A). All four 5′ substrates underwent the first step of splicing at comparable efficiencies as evidenced by the levels of lariat species (Fig. 4B). However, only RNA 5A permitted efficient bimolecular exon ligation (lanes 2 and 3), with the amount of free 5′ exon decreasing as ligated exons appeared. RNAs 5B–5D all underwent bimolecular exon ligation at least 15-fold less efficiently than RNA 5A (Fig. 4B). These data indicate that the additional sequences downstream of the PPT in the 5′ substrate inhibit bimolecular exon ligation. Interestingly, however, even though RNA 5B contained more nucleotides downstream of the branch than RNA 5C, it reproducibly yielded somewhat higher levels of ligated exons. Thus, a stem–loop structure 28 nt from the branch is not as inhibitory for bimolecular exon ligation as a shorter stretch of RNA that is not predicted to form any particular structure.
Discussion
Our previous study (8) indicated that whatever the mechanism of 3′ splice site choice in the bimolecular system, it imparted a strong preference for use of the 5′-most AG. The present study confirms these results and rules out several possible explanations. Because all AGs in the bimolecular system are provided on a separate RNA from that containing the branch and PPT, no AG is initially “closer” to the branch (and therefore possibly at a higher relative concentration to the active site) than any other in this assay. Thus, if the active site for exon ligation can be accessed freely by any AG regardless of its position in an RNA, one would have expected use of multiple AGs in the tandem repeat-containing substrates (Fig. 1), which were designed specifically to minimize sequence context effects. Therefore, it seems unlikely that use of the 5′-most CAG reflects simply that, being closer to the 5′ end, it is less likely to be masked by internal secondary structures or bound proteins than any downstream CAG.
So why is the 5′-most AG so strongly preferred in the bimolecular system? The effects of 5′-terminal secondary structures (Fig. 2) and circularization of the 3′ substrate (Fig. 3) indicate that the 3′ substrate can gain entry to the splicing machinery only via a free 5′ end (Fig. 5). In another study we showed that the 3′ substrate is not required or apparently recognized by the spliceosome until after lariat formation (19). Thus, entry of the 3′ substrate into the splicing machinery likely does not occur until after both spliceosome assembly and the first chemical step of splicing. This agrees with previous data using cis splicing substrates that showed that the 3′ splice site region remains accessible to RNaseH digestion and, therefore, is not strongly bound by any factors that might predetermine the exact 3′ splice site until after lariat formation (20, 21).
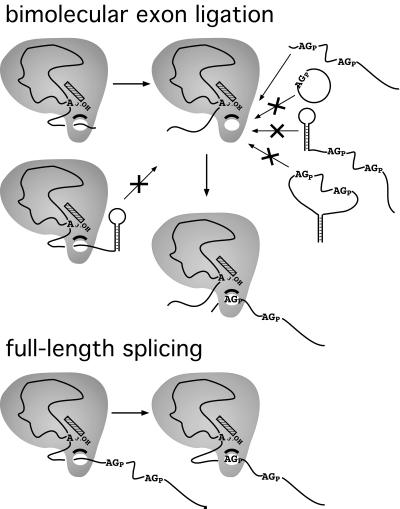
Figure 5.
Summary of 5′ and 3′ substrate structural effects on bimolecular exon ligation. The simplest model accounting for all data regarding 3′ splice site AG selection in bimolecular exon ligation assays (Upper) is one in which access to the active site for exon ligation is limited such that only linear 3′ substrates with no 5′ end structures can gain entry. Effects of structures in the 5′ substrate suggest that this space normally is occupied by intronic RNA downstream of the branch site in cis constructs (Lower).
Numerous proteins have been shown to interact with the region downstream of the branch after lariat formation (10–16). If these proteins totally surround the intronic RNA downstream of the branch on full-length introns, then an empty channel might be generated on our truncated 5′ substrate into which the 3′ substrate could feed via its 5′ end (Fig. 5). Consistent with this idea, when sequences were appended to the 3′ end of the 5′ substrate, bimolecular exon ligation was strongly inhibited (Fig. 4). Interestingly, a 24-nt hairpin was somewhat less inhibitory than a shorter, 22-nt sequence not predicted to form any particular structure. This is consistent with the idea that the likelihood the channel will be occupied by the RNA downstream of the branch increases as the effective length of this RNA increases.
It has been proposed previously that the 3′ splice site AG in cis constructs is identified via some sort of scanning mechanism that initiates at the branch (6, 22–24). Such a mechanism would readily explain why exon ligation usually occurs at the first AG downstream of the branch site in natural introns (5, 25) even when the branch-to-3′ splice site distance is remarkably long (6, 22). However, none of the proposed linear search mechanisms have gained general acceptance because apparently incompatible data exist (see, for example, refs. 7, 15, 26, and 27). For example, tandemly repeated AGs with internal spacing identical to those we employed (i.e., 6 nt between splice sites) do actively compete when placed 13–22 nt downstream of the branch in a yeast intron (27). This is clearly inconsistent with a strict linear search initiating from the branch. One possibility is that two different modes of 3′ splice site selection are used that are dependent on the branch-to-5′-most AG distance. If a viable AG is very close to the branch and is in a favorable sequence context, this AG may encounter by diffusion (15) an AG-binding factor quite rapidly after the first step, and exon ligation could proceed immediately. However, if no AG is found within a certain timeframe, then the spliceosome could switch into a linear search mode. That there may be two modes of 3′ splice site AG selection is supported by observations that the yeast second-step splicing factors, Slu7p, Prp18p, and Prp22p, are required in vitro only as the branch to 3′ splice site distance increases (28–30). All of these proteins are conserved in humans (31–34), suggesting general conservation of the mechanisms of 3′ splice site definition. Interestingly, the branch to 3′ splice site distance is much less variable for introns removed by the minor U12-dependent spliceosome (only 10–15 nt; ref. 35), suggesting that it may use only a subset of the factors required for 3′ splice site definition by the major spliceosome.
The simplest interpretation of the results presented here is that the 5′→3′ directionality of 3′ splice site selection in the bimolecular assay reflects a general, linear search mechanism that normally functions on full-length introns when the 5′-most AG is relatively distant from the branch. Consistent with this idea, the same patterns of 3′ splice site use and exon ligation efficiencies were observed when the tandem repeats used here were physically attached to the PPT of the 5′ substrate (Fig. 1 B and C). Thus, in contrast to the results with tandem repeats 13–22 nt from a yeast branch site (27), CAGs spaced 6 nt apart do not compete when placed at least 35 nt from a mammalian branch (our results). It now might be revealing to examine more systematically how AG-AG spacing affects 3′ splice site choice at different distances from the branch in mammalian introns.
Acknowledgments
We thank Charles Query, Michael Rosbash, Ranjan Sen, and members of the Moore laboratory for critical comments on the manuscript. This work was supported by National Institutes of Health Grant GM 53007 (M.J.M.).
References
- 1.Moore M J, Query C C, Sharp P A. In: The RNA world. Gesteland R F, Atkins J F, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 303–358. [Google Scholar]
- 2.Burge C B, Tuschl T, Sharp P A. In: The RNA World. 2nd Ed. Gesteland R F, Cech T R, Atkins J F, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1998. pp. 525–560. [Google Scholar]
- 3.Senapathy P, Shapiro M B, Harris N L. Methods Enzymol. 1990;183:252–278. doi: 10.1016/0076-6879(90)83018-5. [DOI] [PubMed] [Google Scholar]
- 4.Burge C B, Padgett R A, Sharp P A. Mol Cell. 1998;2:773–785. doi: 10.1016/s1097-2765(00)80292-0. [DOI] [PubMed] [Google Scholar]
- 5.Mount S M. Nucleic Acids Res. 1982;10:459–472. doi: 10.1093/nar/10.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith C W, Chu T T, Nadal-Ginard B. Mol Cell Biol. 1993;13:4939–4952. doi: 10.1128/mcb.13.8.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Umen J G, Guthrie C. RNA. 1995;1:869–885. [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson K, Moore M J. Science. 1997;276:1712–1716. doi: 10.1126/science.276.5319.1712. [DOI] [PubMed] [Google Scholar]
- 9.Moore M J, Query C C. In: RNA-Protein Interactions: A Practical Approach. Smith C, editor. Oxford: IRL; 1998. pp. 75–108. [Google Scholar]
- 10.Teigelkamp S, Newman A J, Beggs J D. EMBO J. 1995;14:2602–2612. doi: 10.1002/j.1460-2075.1995.tb07258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Umen J G, Guthrie C. RNA. 1995;1:584–597. [PMC free article] [PubMed] [Google Scholar]
- 12.Umen J G, Guthrie C. Genes Dev. 1995;9:855–868. doi: 10.1101/gad.9.7.855. [DOI] [PubMed] [Google Scholar]
- 13.Umen J G, Guthrie C. Genetics. 1996;143:723–739. doi: 10.1093/genetics/143.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiara M D, Gozani O, Bennett M, Champion-Arnaud P, Palandjian L, Reed R. Mol Cell Biol. 1996;16:3317–3326. doi: 10.1128/mcb.16.7.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiara M D, Palandjian L, Feld Kramer R, Reed R. EMBO J. 1997;16:4746–4759. doi: 10.1093/emboj/16.15.4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu S, Green M R. EMBO J. 1997;16:4421–4432. doi: 10.1093/emboj/16.14.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore M J, Sharp P A. Science. 1992;256:992–997. doi: 10.1126/science.1589782. [DOI] [PubMed] [Google Scholar]
- 18.Jucker F M, Pardi A. Biochemistry. 1995;34:14416–14427. doi: 10.1021/bi00044a019. [DOI] [PubMed] [Google Scholar]
- 19.Anderson K, Moore M J. RNA. 2000;6:1–10. doi: 10.1017/s1355838200001862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sawa H, Shimura Y. Nucleic Acids Res. 1991;19:3953–3958. doi: 10.1093/nar/19.14.3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwer B, Guthrie C. EMBO J. 1992;11:5033–5039. doi: 10.1002/j.1460-2075.1992.tb05610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith C W, Porro E B, Patton J G, Nadal-Ginard B. Nature (London) 1989;342:243–247. doi: 10.1038/342243a0. [DOI] [PubMed] [Google Scholar]
- 23.Liu Z R, Laggerbauer B, Luhrmann R, Smith C W. RNA. 1997;3:1207–1219. [PMC free article] [PubMed] [Google Scholar]
- 24.Dix I, Russell C S, O'Keefe R T, Newman A J, Beggs J D. RNA. 1998;4:1239–1250. doi: 10.1017/s1355838298981109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langford C J, Gallwitz D. Cell. 1983;33:519–527. doi: 10.1016/0092-8674(83)90433-6. [DOI] [PubMed] [Google Scholar]
- 26.Patterson B, Guthrie C. Cell. 1991;64:181–187. doi: 10.1016/0092-8674(91)90219-o. [DOI] [PubMed] [Google Scholar]
- 27.Luukkonen B G, Seraphin B. EMBO J. 1997;16:779–792. doi: 10.1093/emboj/16.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brys A, Schwer B. RNA. 1996;2:707–717. [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X, Schwer B. Nucleic Acids Res. 1997;25:2146–2152. doi: 10.1093/nar/25.11.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwer B, Gross C H. EMBO J. 1998;17:2086–2094. doi: 10.1093/emboj/17.7.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horowitz D S, Krainer A R. Genes Dev. 1997;11:139–151. doi: 10.1101/gad.11.1.139. [DOI] [PubMed] [Google Scholar]
- 32.Ono Y, Ohno M, Shimura Y. Mol Cell Biol. 1994;14:7611–7620. doi: 10.1128/mcb.14.11.7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohno M, Shimura Y. Genes Dev. 1996;10:997–1007. doi: 10.1101/gad.10.8.997. [DOI] [PubMed] [Google Scholar]
- 34.Chua K, Reed R. Genes Dev. 1999;13:841–850. doi: 10.1101/gad.13.7.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharp P A, Burge C B. Cell. 1997;91:875–879. doi: 10.1016/s0092-8674(00)80479-1. [DOI] [PubMed] [Google Scholar]