Abstract
The highly conserved nature of rRNA sequences throughout evolution allows these molecules to be used to build philogenic trees of different species. It is unknown whether the stability of specific interactions and structural features of rRNA reflects an optimal adaptation to a functional task or an evolutionary trap. In the work reported here, we have applied an in vivo selection strategy to demonstrate that unnatural sequences do work as a functional replacement of the highly conserved binding site of ribosomal protein S8. However, growth competition experiments performed between Escherichia coli isolates containing natural and unnatural S8-binding sites showed that the fate of each isolate depended on the growth condition. In exponentially growing cells, one unnatural variant was found to be equivalent to wild type in competition experiments performed in rich media. In culture conditions leading to slow growth, however, cells containing the wild-type sequence were the ultimate winner of the competition, emphasizing that the wild-type sequence is, in fact, the most fit solution for the S8-binding site.
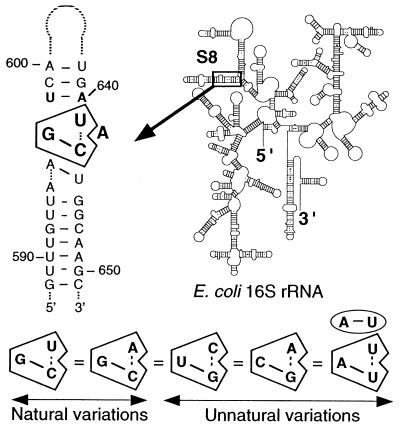
The creation of an artificial phylogeny by using iterative selection techniques is a very powerful tool for analyzing RNA function and obtaining insights into structural requirements for ligand recognition (for review, see refs. 1–3). Artificial phylogeny can greatly aid in the study of rRNA because, despite the tremendous number of rRNA sequences available, the important functional regions of rRNA are extremely well conserved throughout evolution, and therefore covariations of interest are often lacking in these sites. Site-directed mutagenesis, the technique of choice to prove Watson–Crick base pairing, is ineffective in investigating noncanonical features that are suspected to be used frequently to build functional sites in rRNA. This is the case for the rRNA-binding site of prokaryotic ribosomal protein (r-protein) S8, which has an extremely well conserved core sequence to which it binds (4). Protein S8 is one of the crucial proteins in ribosome biosynthesis. It is a primary binding protein required for proper assembly of the 30S ribosomal subunit (5), and it negatively regulates the expression of its own operon (spc) (6). Deletion and structure-probing experiments have demonstrated that the RNA-binding site of r-protein S8 of Escherichia coli (EcS8) is confined to a small region within the bottom stem of a hairpin in 16S rRNA (nucleotides 588–605/633–651). The nucleotides essential for EcS8 binding (Fig. 1) were mapped in the center of this stem (7–10). Other protection sites were identified elsewhere in the 16S rRNA (11), but these additional protected sites are directly dependent on the binding of EcS8 to its anchoring site at 597–599/640–643 and are lost if this anchoring site is disrupted (H.M., unpublished work). Numerous studies performed on the EcS8 site (7–10, 12, 13) have raised the possibility of involvement of noncanonical features essential for S8 binding, but that, however, could not be proven with standard genetical or biochemical means. Selex experiments permitted the isolation of RNA aptamers that recognize EcS8 as efficiently as the wild-type (wt) RNA (4). These results revealed that the core elements essential for EcS8 binding include the four most naturally conserved nucleotides of the binding site (G597, U/A641, A642, C643) (Fig. 1). Three of these residues (nucleotides 597, 643, 641) covary in a manner that suggests a functional and structural relationship. On the basis of this interdependence, nucleotides C597-G643/U641 were proposed to be involved in a triple interaction with an intervening adenine nucleotide at position 642 (4). This base triple is compatible with NMR data (14), although this region appears to be dynamic in solution in the absence of protein and thus might require EcS8 protein to be stabilized.
Figure 1.
Secondary structure of the EcS8-binding site and its location on the secondary structure model of 16S rRNA. The essential nucleotides of the site are shown in bold. The three positions 597–643/641 randomized in this study and capable of forming a triple interaction are boxed. An additional possible triple involving nucleotides 594/644/645 (28) is shown. The four other covariations identified by in vitro selection for the base triple 597–643/641 are presented. The A/U-U triple requires the inverted A-U at 598–640.
One of the most interesting features revealed by the Selex experiments was that unnatural solutions for the triple (U-G/C and C-G/A) bind EcS8 in vitro as does the original solution (G-C/U). It is worth noting that none of these combinations are found among more than 6,000 prokaryotic 16S rRNA sequences in the rRNA data bank (15). Although they could clearly bind S8, it was unclear whether these unnatural sequences could serve as functional replacements for natural sequences in vivo. Indeed, these alternative solutions might have been prevented from emerging during natural evolution, because they would interfere with other functions of the rRNA, or simply because intermediate variants (there are three nucleotides to change in a coordinated way) were not viable. To address such a question requires the testing of S8 binding to unnatural solutions in the full context of a functioning ribosome in vivo. Thus, we developed a strategy of in vivo selection that allowed us to isolate viable variants generated by an artificial phylogeny involving the three positions engaged in the base triple. Growth competition experiments between isolates containing natural and unnatural sequences allowed the selection of the most well-adapted sequence.
Materials and Methods
DNA Templates, Plasmids, and Strains.
Plasmid pKK3535 (16) is a pBR322-derived vector carrying the complete rrnB operon under the control of its natural P1/P2 promoters. pKK3535 containing the mutation 1192-U was used as a template to produce, by PCR, an AvrII-AvrII fragment. This fragment extends from position 271 in the 16S rRNA into the spacer tRNA2Glu gene located downstream. The 1192-U mutation confers spectinomycin resistance on the ribosomes carrying it (17). Strain DHI was used as a host for all transformations, which were performed according to (18). Strain AVS69009, ΔrrnC∷cat+, ΔrrnE, ΔrrnB, ΔrrnH, ΔrrnG∷lacZ+, ΔrrnA, ΔrrnD∷cat+, recA/p(tRNA)70/pHK rrnC, was derived from TA531 (19) and was used to perform control analyses of the phenotypes of the EcS8-binding site variants.
Randomization of Positions 597/641–643 in the EcS8-Binding Site and in Vivo Selection.
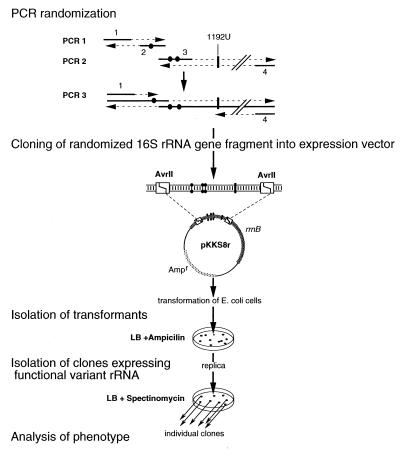
The AvrII fragment encompassing the randomized EcS8-binding region was obtained in a two-step procedure (Fig. 2). In the first step, two distinct PCR amplifications were performed in parallel. The first PCR used primer 1: 5′-GCTAGTAGGTGGGGTAACGGC, encompassing the 5′ AvrII site, and primer 2: 5′-GTTGAGCCCGGGGATTTCACATCTGANTTAACAAACCGC, containing the 5′ side of the EcS8-binding site (with N standing for any of the four nucleotides at position 597 in 16S rRNA). The second PCR used primer 3: 5′-GCGACTTTCACTCACAAACCAGCA, encompassing the 3′ AvrII site, and primer 4: 5′-AAATCCCCGGGCTCAACCTGGGAACTGCATCTGANANTGGCAAGCTTG containing the 3′ side of the EcS8-binding site (with N standing for any of the four nucleotides at 641 and 643). These two fragments correspond to the 5′ and the 3′ halves of the AvrII fragments and overlap (underlined nucleotides in primers 2 and 4) at the level of the EcS8-binding site. In a second step, these two PCR products were gel purified and used in equimolar amounts as overlapping templates for another PCR amplification by using primers 1 and 4. This fragment was digested by AvrII and inserted within the same sites of pKK3535 to give pKKS8r.
Figure 2.
In vivo selection scheme. Primers 1 to 4 are shown with randomized positions 597, 641, and 643 depicted by dots. Position 1192U conferring resistance to spectinomycin is indicated. Details are given in Materials and Methods.
Selection was performed in two steps. Strain DHI transformed with pKKS8r was plated on LB medium with ampicillin (200 μg/ml) to allow expression of the plasmid-encoded rRNAs. The transformants were then replica plated on LB with spectinomycin (100 μg/ml). About 500 clones appearing on spectinomycin after 24 hr were obtained from three independent randomization/selection procedures. Subcloning of the NdeI/BglII and AvrII/AvrII fragments of pKKS8r of certain variants was performed into the pKK3535 vector background. Both NdeI/BglII and AvrII/AvrII fragments encompass the EcS8-binding site and extend on the 5′ or 3′ side, respectively. The NdeI/BglII fragment extends from position −3440 upstream of 16S rRNA to position 704 inside 16S rRNA. The AvrII/AvrII fragment extends from position 271 in 16S rRNA to position 234 downstream the 3′ end of the 16S rRNA gene.
Identification of the EcS8-Binding Site Variations and Growth Rate Determination.
Primer 5′-CTACCCCCCTCTACG, annealing from positions 660 to 674 on 16S rRNA, was used for sequencing DNA from the region 270 to 660 of pKKS8r, which had been extracted from spectinomycin-resistant clones. Sequencing reactions were performed with Sequenase (Amersham Life Science) and extended to read up to position 271 for detection of second site mutations. From the isolated clones, 92 were sequenced and tested individually for their growth rate. Growth rates were measured at 37°C in LB containing spectinomycin (100 μg/ml) or in M9 media (29) plus 0.4% glucose and 0.05 mM thiamine and spectinomycin.
Binding Affinities Measurements.
Kds at equilibrium were measured for each species of rRNA variant with filter-binding assays as described in ref. 9 by using EcS8 protein purified according to ref. 20 and α-32P labeled 16S rRNA fragment 576–765. RNA fragment 576–765, encompassing the EcS8-binding site, was obtained by in vitro transcription with T7 RNA polymerase from a PCR-generated dsDNA. The dsDNA-encoding fragment 576–765 was produced by using as template the plasmid pKKS8r extracted from the selected clones, as 5′ primer 5′-AGACCGGATCCTAATACGACTCACTATAGGGCGCACGCAGGCGGTTTGTT, which contains the BamHI site, the T7 RNA polymerase promoter, and a sequence complementary to nucleotides 576–594 of 16S rRNA, and as 3′ primer 5′-GGCAAGAATTCGCGCACCTGAGCGTCAGTCTT, which contains the EcoRI site and a sequence complementary to nucleotides 746–765 of 16S rRNA. Two picomol of dsDNA served as template for transcription with T7 RNA polymerase in the presence of 20 μCi α-32P ATP (3,000 Ci/mmol) in a 10-μl reaction volume as described in ref. 4. Transcripts were purified by electrophoresis on an 8% polyacrylamide gel under denaturing conditions.
Coculture Competition Experiments.
In vivo competition between the selected clones was performed in 30 ml of LB broth containing spectinomycin (100 μg/ml) and starting with various combinations of growing cells. The starting pool of competitor clones was either the total population from plates where spectinomycin-resistant clones were isolated (in this case all the clones were washed from the plates by using 2 ml LB broth, and 0.005 OD600 of this suspension was used) or various combination of individual clones. In the latter case, the same amount of growing cells determined by absorbance (0.005 OD600) and by dilution plating and counting the colonies was used for each variant species. Once the cultures had reached 0.8 OD600, aliquots were plated on LB agar media for isolation of single clones and subsequent sequencing of the EcS8-binding site area on plasmid pKKS8r. An amount of 0.005 OD600 of isolated clones was used to seed a fresh LB broth medium for several new rounds of competition. These different coculture competition experiments were repeated at least four times each by using different isolates of selected clones.
Results
In Vivo Selection of Clones That Can Use Mutated 16S rRNA.
Three highly conserved positions of 16S rRNA (597–643/641) are essential for EcS8 binding and are proposed to be involved in a base triple (4). Randomization of these three bases was performed by using several steps of PCR amplification, as summarized in Fig. 2. The PCR-produced fragment containing the randomized bases extended from position 271 of 16S rRNA into the spacer tRNA2Glu and was inserted into the rRNA expression vector pKK3535 to give pKKS8r. The inserted fragment also contained the mutation C to U at position 1192, which confers resistance to spectinomycin on the host cell (17). After transformation of E. coli DH1 cells with the randomized sequences in 16S, two consecutive sets of selections were performed. The first selection was on solid medium containing ampicillin to select for plasmid-containing transformants, and the second was on solid medium containing spectinomycin. This second step allowed the selection of strains containing functional plasmid-encoded ribosomes. In the presence of spectinomycin, the wt 30S subunits assembled with chromosomally encoded 16S rRNA are inactive. Our selection strategy relies on the assumption that only variants of the EcS8-binding site that are capable of binding EcS8 and assembling into functional ribosomes are able to sustain normal growth in the presence of spectinomycin.
The growth rates of clones appearing on spectinomycin-containing media were analyzed, and the EcS8-binding site encoded on the plasmid-borne 16S rRNA gene was sequenced. Variations in the EcS8-binding site and their corresponding doubling times (dts) are presented on Table 1. Variants were put into three classes according to their effect on growth rates in LB medium with 100 μg/ml of spectinomycin. Note that the dt of cells containing the pKKS8r plasmid with the wt S8-binding sequence (G-C/U) and the 1192U mutation are increased in the presence of spectinomycin. This effect of spectinomycin was proposed to be caused by its inhibitory effect on the population of sensitive ribosomes (21) because in the cell, about 25% of total ribosomes are synthesized from the host chromosome (ref. 22 and data not shown). This view is confirmed by the fact that spectinomycin has no effect on the growth rate of a pure population of spectinomycin-resistant ribosomes (see below). Variants of the class I type had the same growth rate as the wt (dt = 70 min ± 5), whereas variants of class II and III were moderately (dt = 135 to 165 min ± 10) and severely (dt = more than 300 min ± 20) affected, respectively. Influence of variations in the ratio between chromosomal rRNA and plasmid rRNA expression levels (determined as in ref. 22) was excluded, because this ratio was found constant for all variants tested (all class I and several class II and III variants) (data not shown). Interestingly, variants of class I contained the wt combination (G-C/U) as well as the one most frequently found in nature (G-C/A). Together with these natural combinations, two unnatural ones were found that were also previously identified by Selex experiments (U-G/C and C-G/A). In addition, another combination (C-A/A), not isolated in the Selex experiments, was found to promote wt growth rates. Thus, unnatural solutions to the S8-binding site were able to be used in vivo and to sustain growth in a manner indistinguishable from wt under these experimental conditions.
Table 1.
S8-binding site variations at 597-643/641 and their corresponding growth rates and Kds
| (597-643)/641 | No. isolates | Doubling time, min | Kd | |
|---|---|---|---|---|
| Class I | G-C/U | 3 | 70 | 1 |
| C-G/A | 7 | 70 | 2 | |
| U-G/C | 3 | 70 | 1 | |
| G-C/A | 3 | 70 | 1 | |
| C-A/A | 8 | 70 | 7.5 | |
| Class II | C-G/U | 4 | 160 | 10 |
| C-G/C | 5 | 140 | 10 | |
| C-C/A | 3 | 165 | 50 | |
| U-A/G | 10 | 135 | 40 | |
| U-G/G | 4 | 150 | 100 | |
| U-A/A | 10 | 160 | 100 | |
| C-U/A | 1 | 165 | >100 | |
| Class III | A-C/C | 1 | 300 | 10 |
| C-G/G | 17 | 400 | 100 | |
| U-G/A | 2 | >400 | >100 | |
| G-G/A | 2 | >400 | >100 | |
| A-A/G | 2 | >400 | >100 | |
| G-A/G | 1 | 400 | ND | |
| U-A/C | 1 | >400 | ND | |
| C-A/U | 1 | >400 | ND | |
| U-G/U | 1 | >400 | ND | |
| U-A/C | 1 | >400 | ND | |
| A-A/A | 2 | >400 | ND |
The number of isolates for each variant is indicated (for a total of 92 clones sequenced). Growth rate values reflect the mean of at least three independent dt measurements done with different isolates. Standard deviation is ±25%. Kd values are relative to the wt value of 3 nM for rRNA transcript 576-765 with standard deviation being ±10%. ND, not determined.
Correlation Between in Vivo Selection and in Vitro EcS8 Binding.
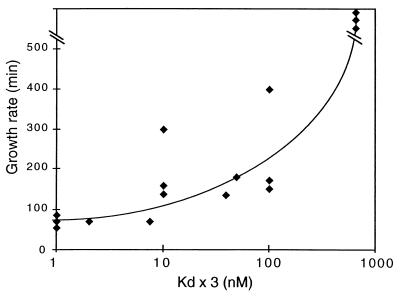
To determine whether a link existed between the phenotype of in vivo-selected variants and their binding affinities in vitro, we measured the affinity of EcS8 for the different selected binding-site variants. The RNA fragments encompassing nucleotides 576 to 764 were generated by PCR by using the pKKS8r vector from the isolated clones as template followed by in vitro transcription. Apparent Kds for EcS8 binding of the 16S rRNA fragments containing nucleotides 576–764 were estimated from saturation curves by using filter-binding assays (9). The results are summarized in Table 1. The variants from class I, which grew with a wt rate, had Kds very similar to that of the wt combination, with the exception of the C-A/A variant, which bound EcS8 with a 7.5-fold higher Kd. Variants from class II showed a Kd enhanced by a factor of 10 to 100, whereas class III variants bound EcS8 with a very low affinity. The only exception was the combination A-C/C, which displayed only a relatively modest increase of Kd (10-fold), as compared with its prolonged growth rate. Thus, there is a good correlation between the growth rate and the corresponding EcS8-binding affinity for each variant (Fig. 3).
Figure 3.
Plot of the growth rate of the variant species isolated vs. the Kd of their 16S rRNA S8-binding site.
Influence of Competitor wt rRNA and Spectinomycin on Selection Process.
We examined two possible mechanisms by which the selection of a particular variant could be influenced. The first is that the wt chromosomally encoded and spectinomycin-sensitive 16S rRNA could act as an intracellular competitor for EcS8 binding during 30S assembly. The second is the possible influence of the spectinomycin-resistance mutation and the presence of the antibiotic in the medium during the selection procedure. Although the 1192 substitution conferring resistance to spectinomycin does not seem to affect normal ribosome function in the absence of spectinomycin (21), it has been suggested that spectinomycin could alter EF-G function with the spectinomycin-resistant ribosome (23). However, our attempts to perform selection in the absence of spectinomycin were not conclusive, possibly because of the presence of the population of ribosomes synthesized from the host chromosome (≈25%). Thus, we used the strain AVS69009, in which all endogenous rRNA genes have been deleted or disrupted (19). In this strain, the pKKS8r-borne rRNA gene was the only source of rRNA for the ribosome. This enabled us to analyze the phenotype of a pure population of variant rRNA in the absence of wt rRNA. All variants from class I and representative examples of class II (C-G/U) and III (C-G/G) were individually tested in this strain in the absence and presence of spectinomycin. All variants, even one from class III, grew in LB medium at the same rate as the wt species (43 ± 2 min). When transformed cells were grown in M9 minimal medium, the dt was increased to 60 ± 4 min for all class I variants, and variants of class II and III showed a moderate increase of their growth rates (68 ± 5 min and 75 ± 5 min, respectively). It is important to note that identical growth-rate values were measured in the presence or absence of spectinomycin. These experiments demonstrate that suboptimal configurations of the S8-binding site can be used by the cell when no intracellular competitors (chromosome-encoded rRNAs) are present. Furthermore, there was no detectable phenotype on rich media. These data suggest that the unnatural combinations do not perturb significantly the translational properties of the ribosomes containing them. Under conditions that cause slow growth (M9 medium), however, a moderate effect on growth rate caused by the unnatural variations allowed the partitioning of the variants into distinct classes similar to those defined in a wt strain in the presence of spectinomycin. The experiments performed in the AVS strain show that the presence of spectinomycin had no effect on phenotype caused by the various combinations of the S8-binding site and therefore should be neutral on selection process. Rather, the loss of the observed phenotype in this strain (compared with DHI strain) for the class II and III variants suggests that the major determinant of the selection process is the competition for ribosome assembly between intracellular 16S rRNA species.
In Vivo Competition Between Natural and Unnatural S8-Binding Sequences.
All base triple mutations identified in the preceding experiments were viable, with a more or less dramatic effect on cell growth. Although five class I variants allowed a wt growth rate, the experiments did not reveal which of the natural or unnatural combinations was the most adapted if they were allowed to compete against each other. To address this question, all the clones of cells (DH1) transformed with pKKS8r selected on spectinomycin plates were scraped from the plates and grown in liquid LB medium in the presence of spectinomycin (100 μg/ml). We chose to take all the selected clones to ensure that we did not miss any particular variant during the preceding analysis. After one to several consecutive passages in liquid medium, the different variants present in the resulting population were sequenced after isolation of individual clones. After the first passage in liquid medium (about 10 doublings), all the variants from class I were found among the most represented species (Table 2A). After two more passages in liquid culture (or about 30 doublings), the unnatural C-A/A variant appeared as the final winner of the competition.
Table 2.
Results of in vivo competition
| Medium (clones used) | 10 doublings | 30 doublings | |
|---|---|---|---|
| A | LB (total clones) : | C-A/A* : 50 | C-A/A* : 100 |
| C-G/A* : 14 | |||
| G-C/U : 15 | |||
| G-C/A* : 8 | |||
| C-G/U : 7 | |||
| Others : 6 | |||
| B | LB (1× clones) : | G-C/U : 25 | G-C/U : 50 |
| C-A/A* : 25 | C-A/A* : 50 | ||
| C-G/A* : 16 | |||
| U-G/C* : 15 | |||
| C-C/A : 5 | |||
| C-G/U : 5 | |||
| Others : 9 | |||
| C | LB (1× class I + | G-C/U : 40 | G-C/U : 50 |
| subclones) : | C-A/A* : 40 | C-A/A* : 50 | |
| C-G/A* : 10 | |||
| U-G/C* : 10 | |||
| D | M9 (1× class I + | G-C/U : 46 | G-C/U : 100 |
| subclones) | U-G/C* : 25 | ||
| C-G/A* : 24 | |||
| C-A/A* : 5 |
Composition of the variant population (positions 597-643/641) in different liquid culture media after 10 and 30 doublings in presence of 100 μg/ml spectinomycin is given. Different combinations of bacteria clones were used for the competition: A, all isolated clones from the isolation spectinomycin plates; B, the same amount of cells of each variant species was used; C, the same amount of cells of each variant from class I or subcloned versions of these variants were used; D, same as in C but in M9 medium. The wt combination is underlined, and class I variations are indicated by an asterisk. Data are mean percentage values obtained from sequencing the S8-binding region on pKKS8r of at least 20 clones from at least 4 independent competition experiments.
To avoid possible bias in the competition because of some strong imbalance existing in the population for certain selected clones, possibly reflecting their amplification/ligation efficiency more than their competitiveness, we performed another competition experiment starting with an identical amount of cells of each isolated clone species. These amounts were determined by OD600 measurement and dilution plating. After the first passage, the results were similar to the previous experiment, but after the third passage, the wt combination (G-C/U) together with the unnatural C-A/A combination appeared as the final cowinners of the competition (Table 2B). These data remained the same after an additional passage. The same results were obtained by starting the competitions with different clone isolates.
Another potential bias in these experiments is the possible influence of second site mutations occurring elsewhere on the plasmid or in the chromosome. No additional mutation was detected by sequencing the region proximal to the randomized triple (nucleotides 660 to 270) in individual clones. The presence of a distal mutation was tested by subcloning a fragment from −3440 upstream of the 16S gene to position 704 inside 16S rRNA. Independently, a fragment extending from position 271 in 16S rRNA to position 234 downstream of the 3′ end of 16S rRNA gene of each class I variant was also subcloned into a new vector. Cells were transformed with these new plasmids, and an identical amount of freshly transformed cells was used for seeding the culture medium for a new competition experiment. After the first passage, all class I variants were still present with the exception of the G-C/A variant, and after three passages, G-C/U, together with the C-A/A variant, was again the cowinner of the competition (Table 2C). These results were very similar to the previous ones, indicating that no secondary site mutation interfered with the competition.
The competition experiments just described were all done in rich liquid medium. We repeated the experiments under slow growth conditions in M9 minimal medium, with glucose as the only source of carbon. As above, the transformed cells obtained after subcloning class I variants were used to seed minimal medium. After the first passage, the same class I variants as those obtained in the preceding selection were recovered. However, the C-A/A variant was clearly disfavored (Table 2D), and it was completely discarded after three passages, leaving the wt combination as the ultimate and sole winner of the competition. This result was reproducibly obtained in five independent experiments. These results stress the strong influence of growth conditions on evolutionary constraints placed on 16S rRNA sequences.
Discussion
Unnatural S8-Binding Sequences Can Serve as Functional Replacements for Natural Sequences in Vivo.
The rRNA-binding site of prokaryotic r-protein EcS8 is subjected to a strong evolutionary pressure. The specificity of recognition is essentially governed by four nucleotides, building a precise structural motif. Three of them, G597, A642, and C643 (according to E. coli nomenclature), are extremely well conserved (≈99.9%), whereas position 641 is most frequently represented by A or, less often, by U (as in E. coli). On the basis of natural phylogeny, no covariation could be detected that would have allowed the demonstration of a particular relationship between these positions. Selex experiments, however, by identifying unnatural covariation combinations, clearly demonstrate a functional and structural interdependency between positions 597, 641, and 643 (4). Therefore, a base triple denoted G-C/U in the case of E. coli was proposed. In addition to revealing unsuspected structural features, these results indicated that alternative combinations, different from those selected in nature, were able to bind EcS8 as efficiently as the natural sequence despite its high evolutionary conservation. Here, we showed that these unnatural combinations are also able to be used by the cell without altering its growth rate on LB broth media at 37°C. We used an in vivo selection assay based on the proposal that only cells containing a plasmid carrying a rrnB operon with both the spectinomycin resistance and base combinations allowing binding of S8 and assembly of functional ribosomes should be able to sustain life in the presence of the antibiotic.
Five combinations that provided a wt phenotype emerged from our experiments: the wt combination (G-C/U), the most frequent natural solution (G-C/A), two combinations that arose from in vitro selection (C/G-A, U-G/C), and another unnatural combination not isolated by in vitro selection (C-A/A). Another solution that was selected in vitro (A-U/U) was not recovered in our in vivo assay (Fig. 1). However, this combination was shown to be context dependent, requiring an inverted A-U base pair at positions 598–640. The fact that only nucleotides 597, 641, and 643 were randomized should account for the failure of this combination to emerge in the in vivo selection. Other alternative solutions were also found to sustain life but exhibited altered growth rates. The dts of the different variants appeared to vary as a function of their affinity for EcS8 (Fig. 3). Therefore, a high-affinity EcS8-anchoring site appeared to be required for optimal growth, most likely because of efficient and correct ribosome assembly considering the central role of EcS8 in this process (5). Thus, when Kd values exceed the wt value by more than 10-fold, the dt exponentially increases with the increase of Kd. These data can be explained by a preferential binding of EcS8 to the wt 16S rRNA synthesized from the chromosome, which acts as a competitor for ribosome assembly. Because these ribosomes are sensitive to spectinomycin, they cannot perform translation, resulting in growth defects. This interpretation is supported by the observation that mutations that strongly affect affinity do not impair the growth phenotype in the strain having no chromosomal rrn operons. In this situation, rRNA assembly is not submitted to intracellular competition.
Otherwise, despite the high number of combinations that were selected (23 over 64 possibilities), other sequences were systematically counter selected in the different assays, indicating that some combinations are likely to be lethal for the cells. We can assume that these mutations induce severe defects in the EcS8-binding site and do not permit assembly of active ribosomes.
Unexpectedly, the in vivo system appears to be less stringent than the in vitro selection process, which was designed to select the highest affinity S8-binding RNA aptamers. Indeed, the C-A/A variant was not selected in the Selex experiments, most likely because of its 7.5-fold reduced binding affinity. Because the influence of a second site mutation was excluded by sequencing and subcloning, the requirement for optimal binding of S8 is not the only criterion that governs the in vivo selection process, and other phenomena must compensate for the lower binding affinity. The relative tolerance for poor binding observed in these experiments can probably be accounted for by the growth-rate-dependent control of ribosome biosynthesis. Indeed, the transcription of rRNA under the control of the rrn P1 promoter can be directly modulated by the yield of active ribosomes through feedback signaling involving an NTP-sensing mechanism (24). Thus, mutations decreasing 30S subunit assembly or yielding inactive ribosomes should result in a drop of NTP consumption that acts as a signal for enhancing rRNA transcription, which, in turn, might partly compensate for defects in rRNA affinity for EcS8. Otherwise, it should be stressed that the affinity of the different variants for EcS8 was measured at equilibrium, in the absence of the other r-proteins. In the cell, however, assembly is coupled with rRNA transcription and the addition of the other r-proteins that occurs in a cooperative and ordered manner. These complex interactions may stabilize the EcS8–rRNA interactions and shift the equilibrium toward association by preventing dissociation. Therefore, various kinetics aspects (unexplored in our Kd measurements and Selex experiments) might play an important role, because the association rate, rather than the dissociation rate, is likely the driving component of the process. Because the assembly process of the ribosome is irreversible, it can provide a sink for initial binding complexes that might help to compensate for nonlethal assembly defects in the recognition site.
A strong defect in affinity should also perturb the regulatory loop that is responsible for the coordinated synthesis of r-proteins. Indeed, EcS8 is a regulatory protein involved in the translational control of the spc operon, encoding 10 r-protein genes in addition to itself and secY. It is able to bind to the spc mRNA with a 5-fold reduced affinity vs. 16S rRNA (13). This binding, which occurs on saturation of the 16S rRNA sites, inhibits the translation of all proteins of the spc operon. If EcS8 affinity for the variant rRNA becomes significantly lower than for the spc mRNA, it will preferentially bind to the mRNA, thus increasing the repression of the spc operon, and will result in an imbalance between rRNA transcription and ribosomal protein translation.
Evolutionary Competition Between Clones Containing Natural and Unnatural S8-Binding Sequences.
Analysis of individual clones indicated that growth can be maintained at nearly wt levels with unnatural S8-binding sites. As expected, when the different clones containing unnatural combinations were challenged together with the natural sequence in coculture in rich media, the unnatural combinations with increased dts were rapidly evicted after a few generation times (10 doublings). However, when this competition experiment was continued for more generations (30 doublings), the unnatural variant C-A/A was the cowinner with the wt solution, G-C/U. This result was unexpected because of the suboptimal affinity of this variant for EcS8, as compared with the other combinations that bind EcS8 with the same Kd. The eviction of these other variants (including G-C/A, the most frequent solution found in natural 16S rRNA-like sequences) addresses puzzling questions. Nevertheless, it should be stressed that in our selection and competition experiments, only one partner was allowed to vary (the 16S rRNA). In contrast, natural evolution is the result of a long process in which the protein and the RNA targets (rRNA and mRNA) covary by mutual adaptation. Thus, the C-A/A solution, together with the naturally evolved sequence, presents the best adapted solutions that fulfill all the various requirements including EcS8 association, precise structural adjustment, subsequent assembly steps, functioning of the ribosome, and interplay with regulatory loops.
Strikingly, when the selection was performed on minimal medium, the C-A/A combination failed to compete with the wt solution. Most likely, the mechanism(s) that compensate(s) for the lower S8-binding affinity in rich medium become(s) insufficient in minimal medium. Indeed, the rate of stable RNA synthesis is known to decrease with growth rate (25), and the stimulation of rRNA transcription, which occurs in response to protein synthesis inhibition, is reduced in minimal medium (26). Thus, endogenous competition between chromosomally encoded G-C/U and plasmid-encoded C-A/A rRNA is probably enhanced in slow growth rate conditions to the detriment of the lower-affinity C-A/A rRNA. Another consequence of slow growth rate might be to favor binding of EcS8 to the spc mRNA. This binding might even be facilitated by the decreased occupancy of the mRNA by the ribosomes in slow growth conditions (25), thus resulting in an increased translation repression of the spc operon. Such a similar enhancement of repression at low growth rate was described in the case of the threonyl–tRNA synthetase gene, which is also negatively autoregulated at the translational level (27). Thus, the loss of the C-A/A variant in response to growth-rate limitation would result essentially from the inability of this variant (because of its lower binding affinity) to adapt to the regulatory loops.
The most striking result of this study is that unnatural sequences were able to be used as functional replacements of the wt sequence that emerged from natural evolution. Although the affinity of EcS8 for its rRNA-binding site was found to be an essential determinant of selection, this was not an absolute requirement, because a suboptimal solution could provide wt growth and was able to challenge the natural solution in competition assays in rich medium. Importantly, however, the wt solution appeared to be the best adapted solution under suboptimal growth conditions. Thus, our in vivo selection assay, as a paradigm of the evolutionary process, revealed the strong selection pressure imposed on ribosome assembly in the prokaryotic cell and emphasized the complex interplay of the multiple factors governing ribosome biosynthesis, assembly, and function.
Acknowledgments
We thank C. Cachia for EcS8; A. Vila (Brown University, Providence, RI) for the AVS69009 strain; P. Romby and M. Springer for helpful discussions; M. O'Connor and A. E. Dahlberg for valuable suggestions; and M. Dreyfus and S. Lodmell for comments and critical reading of the manuscript. This work was supported in part by National Institutes of Health grant GM24751 to C.L.S.
Abbreviations
- wt
wild type
- dt
doubling time
- r-protein
ribosomal protein
References
- 1.Gold L, Polisky B, Uhlenbeck O, Yarus M. Annu Rev Biochem. 1995;64:763–797. doi: 10.1146/annurev.bi.64.070195.003555. [DOI] [PubMed] [Google Scholar]
- 2.Uphoff K W, Bell S D, Ellington A D. Curr Opin Struct Biol. 1996;6:281–288. doi: 10.1016/s0959-440x(96)80045-5. [DOI] [PubMed] [Google Scholar]
- 3.Baskerville S, Frank D, Ellington A D. RNA Structure and Function. Plainview, NY: Cold Spring Harbor Lab. Press; 1998. pp. 203–251. [Google Scholar]
- 4.Moine H, Cachia C, Westhof E, Ehresmann B, Ehresmann C. RNA. 1997;3:255–268. [PMC free article] [PubMed] [Google Scholar]
- 5.Held W A, Ballou B, Mizushima S, Nomura M. J Biol Chem. 1974;249:3103–3111. [PubMed] [Google Scholar]
- 6.Dean D, Yates J L, Nomura M. Nature (London) 1981;289:89–91. doi: 10.1038/289089a0. [DOI] [PubMed] [Google Scholar]
- 7.Gregory R J, Zimmermann R A. Nucleic Acids Res. 1986;14:5761–5776. doi: 10.1093/nar/14.14.5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mougel M, Eyermann F, Westhof E, Romby P, Expert-Bezançon A, Ebel J P, Ehresmann B, Ehresmann C. J Mol Biol. 1987;198:91–107. doi: 10.1016/0022-2836(87)90460-8. [DOI] [PubMed] [Google Scholar]
- 9.Mougel M, Allmang C, Eyermann F, Cachia C, Ehresmann B, Ehresmann C. Eur J Biochem. 1993;215:787–792. doi: 10.1111/j.1432-1033.1993.tb18093.x. [DOI] [PubMed] [Google Scholar]
- 10.Allmang C, Mougel M, Westhof E, Ehresmann B, Ehresmann C. Nucleic Acids Res. 1994;22:3708–3714. doi: 10.1093/nar/22.18.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Powers T, Noller H F. RNA. 1995;1:194–209. [PMC free article] [PubMed] [Google Scholar]
- 12.Gregory R J, Cahill P B F, Thurlow D L, Zimmerman R A. J Mol Biol. 1988;204:295–307. doi: 10.1016/0022-2836(88)90577-3. [DOI] [PubMed] [Google Scholar]
- 13.Wu H, Jiang L, Zimmermann R A. Nucleic Acids Res. 1994;22:1687–1695. doi: 10.1093/nar/22.9.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalurachchi K, Nikonowicz E P. J Mol Biol. 1998;280:639–654. doi: 10.1006/jmbi.1998.1915. [DOI] [PubMed] [Google Scholar]
- 15.Maidak B L, Cole J R, Parker C T, Jr, Garrity G M, Larsen N, Li B, Lilburn T G, McCaughey M J, Olsen G J, et al. Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brosius J, Dull T, Sleeter D, Noller H F. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 17.Sigmund C D, Ettayebi M, Morgan E. Nucleic Acids Res. 1984;12:4653–4663. doi: 10.1093/nar/12.11.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanahan D, Jessee J, Bloom F R. Methods Enzymol. 1991;204:63–113. doi: 10.1016/0076-6879(91)04006-a. [DOI] [PubMed] [Google Scholar]
- 19.Asai T, Zapoprojets D, Squires C, Squires C L. Proc Natl Acad Sci USA. 1999;96:1971–1976. doi: 10.1073/pnas.96.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cachia C, Flamion P J, Schreiber J P. Biochimie. 1991;73:607–610. doi: 10.1016/0300-9084(91)90029-z. [DOI] [PubMed] [Google Scholar]
- 21.Makosky P C, Dahlberg A E. Biochimie. 1987;69:885–889. doi: 10.1016/0300-9084(87)90216-1. [DOI] [PubMed] [Google Scholar]
- 22.Sigmund C D, Ettayebi M, Borden A, Morgan E. Methods Enzymol. 1988;164:673–690. doi: 10.1016/s0076-6879(88)64077-8. [DOI] [PubMed] [Google Scholar]
- 23.Bilgin N, Richter A A, Ehrenberg M, Dahlberg A E, Kurland C G. EMBO J. 1990;9:735–739. doi: 10.1002/j.1460-2075.1990.tb08167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaal T, Bartlett M S, Ross W, Turnbough C L, Gourse R L. Science. 1997;278:2092–2097. doi: 10.1126/science.278.5346.2092. [DOI] [PubMed] [Google Scholar]
- 25.Bremer H, Dennis P P. In: Escherichia coli and Salmonella typhimurium. Cellular and Molecular Biology. Neidhardt F C, editor. Vol. 2. Washington, DC: American Society for Microbiology; 1987. pp. 1527–1542. [Google Scholar]
- 26.Shen V, Bremer H. J Bacteriol. 1977;130:1098–1108. doi: 10.1128/jb.130.3.1098-1108.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Comer M M, Dondon J, Graffe M, Yarchuk O, Springer M. J Mol Biol. 1996;261:108–124. doi: 10.1006/jmbi.1996.0445. [DOI] [PubMed] [Google Scholar]
- 28.Kalurachchi K, Uma K, Zimmermann R A, Nikonowicz E P. Proc Natl Acad Sci USA. 1997;94:2139–2144. doi: 10.1073/pnas.94.6.2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Cold Spring Harbor Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]