Abstract
Nucleotide excision repair (NER) is one of several DNA repair pathways that are universal throughout phylogeny. NER has a broad substrate specificity and is capable of removing several classes of lesions to the DNA, including those that accumulate upon exposure to UV radiation. The loss of this activity in NER-defective mutants gives rise to characteristic sensitivities to UV that, in humans, is manifested as a greatly elevated sensitivity to exposure to the sun. Xeroderma pigmentosum (XP), Cockaynes syndrome (CS), and trichothiodystrophy (TTD) are three, rare, recessively inherited human diseases that are linked to these defects. Interestingly, some of the symptoms in afflicted individuals appear to be due to defects in transcription, the result of the dual functionality of several components of the NER apparatus as parts of transcription factor IIH (TFIIH). Studies with several model systems have revealed that the genetic and biochemical features of NER are extraordinarily conserved in eukaryotes. One system that has been studied very closely is the budding yeast Saccharomyces cerevisiae. While many yeast NER mutants display the expected increases in UV sensitivity and defective transcription, other interesting phenotypes have also been observed. Elevated mutation and recombination rates, as well as increased frequencies of genome rearrangement by retrotransposon movement and recombination between short genomic sequences have been documented. The potential relevance of these novel phenotypes to disease in humans is discussed.
DNA REPAIR AND DISEASE
The maintenance of the integrity of DNA is of a paramount biological importance as it serves as the central repository for all genetic information. A network of biochemical pathways exists in all cells to maintain the informational and structural fidelity of DNA. These pathways are characterized, in part, by the types of modifications to the DNA to which they respond. The dissection of these pathways has involved a coordinated biochemical, genetic, and molecular biological approach where specific enzymatic defects are correlated with mutations at particular chromosomal loci, and distinctive cellular phenotypes. In humans, the specific DNA repair defects have been correlated with particular inherited diseases. Hereditary nonpolyposis colorectal cancer, for example, is a disorder associated with mutations in genes encoding components of the mismatch repair apparatus that lead to profound defects in cellular mismatch repair, and increases in genome instability [1]. Similarly, a trio of phenotypically disparate diseases, XP, CS, and TTD have been connected with mutations in genes that encode subunits of the NER and TFIIH apparatus, that confer measurable defects in NER and transcription [2, 3, 4]. The broad symptomology of these diseases most likely reflects the multiple biochemical defects caused by the mutations, some of which have not been extensively explored. This work focus on some of the less described effects of mutations in the NER/TFIIH apparatus on genome stability, and their potential implications with respect to human disease.
NUCLEOTIDE EXCISION REPAIR
NER plays a critical role in the maintenance of genomic integrity because of its broad substrate specifity. It plays an important role in the removal of such diverse lesions as UV light induced photoproducts as well as chemically induced bulky adducts, crosslinks, and oxidized bases. NER has been functionally conserved throughout phylogeny, although the apparatus in prokaryotes and eukaryotes is substantially different. NER in eukaryotes involves the concerted action of up to 30 proteins and has been reconstituted in vitro [5, 6]. NER occurs in the context of two distinct pathways related to the way the lesions are identified. General genome repair (GGR) responds to genome-wide damage, which in mammals is recognized by the XPC/HHR23B complex [7]. Transcription coupled repair (TCR) is focused exclusively on the repair of lesions on the transcribed strand of actively transcribed genes [8], and is thought to involve recognition of RNA polymerase II stalled at a lesion by the CSA and CSB proteins [9, 10]. The XPA protein then orchestrates the assembly of a complex including the single-stranded DNA binding protein RPA and the core of the TFIIH complex at the site of the lesion [11]. This complex unwinds the DNA surrounding the lesion in an ATP-dependent process that involves the helicase activities of the XPB and XPD proteins, components of the TFIIH core complex [12, 13]. Interestingly, only the helicase activity of XPB is required for TFIIH-mediated melting of promoter DNA during the initiation of transcription, while XPD helicase activity plays a minor role in RNA polymerase II promoter escape [14]. Two structure-specific endonucleases, the ERCC1/XPF heterodimer, that cleaves on the 3′ side of the lesion, and XPG, that cleaves on the 5′ side of the lesion, are responsible for the removal of a 24–32 base oligonucleotide containing the lesion [15, 16]. The resulting gap is filled by DNA polymerases δ and ɛ, and repair is completed by ligation [5].
NER PHOTOSENSITIVITY SYNDROMES
XP, CS, and TTD have been ascribed to changes in NER and TFIIH function. Complementation analysis, conducted by analyzing the phenotypes of cells derived from the fusion of cells from different patients, identified several complementation groups for each disease [11]. Seven complementation groups, designated XP-A through XP-G, have been identified for XP. Two complementation groups, CS-A and CS-B, have been identified for CS. Three complementation groups, XP-B, XP-D, and XP-G, have been determined for the combined CS/XP patients. Three complementation groups, XP-B, XP-D, and TTD-A, have been identified for TTD. Most of the genes corresponding to these complementation groups have been cloned and the mutations responsible for disease in individual kindreds have been identified.
XP, CS, and TTD patients have distinct symptoms that likely reflect the participation of the mutant proteins in multiple biochemical processes. In patients, symptoms can often be correlated with specific biochemical defects. An example for each disease is listed below. XP patients exhibit distinct alterations of the texture and pigmentation of the skin that are related to exposure to the sun. They are also enormously predisposed to both sunlight-induced and internal cancers, consistent with defects in NER [17]. For example, an 80% reduction in cellular NER activity in an XP patient homozygous for a particular XPF mutation correlates well with the acute reactions to sun exposure and skin cancer that were also observed [18]. CS patients present symptoms of poor growth and developmental deformity that are most likely due to defective transcription. Accordingly, extracts of cells from patients with particular CSA and CSB mutations have been shown to support reduced levels of RNA polymerase II mediated transcription, as have cells from patients with combined XP/CS disease that are due to a mutation in XPB [19]. TTD patients suffer from brittle hair and nails, scaly skin and developmental abnormalities that are probably related to defects in NER and transcription. In support of this, fever inducible hair loss was correlated with temperature sensitive transcription and NER in the cells of patients with specific XPD mutations [20].
STUDIES WITH MODEL SYSTEMS
The extraordinary level of conservation of the NER and TFIIH apparatus among eukaryotes permits the extensive use of model systems to better explore the genetic and biochemical control of theses processes. Studies in rodents, flies, plants, and fungi have all contributed to the understanding of the role played by the NER and TFIIH apparatus in eukaryotes [21, 22, 23, 24, 25, 26]. However, trangenic mice and budding yeast have been particularly helpful in studying NER and TFIIH at the molecular level, as well as their effects on the phenotype of the organism. For example, in a recent study, isogenic strains of mice homozygous for null alleles of XPA or CSB were found to be extremely sensitive to a carcinogen that forms bulky adducts on DNA, while an XPC null strain was not [27]. This suggests that XPA-dependent and CSB-dependent TCR is critical for survival to exposure to an important class of carcinogens, while XPC-dependent GGR is not. Such studies are very important in determining how mammals respond to particular types of DNA damage. Mice have also been used to further explore the phenotypic effects of NER mutations known to cause disease in humans. For example, a strain of mouse was created that was homozygous for an XPD mutation analogous to the one found in several NER-deficient TTD patients [28]. The mice exhibited hypersensitivity to UV exposure and a cellular NER defect that was very similar to the TTD symptomology but, also exhibited a marked propensity toward UV-induced and carcinogen-induced skin cancer that was not observed in patients. This suggests the possibility that the NER defects observed in TTD patients could lead to cancer, an observation of potential clinical importance.
The study of NER and TFIIH in budding yeast and mammalian cells has been essentially contemporaneous. The observation that NER and transcription are genetically and biochemically linked occurred nearly simultaneously in the two systems, significantly accelerating the description of eukaryotic NER and transcription initiation at the molecular level [29, 30]. The degree of similarity between the apparatus in humans and yeast is extraordinary, permitting investigators to examine the impact of disease causing alleles of human NER genes on yeast. In one study, two TTD alleles of XPD were unable to complement the lethal effect of a null allele of the RAD3 gene, the budding yeast homolog of XPD, while wild-type and helicase-defective alleles of XPD were able to complement [31]. Since the essential function of Rad3/XPD is thought to be its role in transcription initiation, the authors concluded that the TTD alleles confer a transcription defect, and, therefore, that the disease may be due to defective transcription of a critical gene, or genes.
NER, TFIIH, AND THE MAINTENANCE OF GENOME STABILITY IN BUDDING YEAST
The careful study of NER in budding yeast has revealed its impact on cellular processes yet to be recognized in other organisms. These unexpected relationships could provide insight into clinically important roles for the human NER apparatus that have yet to be carefully explored, especially when the disease phenotype is complex as in CS and TTD. An important example is provided by the RAD1 and RAD10 genes, homologs of XPF and ERCC1. Null mutations in RAD1 and RAD10 confer profound defects in several different types of homologous recombination [32, 33]. Studies analyzing the recombinational repair of defined double-strand breaks (DSBs) revealed that RAD1 and RAD10 were required to remove nonhomologous sequences from the ends of recombining molecules, an important step in the creation of certain genome rearrangments [34]. RAD1 and RAD10 also play a critical role in recombination between short sequences, which is likely to be vital for the maintenance of genome stability [35]. All of these effects are apt to be related to the role of the Rad1-Rad10 nuclease in the cleavage of important recombination intermediates [34, 35, 36].
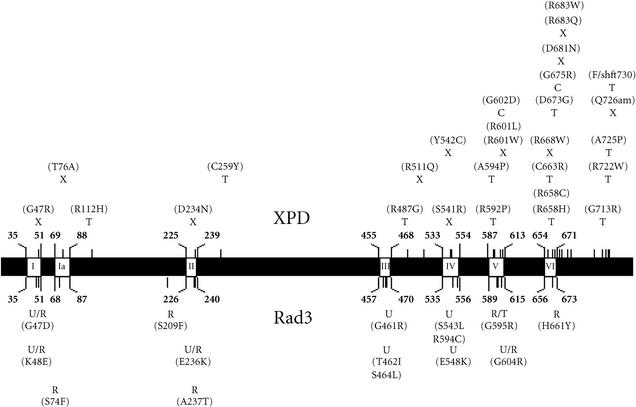
The TFIIH machinery has also been implicated in the maintenance of genome stability. Mutant alleles of RAD3, SSL1, and SSL2 (homologs of XPD, P44, and XPB) have been isolated that confer elevated rates of mutation and recombination. Several rad3 mutants have been isolated that exhibit increased rates of mutation and/or recombination but not defective transcription nor NER, indicating that Rad3 possesses important cellular activities that are independent of transcription and NER [37, 38]. Interestingly, a pair of these mutant alleles, rad3-101 (S74F) and rad3-102 (H661Y), accumulate DSBs and are synthetically lethal in combination with mutations in recombinational repair genes, strongly suggesting that Rad3 influences either the creation of DSBs, or their processing by homologous recombination [38]. Another allele of RAD3, rad3-G595R, blocks the degradation of DSBs, which stimulates genome rearrangement by recombination between short repeated sequences [35, 36, 37, 38, 39, 40, 41]. The analogous mutations in XPD are of potential medical importance as they would be within regions that encode conserved helicase domains of the protein that are mutated in XP, TTD, and XP/CS patients (Figure 1), [42, 43, 44]. Since specific mutations in SSL1 and SSL2 that encode additional members of the heteromeric core of budding yeast TFIIH [29] also stimulate short-sequence recombination (SSR) by blocking the degradation of DSBs [40, 41], it appears that TFIIH acts to maintain genome stability by restricting recombination between repetitive DNA sequences. We speculate that SSR could potentially contribute to the symptoms observed in XP, XP-CS, and TTD patients, as SSR in the human genome has been found to lead to a variety of diseases [45].
Figure 1.
Amino acid changes in the human XPD and budding yeast Rad3 helicases that lead to phenotypic changes. The primary amino acid sequences of human XPD and yeast Rad3 are represented by the black bar. The seven conserved helicase domains are represented by white boxes and are named according to the accepted nomenclature [42]. The positions of these domains in the primary sequence are listed in bold print as described in [46]. Positions of altered residues in the primary sequence of XPD in XP, XP/CS, and TTD patients [44] are marked with a light hash mark above the black bar. The corresponding diseases are marked with an X for XP, a C for XP/CS, and a T for TTD. The amino acid changes are listed in parentheses. Changes in the Rad3 primary sequence [38, 39] are represented by hash marks below the black bar. The corresponding phenotypic changes are denoted by a U for UV sensitive, an R for elevated recombination, and/or mutation, and a T for transcription defective. Corresponding amino acid changes are listed in parentheses.
Recently, it was revealed that by restricting the movement of the retrotransposon Ty1, TFIIH plays an additional, critical role in the maintenance of genome stability in budding yeast. Ty1 can move either by integrase mediated transposition, or by insertion of Ty cDNA into existing Ty or δ elements (the Ty1 long-terminal repeat) by homologous recombination [47]. Components of TFIIH were discovered to play a role in restricting Ty mobility when specific rad3 and ssl2 mutants with increased levels of Ty1 movement were isolated [48]. Other rad3 and ssl2 mutants that confer extreme UV sensitivity do not stimulate Ty mobility, separating the NER and Ty maintenance functions of these genes. The increases in Ty mobility were correlated with a substantial elevation of the steady-state level of Ty1 cDNA, suggesting either that the rad3-RTT (regulator of Ty transposition) and ssl2-RTT mutations increase the synthesis of Ty1 cDNA, or decrease its degradation. Importantly, the level of Ty1 RNA and Ty1 proteins remained unchanged in the rad3 and ssl2 mutants. Another interesting observation is that other NER genes, such as RAD2 and RAD1 do not appear to be involved in Ty transposition [49].
In a subsequent study, the increased level of Ty1 cDNA was found not to be due to increased cDNA synthesis suggesting that TFIIH plays a role in the degradation of Ty1 cDNA [41]. This same study linked TFIIH-control of SSR and Ty1 movement by showing that rad3-G595R and ssl2-RTT increase both SSR and Ty1 movement while increasing the stability of both Ty1 cDNA and DSBs. It remains unclear whether TFIIH plays a direct or an indirect role in the degradation of the ends of DNA molecules, however, the helicase activities of Rad3 and Ssl2 suggest that TFIIH could play a role in opening the ends of DNA molecules, thereby facilitating their processing by exonucleases.
Blocking the degradation of DSBs and Ty1 cDNAs could enhance Ty1 movement and SSR in multiple ways. It could preserve sequences at the ends of the molecules that are important for SSR and Ty1 movement. For example, a DSB in a short repetitive sequence creates ends that are never far from the border of homology with potential donor repeats, such that any erosion of the ends would significantly decrease their ability to pair and recombine. Similarly, loss of the sequences at the ends of Ty1 cDNA would block transposition by Ty1 integrase. Another way (that increasing the stability of DNA ends might elevate SSR and Ty mobility) could be by prolonging the signal that elicits the DNA damage checkpoint. Pausing the cell cycle for a protracted period may improve the likelihood that the ends of DSBs in short repeats can find homologous sequences, or that the Ty1 preintegration complex can complete a transposition event. Changes in checkpoint activity have been shown to affect the frequencies of other genome rearrangements [50]. These and other mechanisms could be working simultaneously to increase genome rearrangement in rad3 and ssl2 mutant budding yeast. By extension, similar defects in the cells of XP, XP-CS, or TTD patients with mutations in the XPD or XPB genes might contribute to these diseases.
Acknowledgments
ACKNOWLEDGMENTS
This research was sponsored in part by the National Cancer Institute, Department of Health and Human Services. A. M. Bailis is funded by Public Health Service Grant GM57484.
References
- Parsons R, Li G M, Longley M J, et al. Hypermutability and mismatch repair deficiency in RER+ tumor cells. Cell. 1993;75(6):1227–1236. doi: 10.1016/0092-8674(93)90331-j. [DOI] [PubMed] [Google Scholar]
- Eveno E, Bourre F, Quillet X. Differential removal of ultraviolet photoproducts in genetically related xeroderma pigmentosum and trichothiodystrophy diseases. Cancer Res. 1995;55(19):4325–4332. [PubMed] [Google Scholar]
- Cooper P K, Nouspikel T, Clarkson S G, Leadon S A. Defective transcription-coupled repair of oxidative base damage in Cockayne syndrome patients from XP group G. Science. 1997;275(5302):990–993. doi: 10.1126/science.275.5302.990. [DOI] [PubMed] [Google Scholar]
- Coin F, Bergmann E, Tremeau-Bravard A, Egly J M. Mutations in XPB and XPD helicases found in xeroderma pigmentosum patients impair the transcription function of TFIIH. EMBO J. 1999;18(5):1357–1366. doi: 10.1093/emboj/18.5.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aboussekhra A, Biggerstaff M, Shivji M K, et al. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell. 1995;80(6):859–868. doi: 10.1016/0092-8674(95)90289-9. [DOI] [PubMed] [Google Scholar]
- Guzder S N, Habraken Y, Sung P, Prakash L, Prakash S. Reconstitution of yeast nucleotide excision repair with purified Rad proteins, replication protein A, and transcription factor TFIIH. J Biol Chem. 1995;270(22):12973–12976. doi: 10.1074/jbc.270.22.12973. [DOI] [PubMed] [Google Scholar]
- Sugasawa K, Ng J M, Masutani C, et al. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol Cell. 1998;2(2):223–232. doi: 10.1016/s1097-2765(00)80132-x. [DOI] [PubMed] [Google Scholar]
- Mellon I, Spivak G, Hanawalt P C. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51(2):241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- Henning K A, Li L, Iyer N, et al. The Cockayne syndrome group A gene encodes a WD repeat protein that interacts with CSB and a subunit of RNA polymerase II TFIIH. Cell. 1995;82(4):555–564. doi: 10.1016/0092-8674(95)90028-4. [DOI] [PubMed] [Google Scholar]
- van Gool A J, Citterio E, Rademakers S, et al. The Cockayne syndrome B protein, involved in transcription-coupled DNA repair, resides in an RNA polymerase II-containing complex. EMBO J. 1997;16(19):5955–5965. doi: 10.1093/emboj/16.19.5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer J, Hoeijmakers J H. Nucleotide excision repair and human syndromes. Carcinogenesis. 2000;21(3):453–460. doi: 10.1093/carcin/21.3.453. [DOI] [PubMed] [Google Scholar]
- Winkler G S, Araujo S J, Fiedler U, et al. TFIIH with inactive XPD helicase functions in transcription initiation but is defective in DNA repair. J Biol Chem. 2000;275(6):4258–4266. doi: 10.1074/jbc.275.6.4258. [DOI] [PubMed] [Google Scholar]
- van Vuuren A J, Vermeulen W, Ma L, et al. Correction of xeroderma pigmentosum repair defect by basal transcription factor BTF2 (TFIIH) EMBO J. 1994;13(7):1645–1653. doi: 10.1002/j.1460-2075.1994.tb06428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradsher J, Coin F, Egly J M. Distinct roles for the helicases of TFIIH in transcript initiation and promoter escape. J Biol Chem. 2000;275(4):2532–2538. doi: 10.1074/jbc.275.4.2532. [DOI] [PubMed] [Google Scholar]
- Sijbers A M, de Laat W L, Ariza R R, et al. Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell. 1996;86(5):811–822. doi: 10.1016/s0092-8674(00)80155-5. [DOI] [PubMed] [Google Scholar]
- O'Donovan A, Davies A A, Moggs J G, West S C, Wood R D. XPG endonuclease makes the 3′ incision in human DNA nucleotide excision repair. Nature. 1994;371(6496):432–435. doi: 10.1038/371432a0. [DOI] [PubMed] [Google Scholar]
- Kraemer K H, Lee M M, Scotto J. DNA repair protects against cutaneous and internal neoplasia: evidence from xeroderma pigmentosum. Carcinogenesis. 1984;5(4):511–514. doi: 10.1093/carcin/5.4.511. [DOI] [PubMed] [Google Scholar]
- Sijbers A M, van Voorst Vader P C, Snoek J W, Raams A, Jaspers N G, Kleijer W J. Homozygous R788W point mutation in the XPF gene of a patient with xeroderma pigmentosum and late-onset neurologic disease. J Invest Dermatol. 1998;110(5):832–836. doi: 10.1046/j.1523-1747.1998.00171.x. [DOI] [PubMed] [Google Scholar]
- Dianov G L, Houle J F, Iyer N, Bohr V A, Friedberg E C. Reduced RNA polymerase II transcription in extracts of Cockayne syndrome and xeroderma pigmentosum/Cockayne syndrome cells. Nucleic Acids Res. 1997;25(18):3636–3642. doi: 10.1093/nar/25.18.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen W, Rademakers S, Jaspers N G, et al. A temperature-sensitive disorder in basal transcription and DNA repair in humans. Nat Genet. 2001;27(3):299–303. doi: 10.1038/85864. [DOI] [PubMed] [Google Scholar]
- de Boer J H, Hoeijmakers J. Cancer from the outside, aging from the inside: mouse models to study the consequences of defective nucleotide excision repair. Biochimie. 1999;81(1-2):127–137. doi: 10.1016/s0300-9084(99)80045-5. [DOI] [PubMed] [Google Scholar]
- Sekelsky J J, Hollis K J, Eimerl A I, Burtis K C, Hawley R S. Nucleotide excision repair endonuclease genes in Drosophila melanogaster . Mutat Res. 2000;459(3):219–228. doi: 10.1016/s0921-8777(99)00075-0. [DOI] [PubMed] [Google Scholar]
- Gallego F, Fleck O, Li A, Wyrzykowska J, Tinland B. AtRAD1, a plant homologue of human and yeast nucleotide excision repair endonucleases, is involved in dark repair of UV damages and recombination. Plant J. 2000;21(6):507–518. doi: 10.1046/j.1365-313x.2000.00694.x. [DOI] [PubMed] [Google Scholar]
- Lee S K, Yu S L, Alexander H, Alexander S. A mutation in repB, the Dictyostelium homolog of the human xeroderma pigmentosum B gene, has increased sensitivity to UV-light but normal morphogenesis. Biochim Biophys Acta. 1998;1399(2-3):161–172. doi: 10.1016/s0167-4781(98)00103-1. [DOI] [PubMed] [Google Scholar]
- McCready S J, Osman1 F, Yasui A. Repair of UV damage in the fission yeast Schizosaccharomyces pombe . Mutat Res. 2000;451(1-2):197–210. doi: 10.1016/s0027-5107(00)00050-6. [DOI] [PubMed] [Google Scholar]
- Prakash S, Prakash L. Nucleotide excision repair in yeast. Mutat Res. 2000;451(1-2):13–24. doi: 10.1016/s0027-5107(00)00037-3. [DOI] [PubMed] [Google Scholar]
- Wijnhoven S W, Kool H J, Mullenders L H, Slater R, van Zeeland A A, Vrieling H. DMBA-induced toxic and mutagenic responses vary dramatically between NER-deficient Xpa, Xpc and Csb mice. Carcinogenesis. 2001;22(7):1099–1106. doi: 10.1093/carcin/22.7.1099. [DOI] [PubMed] [Google Scholar]
- de Boer J, van Steeg H, Berg R J, et al. Mouse model for the DNA repair/basal transcription disorder trichothiodystrophy reveals cancer predisposition. Cancer Res. 1999;59(14):3489–3494. [PubMed] [Google Scholar]
- Feaver W J, Svejstrup J Q, Bardwell L. Dual roles of a multiprotein complex from S. cerevisiae in transcription and DNA repair. Cell. 1993;75(7):1379–1387. doi: 10.1016/0092-8674(93)90624-y. [DOI] [PubMed] [Google Scholar]
- Drapkin R, Reardon J T, Ansari A, et al. Dual role of TFIIH in DNA excision repair and in transcription by RNA polymerase II. Nature. 1994;368(6473):769–772. doi: 10.1038/368769a0. [DOI] [PubMed] [Google Scholar]
- Guzder S N, Sung P, Prakash S, Prakash L. Lethality in yeast of trichothiodystrophy (TTD) mutations in the human xeroderma pigmentosum group D gene. Implications for transcriptional defect in TTD. J Biol Chem. 1995;270(30):17660–17663. doi: 10.1074/jbc.270.30.17660. [DOI] [PubMed] [Google Scholar]
- Schiestl R H, Prakash S. RAD1, an excision repair gene of Saccharomyces cerevisiae, is also involved in recombination. Mol Cell Biol. 1988;8(9):3619–3626. doi: 10.1128/mcb.8.9.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl R H, Prakash S. RAD10, an excision repair gene of Saccharomyces cerevisiae, is involved in the RAD1 pathway of mitotic recombination. Mol Cell Biol. 1990;10(6):2485–2491. doi: 10.1128/mcb.10.6.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman-Lobell J, Haber J E. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1 . Science. 1992;258(5081):480–484. doi: 10.1126/science.1411547. [DOI] [PubMed] [Google Scholar]
- Bailis A M, Maines S. Nucleotide excision repair gene function in short-sequence recombination. J Bacteriol. 1996;178(7):2136–2140. doi: 10.1128/jb.178.7.2136-2140.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell A J, Bardwell L, Tomkinson A E, Friedberg E C. Specific cleavage of model recombination and repair intermediates by the yeast Rad1-Rad10 DNA endonuclease. Science. 1994;265(5181):2082–2085. doi: 10.1126/science.8091230. [DOI] [PubMed] [Google Scholar]
- Montelone B A, Hoekstra M F, Malone R E. Spontaneous mitotic recombination in yeast: the hyper-recombinational rem1 mutations are alleles of the RAD3 gene. Genetics. 1988;119(2):289–301. doi: 10.1093/genetics/119.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J M, Montelone B A, Siede W, Friedberg E C. Effects of multiple yeast rad3 mutant alleles on UV sensitivity, mutability, and mitotic recombination. J Bacteriol. 1990;172(12):6620–6630. doi: 10.1128/jb.172.12.6620-6630.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailis A M, Maines S, Negritto M T. The essential helicase gene RAD3 suppresses short sequence recombination in Saccharomyces cerevisiae . Mol Cell Biol. 1995;15(8):3998–4008. doi: 10.1128/mcb.15.8.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maines S, Negritto M C, Wu X, Manthey G M, Bailis A M. Novel mutations in the RAD3 and SSL1 genes perturb genome stability by stimulating recombination between short repeats in Saccharomyces cerevisiae . Genetics. 1998;150(3):963–976. doi: 10.1093/genetics/150.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B S, Bi L, Garfinkel D J, Bailis A M. Nucleotide excision repair/TFIIH helicases RAD3 and SSL2 inhibit short-sequence recombination and Ty1 retrotransposition by similar mechanisms. Mol Cell Biol. 2000;20(7):2436–2445. doi: 10.1128/mcb.20.7.2436-2445.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya A E, Koonin E V, Donchenko A P, Blinov V M. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989;17(12):4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montelone B A, Malone R E. Analysis of the rad3-101 and rad3-102 mutations of Saccharomyces cerevisiae: implications for structure/function of Rad3 protein. Yeast. 1994;10(1):13–27. doi: 10.1002/yea.320100103. [DOI] [PubMed] [Google Scholar]
- Lehmann A R. The xeroderma pigmentosum group D (XPD) gene: one gene, two functions, three diseases. Genes Dev. 2001;15(1):15–23. doi: 10.1101/gad.859501. [DOI] [PubMed] [Google Scholar]
- Deininger P L, Batzer M A. Alu repeats and human disease. Mol Genet Metab. 1999;67(3):183–193. doi: 10.1006/mgme.1999.2864. [DOI] [PubMed] [Google Scholar]
- Weber C A, Salazar E P, Stewart S A, Thompson L H. ERCC2: cDNA cloning and molecular characterization of a human nucleotide excision repair gene with high homology to yeast RAD3 . EMBO J. 1990;9(5):1437–1447. doi: 10.1002/j.1460-2075.1990.tb08260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon G, Burkett T J, Garfinkel D J. Efficient homologous recombination of Ty1 element cDNA when integration is blocked. Mol Cell Biol. 1994;14(10):6540–6551. doi: 10.1128/mcb.14.10.6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B S, Lichtenstein C P, Faiola B, et al. Posttranslational inhibition of Ty1 retrotransposition by nucleotide excision repair/transcription factor TFIIH subunits Ssl2p and Rad3p. Genetics. 1998;148(4):1743–1761. doi: 10.1093/genetics/148.4.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattray A J, Shafer B K, Garfinkel D J. The Saccharomyces cerevisiae DNA recombination and repair functions of the RAD52 epistasis group inhibit Ty1 transposition. Genetics. 2000;154(2):543–556. doi: 10.1093/genetics/154.2.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasullo M, Bennett T, AhChing P, Koudelik J. The Saccharomyces cerevisiae RAD9 checkpoint reduces the DNA damage-associated stimulation of directed translocations. Mol Cell Biol. 1998;18(3):1190–1200. doi: 10.1128/mcb.18.3.1190. [DOI] [PMC free article] [PubMed] [Google Scholar]