The participation of reactive oxygen intermediates (ROI) and reactive nitrogen intermediates (RNI) in intracellular signaling is widely documented. Nonetheless, it has been difficult to understand how their involvement meets the requirement of signaling for specificity, and hence, to accept that their role could be physiologic. The problem can be resolved by adducing three postulates: (a) to execute and coordinate multiple adaptations to a changing environment, intracellular signaling requires several different types of specificity; (b) based on the chemistry of the mediators, types of specificity can be distinguished by whether an intermediate in a signaling pathway regulates additional pathways that are physically noncontiguous with the one in which it was formed, and if it does so, whether it acts like an on/off switch or a rheostat; and (c) a given mediator can have specificity of more than one type.
Type I specificity involves intermediary interactions that are initiated noncovalently based on complementarity of molecular shape. Members of a type I signaling pathway are colocalized such that information is private to a given pathway and flows in a linear manner. Type II specificity also involves initial interactions that are based on complementarity, but the mediators diffuse to switch on or off some pathways that are not colocalized with the pathway that generates them. Type III specificity involves covalent interactions as the initial interaction of one signaling intermediate with another. Mediators exerting type III specificity diffuse to affect multiple targets that are not colocalized, where their major function is to regulate how these pathways respond to their own individual agonists. Mediators acting with specificity of types II and III convey information publicly, that is, across much of the cell. ROI and RNI can serve as agonists to activate pathways that operate with type I or type II specificity, but for the most part, endogenous ROI and RNI behave with specificity of the third kind.
Looking back and taking stock
More than ten years ago, the following “perspective and proposal” was offered in a survey of the rising tide of NO biology:
Our understanding of cell-cell communication is dominated by a single paradigm: signaling is accomplished by molecules that bind noncovalently to specific receptors through complementarity of shape. The most surprising insight to arise from NO research is that there exists a fundamentally different form of intercellular signaling. In this new system, the messenger molecule reacts with its targets covalently . . . The latter system may prove to be as ubiquitous and physiologically important as the former . . . It has only recently been appreciated that reactive oxygen intermediates have broad potential to act as secretagogues, enzyme activators and regulators of transcription, along with their more familiar roles as enzyme inactivators, antiseptics, cytotoxins, and mutagens. Close biochemical and biological parallels between reactive nitrogen intermediates and reactive oxygen intermediates prompt the hypothesis that NO may share the ability of ROI to activate proteases, protein tyrosine kinases, protein kinase C, fos, jun, and NF-κB (1).
In the intervening years, these predictions were fulfilled for both intercellular and intracellular signaling. The list of signaling molecules known to be regulated by ROI and/or RNI has expanded far beyond the original examples — soluble guanylyl cyclase for RNI (2–4) and NF-κB (5) and activation protein-1 (6) for ROI — to include ion channels and transporters, G protein–coupled receptors, small GTPases, phosphatases, kinases, proteases, metabolic enzymes, cytoskeletal elements, translation regulators, cell-cycle control factors, transcription factors, histone (de)acetylases, and DNA methylases (7–16). Through these reactions, ROI and RNI help regulate the development of plasmodia (17), flies (18), frogs (19), and mice (20); cells’ motility, matrix (21), interconnections (22), secretion, respiration, metabolism, gene expression, replicative cycle, and apoptosis; and clocks controlling circadian rhythms and senescence (7–16). Examples in one cell type potentially pertain to others, because enzymes producing ROI and RNI are expressed and/or inducible in nearly every type of nucleated mammalian cell as well as in other animals, plants, and prokaryotes. Reviews marshal abundant evidence (7–16).
A persisting paradox
However, something is missing from this story: a conceptual framework for comprehending the role of small, diffusible, chemically reactive molecules in cell signaling. As matters stand, lists of examples do not add up to understanding, and facts accumulate without being incorporated into the thinking of many in the field.
Specificity is the currency of cell signaling. The rapid formation, diffusibility, and short half-lives of ROI and RNI commend them as second messengers. However, the very concept of second messengers suggests a lack of spatial resolution and target restriction that runs counter to contemporary notions of specificity in signaling.
There is a second problem. Many years after the key facts were in, it still seems paradoxical that ROI and RNI participate in homeostatic regulation of physiologic processes, yet the same molecules kill cells (foreign and self) to protect the host from infection (12). Killing is the end result of covalent modifications that lead to widespread malfunction. Signaling reactions are generally reversible; death is not. If a molecule is reactive enough to be lethal, how can it be specific and reversible enough to participate in signaling?
By 1992 it appeared that what separates signaling from killing by ROI and RNI is chiefly the tempo and extent of their production (1). A first-pass paradigm to resolve the paradox described above was to posit that the low-output NO synthases (NOS1 and NOS3) were dedicated to signaling under the control of elevations of intracellular Ca2+, while the high-output isoform, NOS2 or iNOS, being inducible and independent of elevated intracellular Ca2+, was responsible for killing (23). Similarly, a variety of sources of ROI, including the NADPH oxidases of nonmyeloid cells (known as NOXs) (24), were involved in signaling, while the phagocyte oxidase (phox) was dedicated to killing. However, recent experiments have taught us that iNOS and phox can carry out signaling functions (25, 26). Thus, the paradox is not dispelled by pigeonholing the enzymes that generate the products. What we need is a principle according to which cells’ use of ROI and RNI in signaling can be seen as an adaptive solution to the problem of specificity, and killing can be viewed as a special case of signaling.
The present commentary suggests that the answer to the apparent paradox of homeostatic signaling by ROI and RNI lies, as is usual with paradoxes, in taking a less constrained view of its central concept. Below it is recounted how the paramount concept of specificity in intracellular signaling evolved from an earlier idea that persists in an uneasy relationship with its descendant. Neither of these earlier views readily accommodates ROI and RNI as signaling intermediates. The discord is eased by the concept that there are several individually essential and mutually complementary kinds of specificity in intracellular signaling.
Specificity of three kinds
When most of us think about cell signaling, we envision oligomolecular interactions among signaling intermediates. A given signaling intermediate reacts with a small number of other intermediates to which it binds transiently and noncovalently as a ligand, receptor, adapter, enzyme, cosubstrate, or cofactor by virtue of complementarity of shape. Shape in this context includes not just the disposition of mass in space but also the accompanying distribution of charge and hydrophobicity. Complementarity of shape can be defined as the set of properties that determine that two distinct types of molecules will approach each other significantly more closely and for a significantly longer time than the average for all pairwise combinations of molecules in the system under consideration (here, a cell). Most of the signaling intermediates with this type of specificity are macromolecules. The great diversity of macromolecular shapes and the highly restricted distribution of any given shape dictate that this type of specificity is oligomolecular in its range. Binding may lead to a covalent modification, as when a protein kinase attaches a phosphate to its substrate, but the associations that precede covalent modification are determined by the fit between enzyme and substrate. Similarly, when signaling paths involve dephosphorylation or proteolysis, the breaking of a covalent bond is preceded by a specific handshake between two or more molecules based on shape. Related to this notion of specificity is a view of information flow in signaling as being linear (continuous), whether the lines are simple, branched, or webbed (27). The information is relayed privately, that is, shielded from most other signaling pathways in the same cell.
Progress in genomics and proteomics, along with discoveries that many signaling pathways are conserved among disparate organisms, makes it reasonable to anticipate that most oligomolecular signaling pathways will soon be identified. It is already apparent that a list of such pathways will not add up to understanding physiology within a cell. Oligomolecular networks link pathways together (27). However, it is difficult to imagine that a cell could coordinate all its responses solely by relying on the linkage of each of several hundred pathways to a subset of the others. If the pass-through of a signal from path 1 to path 3 depended on simultaneous activity in path 2, the linkage between 1 and 3 would be erratic. Signal strength would fade and error would mount as the number of relays increased.
This line of reasoning suggests a need for signals that can reach relatively large parts of a cell at nearly the same time, react with enough different molecules to influence many signaling paths directly or indirectly and thereby link the cell’s behavior to its recent commitments and present status. A signal perceived nearly simultaneously by individually distinct signaling pathways across a cell can be described as public.
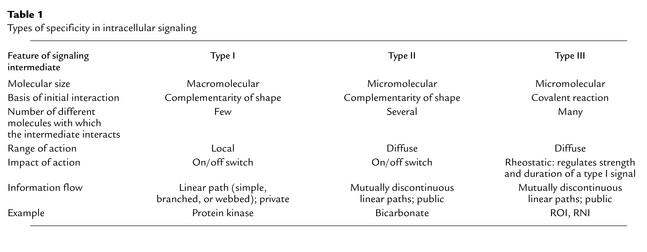
As illustrated in Figure 1 and Table 1, macromolecular signaling intermediates such as kinases and phosphatases act with oligomolecular specificity to signal locally in a cell in a linear, private manner. Small, diffusible molecules such as cAMP, Ca2+, AMP, bicarbonate, and inositol triphosphate signal to activate some pathways locally and others at a distance from the site of their generation or import, still in an oligomolecular manner. Diffusion enables these mediators to activate several pathways at once that are not themselves spatially linked, in which case information can flow in a discontinuous manner. ROI and RNI, it is proposed, often signal with specificity of a third kind. They can act both locally and distantly to tune responses to agonists, a role for which they are suited not only by their small size and diffusibility but also by their reactive chemistry. In fact, chemical reactivity is their distinctive feature as signaling molecules, because it imparts submolecular (atomic) specificity and therewith multimolecular specificity. The combination of diffusibility and multimolecular reactivity suits ROI and RNI to the role of public mediators of intracellular signaling.
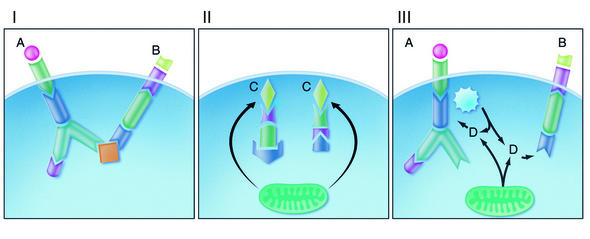
Figure 1.
Distinct types of specificity in intracellular signaling. Panel I: Two first messengers, A and B, each initiate a separate, private, linear signaling pathway that proceeds with type I specificity via oligomolecular handshakes. A or B may also arise intracellularly and activate a pathway that is entirely intracellular. Panel II: A micromolecular first messenger, C, may originate outside or inside the cell. C is shown arising from mitochondria, but there may be other intracellular origins, including type I pathways. C diffuses to various locations in the cell and initiates several signaling pathways. Where C is bicarbonate and the next component of the pathway is bicarbonate-activated adenylyl cyclase, cAMP diffuses a short distance to the next mediator, such as protein kinase A. Panel III: Private pathways with an associated enzymatic source of ROI or RNI (jagged circle), mitochondria, other organelles, cytosolic enzymes, and/or extracellular sources furnish D, a diffusible, micromolecular mediator that reacts covalently with submolecular specificity to modulate the activity of diverse other pathways. Panels II and III illustrate ways in which information can be shared publicly via discontinuous pathways. ROI and RNI act like D. In some cases, ROI and RNI may also act like A or C, except that activation of the pathway begins with a covalent reaction. Photo credit: Thom Graves.
Table 1.
Types of specificity in intracellular signaling
When a cell responds to an agonist, it commits resources to a particular program of gene expression and posttranslational behavior with distinct metabolic consequences. Multiple incoming signals call for multiple resource commitments. A cell will collapse in chaos if it fails to prioritize its commitments and balance them with its resources. It is proposed that signaling with specificity of types II and III allows integration by recruiting, timing, and tuning oligomolecular signaling paths via diffusible reactants whose levels report the cell’s recent history (28) and current state. Thus, a major homeostatic role of ROI and RNI may be to link the behavioral and differentiative commitments of a cell to its metabolic budget. When public mediators are produced in response to a cell’s commitments and in proportion to its resources (29), they can serve to integrate multiple oligomolecular signaling events that would otherwise anarchically compete. It is only the exaggerated production of ROI and RNI that leads to maladaptive signaling in the producing cell or a neighboring or ingested target cell, resulting in disease or defense, depending on the context.
Terms of discourse
Some scientists write as if the terms ROI and RNI lack precise definitions, or conversely, as if all members of each class fit one definition. ROI and RNI are sets of related molecules with individually distinct chemical and biological properties. ROI refers to all oxidation and excitation states of O2, from superoxide (O2.–) up to but excluding water, that arise in physiological environments, including singlet oxygen (1O2*), ozone (O3) (30), hydrogen peroxide (H2O2), hypohalites, and hydroxyl radical (OH.). RNI refers to all oxidation states and reactive adducts of nitrogenous NOS products, from NO up to but excluding nitrate (NO3–), that arise in physiological settings, including nitroxyl (NO–), nitrosonium (NO+), higher oxides of nitrogen, S-nitrosothiols (RSNOs), peroxynitrite (OONO–), and dinitrosyl iron complexes. No two ROI have identical biological properties, nor do any two RNI. Their distinctive properties arise from differences in such features as reactivity, half-life, and lipid solubility.
Some discussions of ROI and RNI assume that RNI cause only nitrosative reactions, whereas only ROI cause oxidative reactions. Oxidative reactions include carbonylations, hydroxylations, peroxidations, or oxidation of sulfhydryls to disulfides or sulfenic, sulfinic, or sulfonic acids. Nitrosative reactions include nitrosylation of sulfhydryls or metals and nitrations of tyrosine residues. Actually, various ROI can oxidize or reduce, and RNI can lead to both nitrosative and redox reactions. It is critical that many of these reactions are reversible, such as formation of methionine sulfoxides and cysteinyl nitrosyls, disulfides, and sulfenic acids.
With respect to the biologic actions of ROI and RNI, some authors use the terms oxidative stress or nitrosative stress in a neutral sense indistinguishable from perturbation. However, most use it to mean reactions that threaten or cause harm. The frame of reference of the present discussion is that ROI and RNI are routinely produced throughout the aerobic biome. Evolution has capitalized on their properties to put them to use as signaling molecules, including in the special case of host defense. From this perspective, the molecules usually referred to as antioxidant and antinitrosative defenses spend most of their time acting as integral parts of homeostatic signaling systems. As with any aspect of physiology, production of ROI and RNI can become excessive to the point that it is maladaptive, if not for the producing cell, then for a target cell. In those circumstances, the systems that catabolize ROI and RNI, or reverse their effects, act as defenses.
Second messengers and second thoughts
The modern era of cell signaling had its beginnings in work that Earl Sutherland summarized in his Nobel address over 30 years ago (31). Sutherland brought the study of hormone action from the level of the organism or organ to the level of the individual cell, thereby helping to open the field of signal transduction. With cAMP as the first example, he introduced the notion of the diffusible second messenger that transduces the signal from the hormone (agonist), or first messenger. He established that an important mechanism of signal transduction is the regulation of enzyme activity by the reversible action of phosphokinases and phosphatases, regulated in part by cAMP. Finally, he pointed out that the functions of guanylyl cyclase and cGMP might be analogous to those of adenylyl cyclase and cAMP. His prescience was rewarded by the discovery that endothelial cell–derived NO activates vascular smooth muscle cell soluble guanylyl cyclase to produce cGMP in a vasoregulatory path, work recognized by the Nobel Prize for Physiology or Medicine in 1998 (32).
However, there was a problem with the second messenger concept: specificity. Sutherland proposed that a given second messenger signaled with specificity because different types of cells exclusively contained different types of enzymes responsive to the same second messenger. That is, cell type A contained cAMP-responsive enzyme A, while cell type B contained cAMP-responsive enzyme B. However, it soon became clear that one and the same cell contains large numbers of signaling systems responding to cAMP. Moreover, cAMP usually activates the same enzyme — protein kinase A — in response to different first messengers. Thus, the second messenger principle could no longer account for specificity. Another concept was necessary to explain how different agonists can act on a single cell to elicit agonist-specific responses.
The rise of oligomolecular signal transduction
The answer lay in the growing appreciation of oligomolecular signaling pathways. Work by many investigators, epitomized by the contributions of Paul Greengard, James Darnell, Anthony Pawson, Joan Massague, Joseph Schlessinger, and Louis Cantley, among others, established that much of cell signaling depends on specific intermolecular cascades executed by protein-protein and protein-lipid interactions. Constitutive molecular shapes such as src homology 2 (SH2), SH3, polyproline, or pleckstrin homology domains, and acquired molecular shapes such as those conferred by (de)phosphorylation dictate the transient handshakes between signaling intermediates. The concept of molecular handshakes revealed how signaling achieves spatial constraint and agonist specificity. For example, with Janus kinases (JAKs) and signal transducers and activators of transcription (STATs), one can trace a path from a cytokine receptor at the plasma membrane to transcriptional regulation in the nucleus without recourse to small, diffusible second messengers, although some of the proteins must diffuse (33). Co-isolation of interacting proteins (signalosomes) demonstrated that specific oligomolecular interactions may anticipate receipt of the signals they help transduce, further reducing the role of diffusional encounters and improving the pathway’s privacy.
Inklings arose that oligomolecular specificity may not be the whole story, but these concerns were muted. For example, it became clear that many agonists share the same oligomolecular signaling paths, yet evoke distinct responses. The explanation was offered that all agonists activate several oligomolecular pathways. Since we don’t yet know all the paths activated by any agonist, the possibility cannot be excluded that agonists eliciting distinct responses always activate unique combinations of pathways. An extension of this argument was the postulate that signaling specificity is ultimately manifest by unique combinatorial assemblies of variously modified transcription factors on gene promoters and enhancers.
The combinatorial argument raised its own challenges. Shared transcription factors could be titrated out by reactions at one promoter, decreasing activation at others. Thus, transduction of each signal could potentially be interrupted by, or preclude a response to, the next. Competitive principles such as “first-come, first-served” or “winner-takes-all” would dictate how a cell would respond in the complex situations subsumed under the terms physiology and pathophysiology. It was not clear how the cell might respond if dozens of signals of comparable strength were received simultaneously, or if successive signals reflected changing circumstances, even though an earlier signal had committed the cell to a course of behavior that was no longer adaptive.
More second thoughts about second messengers: denying diffusion
Once molecular handshakes were considered to account for specificity in intracellular signaling, an effort was made to incorporate second messengers into the same vision of private information flow. It was argued that second messengers are not, operationally speaking, freely diffusible. This viewpoint was based on the discovery that intracellular elevations of second messengers are sometimes most pronounced in circumscribed subcellular localizations. For example, cAMP sometimes rises highest near an activated receptor (34) where an anchoring protein tethers adenylyl cyclase, protein kinase A, and the kinase’s downstream targets in a signalosome (35). Similarly, Ca2+ sometimes rises in plumes near activated channels (36). For lipid second messengers as well, confined localization was demonstrated at discrete plasma membrane sites near points of signal initiation (37). To explain restricted diffusion, it was proposed that enzymes or channels capable of metabolizing or exporting the second messengers were recruited to the signaling site via protein-protein or protein-lipid interactions with oligomolecular specificity, building a moat around the receptor across which second messengers could not readily escape (38).
Questions remained. What about the presence of numerous other potentially responsive signaling components within the second-messenger high-rise zone? What about the functional import of lesser but still substantial elevations of cAMP or Ca2+ throughout other parts of the cell? In fact, recent evidence reinforces that Ca2+ elevations and oscillations act as signals not just locally but also broadly across the cell, including in the nucleus (39–41). At the same time, the identification of additional diffusible mediators, described below, added fresh evidence that public intracellular signals exist and have fundamental utility.
Messengers from metabolism
The yeast SNF1 protein kinase cascade and its homologues in mammals and plants control aspects of intermediary metabolism by regulating transcription factors that act on genes whose products affect the production or fate of glucose, fatty acids, and cholesterol, among others. These kinases are activated by an increase in the ratio of AMP to ATP, such that they were described as a fuel gauge for the cell (42). Thus, a small, diffusible molecule, AMP, acts as a signal that reports metabolic status and exerts widespread control over gene expression by diffusing to its targets from distant points of origin.
Next came the stunning discovery that not all adenylyl cyclases are transmembrane proteins controlled by G protein–coupled receptors responsive to diverse agonists. In fact, most mammalian cells appear to contain adenylyl cyclases in most subcellular compartments that are all activated by the same agonist: bicarbonate (43–45). Bicarbonate, formed almost instantaneously by carbonic anhydrase, reports the ambient levels or intracellular production of CO2, a direct read-out of intermediary metabolism. As a public mediator, bicarbonate can activate mutually noninteractive signaling pathways simultaneously at diverse subcellular localizations.
Another messenger from metabolism is NAD(P)H or NAD(P)+ in those situations where one redox state of the nucleotide, but not the other, serves as an allosteric activator. For example, NADH and NADPH, but not NAD+ or NADP+, sustain the transcriptional activity of Clock:BMAL1 and NPAS2:BMAL1, regulators of cell clock–dependent gene expression (46). Likewise, NADH, but not NAD+, activates the transcriptional corepressor CtBP and thereby helps control development and cell cycle (47). Similarly, some bacteria gauge their nutritional status in part by monitoring the level of charged tRNAs. When a deficiency of charged tRNAs stalls ribosomes, an enzyme is activated that produces guanosine 3′-disposphate, 5′-triphosphate, or guanosine 3′,5′-bispyrophosphate. The nucleotide diffuses to and binds RNA polymerase, altering its specificity with respect to the transcription of numerous genes, thus helping to mediate the stringent response (48).
In the foregoing discussion, signaling has been distinguished from the metabolic regulation that occurs when substrates and products exert positive or negative control on the enzymes that use or make them. However, the distinction is somewhat artificial. Thus, messengers from metabolism also coordinate cellular behavior via pleiotropic signaling pathways when they serve as a cosubstrate for an upstream component that is responsive to physiologic variations in the concentration of the messenger. For example, compared with most protein kinases, mammalian target of rapamycin (mTOR) has a remarkably high Michaelis constant for ATP (∼1 mM) (49). This helps mTOR respond to changes in ATP concentration in the physiologic range to regulate ribosome biogenesis and protein translation (49). Similarly, silent information regulator 2 (SIR2) is sensitive to levels of NAD+ when it deacetylates the master transcriptional regulator p53 (50), a reaction that is proposed to link metabolism to aging. The NADPH/NADP+ ratio also controls the redox state of thioredoxin. Thioredoxin in turn regulates enzymes such as ribonucleotide reductase, and transcription factors such as NF-κB, with a major impact on cellular commitments (51).
ROI and RNI: both first messengers and signaling intermediates
As the literature swelled with examples of intracellular signaling triggered or affected by ROI and RNI, it became increasingly difficult to know how to relate these findings to the classical model of second messengers or to its successor, the contemporary model of molecular handshakes. Several recent studies ratcheted the problem to a new level.
First, Stamler, Lamas, and Fang collected nearly 100 examples of proteins that are regulated posttranslationally by nitrosylation (13). Most were intracellular, and there were examples from almost every functional class. Second, Jaffrey et al. confirmed that at least some of these proteins are nitrosylated in healthy animals (52). Third, using macrophages from mice deficient in iNOS, phox, or both, Ehrt et al. demonstrated that iNOS and phox exert widespread effects on gene expression in activated macrophages (25). Macrophage activation by a cytokine (IFN-γ) and a microbe (the tubercle bacillus) led to altered expression of about 25% of the monitored macrophage genome and about 40% of the monitored transcriptome. Of the genes affected by macrophage activation, 58% were significantly impacted in their expression by iNOS and/or phox. Remarkably, there was functional selectivity among the genes affected by iNOS and phox. Genes involved in immunity and inflammation were almost completely spared (that is, regulated by macrophage activation independently of iNOS and phox), while many others were suppressed in an iNOS- and/or phox-dependent manner. The authors hypothesized that “a self-induced state of metabolic stringency within the activated, infected macrophage may necessitate suppression of the expression of those genes not essential to the emergency at hand” (25). Taking the three reports together, and considering that production of ROI and RNI is nearly ubiquitous in the aerobic biome, it is difficult to escape the conclusion that the impact of ROI and RNI on intracellular signal transduction is physiologic and widespread.
Investigators have offered several different characterizations of intracellular signaling by ROI/RNI. First, ROI and RNI have been described as first messengers triggering private pathways. For example, Zheng et al. described OxyR as a bacterial transcription factor acting as an H2O2 receptor by undergoing oxidation and allosteric activation, leading to expression of antioxidant defenses and other adaptations to oxidative stress (53). Helmann and colleagues extended the concept to include not just cysteine-dependent proteins but also metalloproteins as bacterial peroxide sensors (54, 55). Delaunay et al. presented a more complex version of the same concept, in which a specific enzyme serves as an H2O2 receptor in yeast. The enzyme undergoes allosteric changes when oxidized and then specifically transfers the oxidized status and a corresponding allosteric change to a transcription factor (56) in a manner analogous to a kinase cascade causing successive phosphorylations. For RNI, the classic example has already been mentioned: extracellular NO acts as a first signal to activate intracellular soluble guanylate cyclase, triggering a phosphorylation cascade (2–4). In other examples, Kim et al. (57) argued that OxyR serves not only as an H2O2 receptor but also as an NO receptor, undergoing either nitrosylation or oxidation. Demple and Demple showed that another bacterial transcription factor, SoxR, can act as an NO receptor. SoxR’s iron-sulfur clusters are nitrosylated by NO, leading to a conformational change in the protein that allows it to transactivate the gene encoding another transcription factor, SoxS. SoxS in turn controls induction of antioxidant enzymes (58). Thus, ROI and RNI can signal by initiating pathways that operate with type I specificity. However, in these situations, ROI and RNI themselves are the agonists, not the downstream mediators.
In another characterization of signaling by RNI, nitrosylation has been likened to phosphorylation (13). However, phosphorylation and dephosphorylation convey specificity because cells express an array of kinases and phosphatases, each of which binds transiently to a restricted set of client proteins. In contrast, there does not appear to be a corresponding array of nitrosylases or denitrosylases individually dedicated to modifying specific client proteins. Thus, reversible nitrosylation of signaling intermediates that have oligomolecular specificity is not by itself an indication that RNI act with type I specificity.
Describing a very different set of circumstances, Reth wrote that H2O2 acts as a second messenger (16). He described “a cloud of H2O2” emanating from receptors whose activation by other agonists is coupled both to phosphorylation and to ROI production (16). The best-described impact of H2O2 on signaling initiated by other agonists is that H2O2 transiently inactivates tyrosine phosphatases via reversible oxidation of their active-site cysteinyl residues (59–61) and thereby augments phosphokinase-mediated signal transmission. Because the specificity is imparted by the receptor and is not preserved by the H2O2, H2O2 in this situation is not a second messenger in Sutherland’s sense. Rather, H2O2 is acting in this case as a secondary messenger, modifying the extent or duration of a reaction initiated by another signal. This regulatory role, executed through a reversible covalent interaction, typifies type III specificity.
Carbon monoxide (CO) is another example of a mediator that can signal with type III specificity. Heme synthesis is under circadian control (62). Heme oxygenase, an enzyme with constitutive as well as ROI- and RNI-induced isoforms, breaks down heme to give rise to CO. CO activates a mammalian transcription factor, NPAS2, that regulates circadian rhythm by binding covalently to the heme in its PAS domain (63).
In a special case analogous to ATP acting as a signal to mTOR, O2 acts a covalent mediator by serving as a substrate for two regulatory amino acid hydroxylases that are sensitive to physiologic variations in O2 concentration. Iron-dependent prolyl hydroxylase uses O2 to hydroxylate certain prolyl residues in hypoxia-inducible transcription factor-1α (HIF-1α). This leads to the recognition of HIF-1α by the von Hippel–Lindau tumor suppressor gene product. The result is ubiquitin ligation of HIF-1α and its proteasomal degradation. Another O2-using enzyme hydroxylates an asparagine residue in HIF-1α, preventing p300/CBP from binding there to support HIF-1α’s transactivating activity. Under hypoxic conditions, both hydroxylations are diminished. Activated HIF-1α accumulates, binds HIF-1β, and drives gene transcription (64–68). The process may be regulated or mimicked by ROI and RNI (69).
Submolecular and multimolecular specificity
The specificity of ROI and RNI in signaling can be understood by considering the relationship between chemistry and function. Chemically, their specificity is submolecular, that is, atomic (12); functionally, it is multimolecular and therefore positioned to be integrative.
Atomic specificity is illustrated, in the case of proteins, by the preferred reactivity of ROI and RNI with cysteine sulfhydryls, methionine sulfurs, tyrosine hydroxyls, iron-sulfur clusters, and prosthetic hemes. This reactivity is further affected by the surrounding environment within the protein. For example, peptidyl cysteine sulfhydryls are more readily nitrosylated or oxidized when they are encountered as the thiolate anion, a condition fostered by positively charged amino acid side chains suitably disposed in the vicinity. The reactivity of ROI and RNI with atomic targets in the context of small regions of macromolecules, rather than with a given macromolecule as a unique entity, suggests that the specificity of ROI and RNI for these targets can be described as submolecular.
Because the atomic targets of ROI and RNI are basic building blocks of numerous proteins, the inescapable consequence of submolecular specificity is multimolecular reactivity. For example, as noted by Ehrt et al., “The selective transcriptional re-programming accompanying macrophage activation may depend in part on the relative susceptibility of Cys-dependent transcription factors to nitrosation or oxidation. Transcription factors that depend on Cys residues include NF-κB, activation protein-1, p53 and the most numerous family in the genome, zinc finger proteins” (25). Particular zinc finger proteins, such as protein kinase C and Raf, are sensitized to oxidative activation by binding the redox-active vitamin retinol or one of its derivatives. The retinol facilitates oxidation of zinc-coordinating cysteine sulfhydryls, leading to a conformational change (70). Proteins containing PAS domains represent another large set of possible targets for integrative signaling by ROI and RNI. PAS domain proteins use flavins, hemes, or undefined structures to sense redox potential, energy level, proton motive force, oxygen, or light to regulate diverse aspects of cellular behavior, often through kinase cascades (71). Furthermore, through their impact on the redox state and activity of thioredoxin, ROI and RNI can indirectly control the activity of other enzymes and transcription factors (51).
Production of ROI and RNI can report metabolic status in a cell because their shared precursors are oxygen and NAD(P)H. ROI and RNI can also regulate metabolic status. Besides soluble guanylate cyclase, the most sensitive known NO receptor in mammalian cells is cytochrome oxidase (72). NO physiologically regulates the mitochondrial electron transport chain, and in doing so may be responsible for the mitochondrial electron transport chain serving as a major source of ROI in most mammalian cells (72). At higher levels, RNI can also affect ATP synthesis by inhibiting GAPDH, cis-aconitase, and NADH:ubiquinone and succinate:ubiquinone oxidoreductases (for references, see ref. 25). In this way, ROI and RNI are poised to help control other integrating mediators such as AMP and CO2/bicarbonate. A recently discovered interaction of thioredoxin with carbonic anhydrase (73) suggests another way in which ROI and RNI may influence bicarbonate-triggered production of cAMP.
Antioxidant systems as signaling components
Given that ROI and RNI report and relate the cell’s metabolic state to its signaling apparatus, it is to be expected that catabolic paths operating on ROI and RNI may do so as well. Catalase and superoxide dismutase bear no obvious link to intermediary metabolism. However, other widely conserved antioxidant regulatory systems, such as peroxiredoxins, thioredoxins, methionyl sulfoxide reductases, glutathione, and glutaredoxins, function by means of reversible oxidation of cysteinyl thiols in their active sites. They are linked to glycolysis, the hexose monophosphate shunt, and the citric acid cycle via their dependence on NAD(P)H for reducing equivalents. A striking example is offered by alkylhydroperoxide reductase subunit C (AhpC), a peroxiredoxin. This enzyme and its apparently ubiquitous homologues act to break down H2O2, organic peroxides, and peroxynitrite (74). In Mycobacterium tuberculosis, the reducing power for AhpC is drawn from NADH via components of an α-ketoacid dehydrogenase complex as siphoned off by AhpD, a thioredoxin orthologue (75).
If ROI- and RNI-catabolizing enzymes are integral components of homeostatic signaling, then it must be anticipated that these enzymes themselves will be subject to regulation by other signaling pathways. For example, peroxiredoxin II is phosphorylated and inhibited by Cdc2 (76), a reaction exhibiting type I specificity, while thioredoxin is nitrosylated outside its active site and thereby activated in endothelial cells (77), an example of type III specificity.
When signaling becomes killing
Multimolecular specificity endows ROI and RNI with the ability to kill other cells when high levels impose maladaptive signaling at a global level. This is just one manifestation of a phenomenon that is widespread in biology: the propensity for one organism to exploit or attack another by forcing its opponent to contain or express signaling molecules at levels inappropriate for the time and place (Table 2). Fittingly, this was first shown for cAMP (78, 79).
Table 2.
Examples of forced overexpression of intracellular signals during microbial pathogenesis and host defense
Cells seeking to defend themselves against excessive ROI or RNI would be expected to evolve signaling systems in which the ROI or RNI would be the primary agonist (first messenger). Thus the ability of ROI and RNI to signal with type III specificity predicts the situation, discussed above, in which ROI or RNI initiate pathways, such as the oxyR regulon, that signal with type I specificity. Seen in this light, it is no surprise that many of the pathways in which ROI or RNI act as first messengers serve to induce antioxidant defenses.
Unanswered questions
This essay attempts to present a conceptual framework for integrative signaling and nominates ROI and RNI as some of the mediators that carry it out. More experimental evidence will be needed to evaluate this hypothesis. Experiments in which ROI and RNI are added as reagents will not establish their physiologic importance as convincingly as studies in which endogenous ROI- and RNI-producing and -catabolizing pathways are specifically inhibited by the selective suppression or disruption of the genes encoding them. In practice, lessons from the gene disruptions carried out to date are not fully interpretable. The identities of some of the key enzymes remain elusive, and there may be redundancy among them. Moreover, the phenotype of mutant mice is rarely examined under the range of conditions that exist in the wild. Sources have not been established for the ROI produced in response to and necessary for full signaling by insulin (80, 81), PDGF (82), EGF (83), TGF-β (84), nerve growth factor (85), angiotensin (86), and GM-CSF (87). Until the sources are identified, we are unlikely to understand the mechanisms by which hormones and growth factors activate ROI production. We know that individual ROI and RNI can play distinctive roles (25, 88), but we do not understand how.
Parallels to the immune system
For many years, some immunologists considered that what is now called the innate immune system lay outside the field, because cells of the innate immune system lacked specificity, and specificity is the hallmark of immunity. In retrospect, the issue was not whether specificity is physiologically meritorious, but whether it must necessarily be of only one kind. It is now widely accepted in immunologic circles that survival of an organism in a competitive microbial environment requires specificity of several kinds to detect microbes and coordinate the host’s response with the requisite efficiency (89–91). Adaptive immunity is geared toward recognition of private antigenic specificities. Innate immunity is predicated on binding to shared microbial structures. In highly evolved immune systems, each depends critically on the other. That transformation of outlook inspired the present thoughts.
Acknowledgments
I thank the JCI, in particular Qais Al-Awqati and Brooke Grindlinger, and Roy Soberman, Series Editor for this Perspective series, for their adventurousness in accommodating this article. Special thanks are due Jochen Buck, Heran Darwin, Aihao Ding, Sabine Ehrt, and Lonny Levin for critiques of the manuscript. Preparation of this article was supported in part by NIH grants HL-61241, AG-19520, AI-46382, and AI-56293. The Department of Microbiology and Immunology, Weill Medical College of Cornell University, is supported by the William Randolph Hearst Foundation.
Footnotes
Conflict of interest: The author has declared that no conflict of interest exists.
Nonstandard abbreviations used: reactive oxygen intermediate(s) (ROI); reactive nitrogen intermediate(s) (RNI); phagocyte oxidase (phox); hydrogen peroxide (H2O2); mammalian target of rapamycin (mTOR); carbon monoxide (CO); hypoxia-inducible transcription factor-1α (HIF-1α); alkylhydroperoxide reductase subunit C (AhpC).
References
- 1.Nathan C. Nitric oxide as a secretory product of mammalian cells. FASEB J. 1992;6:3051–3064. [PubMed] [Google Scholar]
- 2.Palmer RM, Ferrige AG, Moncada S. Nitric oxide release accounts for the biological activity of endothelium-derived relaxing factor. Nature. 1987;327:524–526. doi: 10.1038/327524a0. [DOI] [PubMed] [Google Scholar]
- 3.Ignarro LJ, Buga GM, Wood KS, Byrns RE, Chaudhuri G. Endothelium-derived relaxing factor produced and released from artery and vein is nitric oxide. Proc. Natl. Acad. Sci. U. S. A. 1987;84:9265–9269. doi: 10.1073/pnas.84.24.9265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arnold WP, Mittal CK, Katsuki S, Murad F. Nitric oxide activates guanylate cyclase and increases guanosine 3′:5′-cyclic monophosphate levels in various tissue preparations. Proc. Natl. Acad. Sci. U. S. A. 1977;74:3203–3207. doi: 10.1073/pnas.74.8.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schreck R, Rieber P, Baeuerle PA. Reactive oxygen intermediates as apparently widely used messengers in the activation of the NF-kappa B transcription factor and HIV-1. EMBO J. 1991;10:2247–2258. doi: 10.1002/j.1460-2075.1991.tb07761.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amstad PA, Krupitza G, Cerutti PA. Mechanism of c-fos induction by active oxygen. Cancer Res. 1992;52:3952–3960. [PubMed] [Google Scholar]
- 7.Nakamura H, Nakamura K, Yodoi J. Redox regulation of cellular activation. Annu. Rev. Immunol. 1997;15:351–369. doi: 10.1146/annurev.immunol.15.1.351. [DOI] [PubMed] [Google Scholar]
- 8.Beckman KB, Ames BN. The free radical theory of aging matures. Physiol. Rev. 1998;78:547–581. doi: 10.1152/physrev.1998.78.2.547. [DOI] [PubMed] [Google Scholar]
- 9.Dalton TP, Shertzer HG, Puga A. Regulation of gene expression by reactive oxygen. Annu. Rev. Pharmacol. Toxicol. 1999;39:67–101. doi: 10.1146/annurev.pharmtox.39.1.67. [DOI] [PubMed] [Google Scholar]
- 10.Finkel T. Oxygen radicals and signaling. Curr. Opin. Cell Biol. 1998;10:248–253. doi: 10.1016/s0955-0674(98)80147-6. [DOI] [PubMed] [Google Scholar]
- 11.Rhee SG, Bae YS, Lee SR, Kwon J. Hydrogen peroxide: a key messenger that modulates protein phosphorylation through cysteine oxidation. Sci. STKE. 2000;2000:PE1. doi: 10.1126/stke.2000.53.pe1. [DOI] [PubMed] [Google Scholar]
- 12.Nathan C, Shiloh MU. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc. Natl. Acad. Sci. U. S. A. 2000;97:8841–8848. doi: 10.1073/pnas.97.16.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stamler JS, Lamas S, Fang FC. Nitrosylation. The prototypic redox-based signaling mechanism. Cell. 2001;106:675–683. doi: 10.1016/s0092-8674(01)00495-0. [DOI] [PubMed] [Google Scholar]
- 14.Bogdan C. Nitric oxide and the regulation of gene expression. Trends Cell Biol. 2001;11:66–75. doi: 10.1016/s0962-8924(00)01900-0. [DOI] [PubMed] [Google Scholar]
- 15.Droge W. Free radicals in the physiological control of cell function. Physiol. Rev. 2002;82:47–95. doi: 10.1152/physrev.00018.2001. [DOI] [PubMed] [Google Scholar]
- 16.Reth M. Hydrogen peroxide as second messenger in lymphocyte activation. Nat. Immunol. 2002;3:1129–1134. doi: 10.1038/ni1202-1129. [DOI] [PubMed] [Google Scholar]
- 17.Golderer G, Werner ER, Leitner S, Grobner P, Werner-Felmayer G. Nitric oxide synthase is induced in sporulation of Physarum polycephalum. Genes Dev. 2001;15:1299–1309. doi: 10.1101/gad.890501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuzin B, Roberts I, Peunova N, Enikolopov G. Nitric oxide regulates cell proliferation during Drosophila development. Cell. 1996;87:639–649. doi: 10.1016/s0092-8674(00)81384-7. [DOI] [PubMed] [Google Scholar]
- 19.Peunova N, Scheinker V, Cline H, Enikolopov G. Nitric oxide is an essential negative regulator of cell proliferation in Xenopus brain. J. Neurosci. 2001;21:8809–8818. doi: 10.1523/JNEUROSCI.21-22-08809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klein JA, et al. The harlequin mouse mutation downregulates apoptosis-inducing factor. Nature. 2002;419:367–374. doi: 10.1038/nature01034. [DOI] [PubMed] [Google Scholar]
- 21.Gu Z, et al. S-nitrosylation of matrix metalloproteinases: signaling pathway to neuronal cell death. Science. 2002;297:1186–1190. doi: 10.1126/science.1073634. [DOI] [PubMed] [Google Scholar]
- 22.Terman JR, Mao T, Pasterkamp RJ, Yu HH, Kolodkin AL. MICALs, a family of conserved flavoprotein oxidoreductases, function in plexin-mediated axonal repulsion. Cell. 2002;109:887–900. doi: 10.1016/s0092-8674(02)00794-8. [DOI] [PubMed] [Google Scholar]
- 23.Nathan C, Xie QW. Nitric oxide synthases: roles, tolls, and controls. Cell. 1994;78:915–918. doi: 10.1016/0092-8674(94)90266-6. [DOI] [PubMed] [Google Scholar]
- 24.Lambeth JD. Nox/Duox family of nicotinamide adenine dinucleotide (phosphate) oxidases. Curr. Opin. Hematol. 2002;9:11–17. doi: 10.1097/00062752-200201000-00003. [DOI] [PubMed] [Google Scholar]
- 25.Ehrt S, et al. Reprogramming of the macrophage transcriptome in response to interferon-gamma and Mycobacterium tuberculosis: signaling roles of nitric oxide synthase-2 and phagocyte oxidase. J. Exp. Med. 2001;194:1123–1140. doi: 10.1084/jem.194.8.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diefenbach A, Schindler H, Rollinghoff M, Yokoyama WM, Bogdan C. Requirement for type 2 NO synthase for IL-12 signaling in innate immunity. Science. 1999;284:951–955. doi: 10.1126/science.284.5416.951. [DOI] [PubMed] [Google Scholar]
- 27.Jordan JD, Landau EM, Iyengar R. Signaling networks: the origins of cellular multitasking. Cell. 2000;103:193–200. doi: 10.1016/s0092-8674(00)00112-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ingolia NT, Murray AW. Signal transduction. History matters. Science. 2002;297:948–949. doi: 10.1126/science.1075222. [DOI] [PubMed] [Google Scholar]
- 29.Strohman R. Maneuvering in the complex path from genotype to phenotype. Science. 2002;296:701–703. doi: 10.1126/science.1070534. [DOI] [PubMed] [Google Scholar]
- 30.Wentworth P, Jr, et al. Evidence for antibody-catalyzed ozone formation in bacterial killing and inflammation. Science. 2002;298:2195–2199. doi: 10.1126/science.1077642. [DOI] [PubMed] [Google Scholar]
- 31.Sutherland EW. Studies on the mechanism of hormone action. Science. 1972;177:401–408. doi: 10.1126/science.177.4047.401. [DOI] [PubMed] [Google Scholar]
- 32.Smith O. Nobel Prize for NO research. Nat. Med. 1998;4:1215. doi: 10.1038/3182. [DOI] [PubMed] [Google Scholar]
- 33.Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 34.Rich TC, et al. A uniform extracellular stimulus triggers distinct cAMP signals in different compartments of a simple cell. Proc. Natl. Acad. Sci. U. S. A. 2001;98:13049–13054. doi: 10.1073/pnas.221381398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davare MA, et al. A beta2 adrenergic receptor signaling complex assembled with the Ca2+ channel Cav1.2. Science. 2001;293:98–101. doi: 10.1126/science.293.5527.98. [DOI] [PubMed] [Google Scholar]
- 36.Bootman MD, et al. Calcium signalling—an overview. Semin. Cell Dev. Biol. 2001;12:3–10. doi: 10.1006/scdb.2000.0211. [DOI] [PubMed] [Google Scholar]
- 37.Vieira OV, et al. Distinct roles of class I and class III phosphatidylinositol 3-kinases in phagosome formation and maturation. J. Cell Biol. 2001;155:19–25. doi: 10.1083/jcb.200107069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perry SJ, et al. Targeting of cyclic AMP degradation to beta 2-adrenergic receptors by beta-arrestins. Science. 2002;298:834–836. doi: 10.1126/science.1074683. [DOI] [PubMed] [Google Scholar]
- 39.Dolmetsch R. Excitation-transcription coupling: signaling by ion channels to the nucleus. Sci. STKE. 2003;2003:PE4. doi: 10.1126/stke.2003.166.pe4. [DOI] [PubMed] [Google Scholar]
- 40.Dolmetsch RE, Xu K, Lewis RS. Calcium oscillations increase the efficiency and specificity of gene expression. Nature. 1998;392:933–936. doi: 10.1038/31960. [DOI] [PubMed] [Google Scholar]
- 41.Feske S, Giltnane J, Dolmetsch R, Staudt LM, Rao A. Gene regulation mediated by calcium signals in T lymphocytes. Nat. Immunol. 2001;2:316–324. doi: 10.1038/86318. [DOI] [PubMed] [Google Scholar]
- 42.Hardie DG, Carling D, Carlson M. The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu. Rev. Biochem. 1998;67:821–855. doi: 10.1146/annurev.biochem.67.1.821. [DOI] [PubMed] [Google Scholar]
- 43.Chen Y, et al. Soluble adenylyl cyclase as an evolutionarily conserved bicarbonate sensor. Science. 2000;289:625–628. doi: 10.1126/science.289.5479.625. [DOI] [PubMed] [Google Scholar]
- 44.Zippin JH, Levin LR, Buck J. CO(2)/HCO(3)(-)-responsive soluble adenylyl cyclase as a putative metabolic sensor. Trends Endocrinol. Metab. 2001;12:366–370. doi: 10.1016/s1043-2760(01)00454-4. [DOI] [PubMed] [Google Scholar]
- 45.Zippin JH, et al. Compartmentalization of bicarbonate-sensitive adenylyl cyclase in distinct signaling microdomains. FASEB J. 2003;17:82–84. doi: 10.1096/fj.02-0598fje. [DOI] [PubMed] [Google Scholar]
- 46.Rutter J, Reick M, Wu LC, McKnight SL. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science. 2001;293:510–514. doi: 10.1126/science.1060698. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Q, Piston DW, Goodman RH. Regulation of corepressor function by nuclear NADH. Science. 2002;295:1895–1897. doi: 10.1126/science.1069300. [DOI] [PubMed] [Google Scholar]
- 48.Chatterji D, Ojha AK. Revisiting the stringent response, ppGpp and starvation signaling. Curr. Opin. Microbiol. 2001;4:160–165. doi: 10.1016/s1369-5274(00)00182-x. [DOI] [PubMed] [Google Scholar]
- 49.Dennis PB, et al. Mammalian TOR: a homeostatic ATP sensor. Science. 2001;294:1102–1105. doi: 10.1126/science.1063518. [DOI] [PubMed] [Google Scholar]
- 50.Vaziri H, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 51.Arner ES, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 2000;267:6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 52.Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat. Cell Biol. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- 53.Zheng M, Aslund F, Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279:1718–1721. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]
- 54.Mongkolsuk S, Helmann JD. Regulation of inducible peroxide stress responses. Mol. Microbiol. 2002;45:9–15. doi: 10.1046/j.1365-2958.2002.03015.x. [DOI] [PubMed] [Google Scholar]
- 55.Fuangthong M, Helmann JD. The OhrR repressor senses organic hydroperoxides by reversible formation of a cysteine-sulfenic acid derivative. Proc. Natl. Acad. Sci. U. S. A. 2002;99:6690–6695. doi: 10.1073/pnas.102483199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Delaunay A, Pflieger D, Barrault MB, Vinh J, Toledano MB. A thiol peroxidase is an H2O2 receptor and redox-transducer in gene activation. Cell. 2002;111:471–481. doi: 10.1016/s0092-8674(02)01048-6. [DOI] [PubMed] [Google Scholar]
- 57.Kim SO, et al. OxyR: a molecular code for redox-related signaling. Cell. 2002;109:383–396. doi: 10.1016/s0092-8674(02)00723-7. [DOI] [PubMed] [Google Scholar]
- 58.Ding H, Demple B. Direct nitric oxide signal transduction via nitrosylation of iron-sulfur centers in the SoxR transcription activator. Proc. Natl. Acad. Sci. U. S. A. 2000;97:5146–5150. doi: 10.1073/pnas.97.10.5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Denu JM, Tanner KG. Redox regulation of protein tyrosine phosphatases by hydrogen peroxide: detecting sulfenic acid intermediates and examining reversible inactivation. Methods Enzymol. 2002;348:297–305. doi: 10.1016/s0076-6879(02)48648-x. [DOI] [PubMed] [Google Scholar]
- 60.Meng TC, Fukada T, Tonks NK. Reversible oxidation and inactivation of protein tyrosine phosphatases in vivo. Mol. Cell. 2002;9:387–399. doi: 10.1016/s1097-2765(02)00445-8. [DOI] [PubMed] [Google Scholar]
- 61.Xu D, Rovira II, Finkel T. Oxidants painting the cysteine chapel: redox regulation of PTPs. Dev. Cell. 2002;2:251–252. doi: 10.1016/s1534-5807(02)00132-6. [DOI] [PubMed] [Google Scholar]
- 62.Zheng B, et al. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105:683–694. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]
- 63.Dioum EM, et al. NPAS2: a gas-responsive transcription factor. Science. 2002;298:2385–2387. doi: 10.1126/science.1078456. [DOI] [PubMed] [Google Scholar]
- 64.Ivan M, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 65.Jaakkola P, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 66.Epstein AC, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 67.Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 68.Lando D, Peet DJ, Whelan DA, Gorman JJ, Whitelaw ML. Asparagine hydroxylation of the HIF transactivation domain a hypoxic switch. Science. 2002;295:858–861. doi: 10.1126/science.1068592. [DOI] [PubMed] [Google Scholar]
- 69.Semenza GL. HIF-1 and mechanisms of hypoxia sensing. Curr. Opin. Cell Biol. 2001;13:167–171. doi: 10.1016/s0955-0674(00)00194-0. [DOI] [PubMed] [Google Scholar]
- 70.Hoyos B, et al. The cysteine-rich regions of the regulatory domains of Raf and protein kinase C as retinoid receptors. J. Exp. Med. 2000;192:835–845. doi: 10.1084/jem.192.6.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Taylor BL, Zhulin IB. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moncada S, Erusalimsky JD. Does nitric oxide modulate mitochondrial energy generation and apoptosis? Nat. Rev. Mol. Cell Biol. 2002;3:214–220. doi: 10.1038/nrm762. [DOI] [PubMed] [Google Scholar]
- 73.Balmer Y, et al. Proteomics gives insight into the regulatory function of chloroplast thioredoxins. Proc. Natl. Acad. Sci. U. S. A. 2003;100:370–375. doi: 10.1073/pnas.232703799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bryk R, Griffin P, Nathan C. Peroxynitrite reductase activity of bacterial peroxiredoxins. Nature. 2000;407:211–215. doi: 10.1038/35025109. [DOI] [PubMed] [Google Scholar]
- 75.Bryk R, Lima CD, Erdjument-Bromage H, Tempst P, Nathan C. Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein. Science. 2002;295:1073–1077. doi: 10.1126/science.1067798. [DOI] [PubMed] [Google Scholar]
- 76.Chang TS, et al. Regulation of peroxiredoxin I activity by Cdc2-mediated phosphorylation. J. Biol. Chem. 2002;277:25370–25376. doi: 10.1074/jbc.M110432200. [DOI] [PubMed] [Google Scholar]
- 77.Haendeler J, et al. Redox regulatory and anti-apoptotic functions of thioredoxin depend on S-nitrosylation at cysteine 69. Nat. Cell Biol. 2002;4:743–749. doi: 10.1038/ncb851. [DOI] [PubMed] [Google Scholar]
- 78.Wolff J, Cook GH, Goldhammer AR, Berkowitz SA. Calmodulin activates prokaryotic adenylate cyclase. Proc. Natl. Acad. Sci. U. S. A. 1980;77:3841–3844. doi: 10.1073/pnas.77.7.3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Confer DL, Eaton JW. Phagocyte impotence caused by an invasive bacterial adenylate cyclase. Science. 1982;217:948–950. doi: 10.1126/science.6287574. [DOI] [PubMed] [Google Scholar]
- 80.Mahadev K, Zilbering A, Zhu L, Goldstein BJ. Insulin-stimulated hydrogen peroxide reversibly inhibits protein-tyrosine phosphatase 1b in vivo and enhances the early insulin action cascade. J. Biol. Chem. 2001;276:21938–21942. doi: 10.1074/jbc.C100109200. [DOI] [PubMed] [Google Scholar]
- 81.Mahadev K, et al. Hydrogen peroxide generated during cellular insulin stimulation is integral to activation of the distal insulin signaling cascade in 3T3-L1 adipocytes. J. Biol. Chem. 2001;276:48662–48669. doi: 10.1074/jbc.M105061200. [DOI] [PubMed] [Google Scholar]
- 82.Sundaresan M, Yu ZX, Ferrans VJ, Irani K, Finkel T. Requirement for generation of H2O2 for platelet-derived growth factor signal transduction. Science. 1995;270:296–299. doi: 10.1126/science.270.5234.296. [DOI] [PubMed] [Google Scholar]
- 83.Bae YS, et al. Epidermal growth factor (EGF)-induced generation of hydrogen peroxide. Role in EGF receptor-mediated tyrosine phosphorylation. J. Biol. Chem. 1997;272:217–221. [PubMed] [Google Scholar]
- 84.Junn E, et al. Requirement of hydrogen peroxide generation in TGF-beta 1 signal transduction in human lung fibroblast cells: involvement of hydrogen peroxide and Ca2+ in TGF-beta 1-induced IL-6 expression. J. Immunol. 2000;165:2190–2197. doi: 10.4049/jimmunol.165.4.2190. [DOI] [PubMed] [Google Scholar]
- 85.Suzukawa K, et al. Nerve growth factor-induced neuronal differentiation requires generation of Rac1-regulated reactive oxygen species. J. Biol. Chem. 2000;275:13175–13178. doi: 10.1074/jbc.275.18.13175. [DOI] [PubMed] [Google Scholar]
- 86.Griendling KK, Ushio-Fukai M. Reactive oxygen species as mediators of angiotensin II signaling. Regul. Pept. 2000;91:21–27. doi: 10.1016/s0167-0115(00)00136-1. [DOI] [PubMed] [Google Scholar]
- 87.Sattler M, et al. Hematopoietic growth factors signal through the formation of reactive oxygen species. Blood. 1999;93:2928–2935. [PubMed] [Google Scholar]
- 88.Devadas S, Zaritskaya L, Rhee SG, Oberley L, Williams MS. Discrete generation of superoxide and hydrogen peroxide by T cell receptor stimulation: selective regulation of mitogen-activated protein kinase activation and fas ligand expression. J. Exp. Med. 2002;195:59–70. doi: 10.1084/jem.20010659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fearon DT. Seeking wisdom in innate immunity. Nature. 1997;388:323–324. doi: 10.1038/40967. [DOI] [PubMed] [Google Scholar]
- 90.Medzhitov R, Janeway CA., Jr Innate immune induction of the adaptive immune response. Cold Spring Harb. Symp. Quant. Biol. 1999;64:429–435. doi: 10.1101/sqb.1999.64.429. [DOI] [PubMed] [Google Scholar]
- 91.Nathan C. Points of control in inflammation. Nature. 2002;420:846–852. doi: 10.1038/nature01320. [DOI] [PubMed] [Google Scholar]
- 92.O’Brien J, Friedlander A, Dreier T, Ezzell J, Leppla S. Effects of anthrax toxin components on human neutrophils. Infect. Immun. 1985;47:306–310. doi: 10.1128/iai.47.1.306-310.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hanna PC, Kruskal BA, Ezekowitz RA, Bloom BR, Collier RJ. Role of macrophage oxidative burst in the action of anthrax lethal toxin. Mol. Med. 1994;1:7–18. [PMC free article] [PubMed] [Google Scholar]
- 94.Flak TA, Goldman WE. Signalling and cellular specificity of airway nitric oxide production in pertussis. Cell Microbiol. 1999;1:51–60. doi: 10.1046/j.1462-5822.1999.00004.x. [DOI] [PubMed] [Google Scholar]
- 95.Stebbins CE, Galan JE. Structural mimicry in bacterial virulence. Nature. 2001;412:701–705. doi: 10.1038/35089000. [DOI] [PubMed] [Google Scholar]
- 96.Madden JC, Ruiz N, Caparon M. Cytolysin-mediated translocation (CMT): a functional equivalent of type III secretion in gram-positive bacteria. Cell. 2001;104:143–152. doi: 10.1016/s0092-8674(01)00198-2. [DOI] [PubMed] [Google Scholar]