Abstract
DNA damage incurred during the process of chromosomal replication has a particularly high possibility of resulting in mutagenesis or lethality for the cell. The SOS response of Escherichia coli appears to be well adapted for this particular situation and involves the coordinated up-regulation of genes whose products center upon the tasks of maintaining the integrity of the replication fork when it encounters DNA damage, delaying the replication process (a DNA damage checkpoint), repairing the DNA lesions or allowing replication to occur over these DNA lesions, and then restoring processive replication before the SOS response itself is turned off. Recent advances in the fields of genomics and biochemistry has given a much more comprehensive picture of the timing and coordination of events which allow cells to deal with potentially lethal or mutagenic DNA lesions at the time of chromosomal replication.
MECHANISM OF INDUCTION
Irradiation of Escherichia coli with near ultraviolet light (254 nm UV) produces DNA lesions that block DNA replication and pose a dangerous threat to the integrity of the genetic information. DNA damage that blocks replication can result in genomic rearrangements when it resumes from the wrong place, mutagenesis when the incorrect base is incorporated opposite to the lesion, or even cell death when the block to replication cannot be overcome. A large body of work has demonstrated that E coli responds to this challenge by upregulating the expression of several genes which function to repair the DNA lesions, maintain the integrity of the DNA replication fork, and prevent premature cell division. This cellular response to DNA damage produced by UV and other agents has been collectively termed the SOS response, after the international distress signal (see [1]; reviewed in [2, 3]).
The sensor for SOS induction in E coli is a two component repressor/activator system of LexA and RecA that “senses” when the replication fork is impeded from progressing normally. The observation that nonreplicating E coli do not induce a strong SOS response following UV irradiation supports the notion that replication is necessary for signaling the response [4]. Many of the genes induced following DNA damage are regulated by the LexA repressor protein which binds to a 20 base pair consensus sequence in the operator region of the genes, suppressing their expression [5, 6]. Derepression of these genes occurs when RecA binds to single stranded regions of DNA created at replication forks disrupted by DNA damage. RecA bound to single strand DNA becomes conformationally active, serving as a coprotease that promotes the autocatalytic cleavage of the LexA repressor. As the cellular concentration of LexA diminishes, the genes normally suppressed by LexA are more frequently transcribed (see [2, 4] and the references therein).
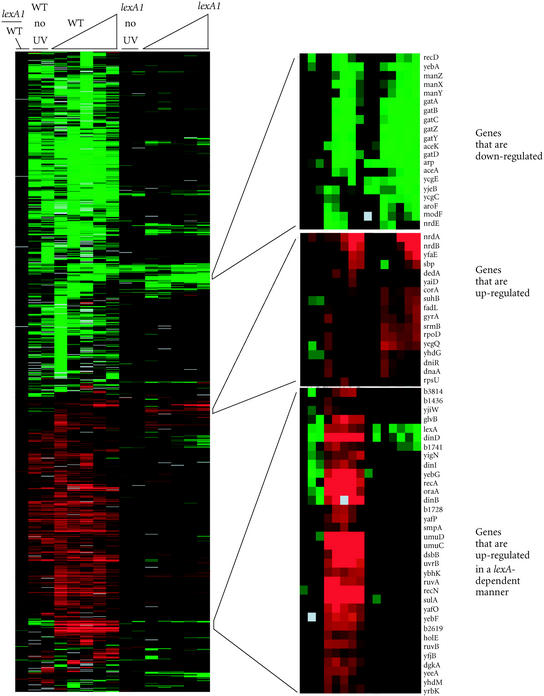
Kenyon and Walker carried out the first systematic search to identify genes that are up-regulated in a recA/lexA-dependent fashion [7]. Through random insertion of a lac reporter gene into the E coli chromosome, they were able to identify promoters which were up-regulated following DNA damage. Subsequent analysis of the up-regulated genes revealed a 20 basepair consensus LexA binding motif, or “SOS box” shared by these genes in their promoter/operator regions [8]. Since these initial studies, several groups, using a variety of strategies, have identified many more LexA-dependent damage inducible genes, as well as some genes that are induced in a LexA-independent manner. [9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21]. In one recent approach, DNA microarrays containing amplified DNA fragments from 95.5% of all open reading frames identified in the E coli genome were used to examine the changes in gene expression during the first hour following UV exposure in both wild-type cells and lexA1 mutants, which are unable to induce genes under LexA control [21]. These DNA microarrays contain PCR amplified DNA fragments of known and predicted genetic sequences printed on the surface of a glass slide. Through the comparative hybridization of two cellular RNA preparations, the relative difference between transcript levels of any gene in these preparations can be determined (Figure 1). These techniques, in total, identified 43 locations on the E coli chromosome that were up-regulated in a lexA-dependent manner after UV irradiation in actively replicating cells. In addition, several transcripts were either down-regulated or degraded following UV irradiation.
Figure 1.
Clustered display of gene expression profiles following UV exposure in wild-type and lexA1 (SOS deficient) Escherichia coli [21]. Increased transcript levels are shown in red, decreased transcript levels are shown in green. The timepoints for irradiated samples (left to right) were 5, 10, 20, 40, and 60 minutes following UV irradiation. The timepoints for unirradiated samples (left to right) were 20 and 60 minutes following UV irradiation. All measurements are relative to time 0. Genes were selected for this analysis if their expression level deviated from time 0 by at least a factor of 2 in at least 3 time points. The colored image was produced using cluster analysis and is publicly available with this data at http://genome-www.stanford.edu/UVirradiation. The color scale ranges from saturated green for log ratios 2.0 and below to saturated red for log ratios 2.0 and above.
The biological function of many of these UV-responsive genes has been studied extensively and, in some cases, their role in the recovery process is well characterized. In this review, we outline some of the recent advances in our understanding of the events that occur after the E coli genome is damaged, focusing particularly on the induction of cellular responses after UV exposure.
MAINTAINING THE REPLICATION FORK
One of the earliest recognized, and most heavily induced, DNA damage inducible gene products is RecA [22]. The act of binding to the single strand DNA regions at blocked replication forks serves not only to “sense” that replication is blocked, but also to maintain the structural integrity of the replication fork itself when progression is impeded. It is well established that recA mutants are extremely hypersensitive to DNA damage. This hypersensitivity correlates with a striking phenomenon called rec-less degradation in which the genomic DNA is rapidly degraded in the presence of DNA damage [22, 23]. The degradation is much more severe when replication is active in the cells at the time of damage and it has been shown to initiate at the replication forks and then degrade progressively back from these points. These observations led Horii and Susuki to propose that RecA plays a role in protecting DNA at replication forks when they are blocked by DNA damage [23].
recA was originally identified and characterized as a gene essential for recombination in E coli [24]. In vitro, purified RecA binds to single strand DNA progressively and then pairs that single stranded DNA with homologous duplex DNA. The product of this reaction creates a RecA protein filament bound to a three strand DNA structure (for reviews see [25, 26, 27]). During recombinational processes, this activity is thought to be critical for bringing together homologous strands from different DNA molecules. During DNA replication, this same biochemical activity of RecA may play a nonrecombinational role in maintaining the DNA replication fork [28, 29, 30]. Semiconservative replication copies both strands of the DNA template concurrently in a 5′ → 3′ direction. While the leading strand can be synthesized continuously, synthesis on the lagging strand template occurs discontinuously, periodically reinitiating as the replication machinery moves processively along the template. The coordination of this process implies that at any given time, the region immediately behind the replication machinery will contain a single stranded region. In the event that replication becomes blocked or contains single strand gaps, the region left behind the replication fork should be an ideal substrate for RecA to bind and protect.
Several aspects of RecA function at replication forks arrested by DNA damage remain to be characterized. For instance, the structure of the blocked replication fork is not well understood. Does it end with the leading strand facing a lesion? Can the lagging strand continue on without the leading strand? It is also not known whether RecA promotes recombination at blocked replication forks, whether it is primarily nonrecombinational at these sites, or whether it is some combination of these two scenarios that depends upon the lesion presented. Finally, during recombinational processes, the act of RecA pairing single stranded DNA with homologous duplex DNA can also be associated with strand exchange and branch migration. These processes have also been proposed to function at damage blocked replication forks, perhaps to allow nucleotide excision repair or translesion DNA polymerases to gain access to the blocking DNA lesion [28, 29, 30, 31]. In vitro experiments suggest that several branch migration proteins are capable of acting on these substrates [32, 33, 34, 35]. However, whether branch migration occurs and which proteins, if any, act on replication structures in vivo has yet to be examined.
INDUCING REPAIR TO CLEAR THE DAMAGE
The production of DNA damage in the genome must be countered with a repair response that clears the DNA prior to the resumption of DNA replication and cell division. In E coli, UV-induced lesions are subject to nucleotide excision repair (NER), catalyzed by the UvrABCD proteins (reviewed in [2]). Three of these genes, uvrA, uvrB, and uvrD, are induced after DNA damage as part of the SOS response and have thus been implicated in promoting damage inducible excision repair. Recent studies have shed new light on the SOS-regulated NER response and have shown that inducible repair of UV lesions is dependent on the up-regulation of UvrA and UvrB but not UvrD.
Crowley and Hanawalt showed that cells treated with the transcription inhibitor rifampicin were deficient in their ability to remove the major UV-induced lesion, the cyclobutane pyrimidine dimer (CPD), from their genome following exposure to 40 J/m2 UV [36]. A similar reduction in repair was observed in lexA3(Ind-) mutants that are unable to induce the SOS response after UV. However, lexA51(Def) mutants that constitutively express the SOS response were not deficient in NER even when treated with rifampicin after UV. Direct measurements of UvrA and UvrB levels in all three cell types correlated with the cell's ability to efficiently remove CPDs from the genome. In wild-type cells, 2-fold induction of both proteins occurred within the first 10 minutes after UV and maximal induction (as compared with the lexA(Def) strain) was reached within the first 40 minutes after UV, a time at which more than 80% of all CPDs are removed. Wild-type cells treated with rifampicin and the lexA3(Ind-) cells maintained low constitutive levels of UvrA and UvrB an hour after UV exposure, consistent with their slower rate of removal of CPDs. The pyrimidine(6-4)pyrimidone photoproduct (6-4 photoproduct), a less prevalent but more distorting UV-induced lesion, was repaired equally well in the presence or absence of SOS induction, indicating that inducible NER is necessary for efficient genomic repair of CPDs, but not 6-4 photoproducts in wild-type cells [36].
To determine the role of UvrD, also known as Helicase II, in promoting SOS-dependent inducible NER, Crowley and Hanawalt studied repair and survival in a uvrD deletion strain that carried a plasmid encoding a wild-type copy of uvrD under control of the T7 promoter. This plasmid construct yielded approximately wild-type levels of UvrD in an undamaged transformant and promoted almost complete complementation of the UV sensitivity and repair deficiencies observed in the nontransformed deletion mutant. Since the levels of uvrD transcript could not be increased from the plasmid promoter after UV, the authors concluded that constitutive levels of UvrD are sufficient for promoting SOS-dependent NER in vivo [37]. It is interesting, therefore, to speculate on a role for UvrD in promoting cellular recovery from UV that is distinct from its role in NER. The observation that uvrD mutations are incompatible with polA (DNA pol I) or rep (Rep helicase) mutations suggests that Helicase II may function in replication in vivo [38, 39]. Recent genetic and biochemical investigations have provided further support for such a role [40, 41, 42, 43]. Its slow rate of up-regulation and modest 2.5-fold induction after UV [21] supports a role for UvrD that occurs later in the damage recovery process, perhaps after the majority of repair has been completed but before replication resumes.
An exciting new finding regarding the inducibility of E coli NER has come from the work of Moolenaar et al who have discovered a second endonuclease, dubbed Cho (UvrC homolog), that catalyzes a UvrB-dependent 3'-incision at a variety of lesions in the presence or absence of UvrC (43a). Cho, the gene product of the ydjQ gene, was discovered on the basis of its homology with the N-terminal region of UvrC, which is required for its 3'-incision activity. The ydjQ gene is a member of the SOS regulon and its gene product is up-regulated with kinetics similar to that of UvrA and UvrB [21]. Given that UvrC is not induced by DNA damage, this discovery suggests that E coli may possess an SOS-dependent subpathway of NER involving the novel Cho endonuclease.
GETTING BY THE LESIONS
The SOS response also upregulates the expression of three DNA polymerases, Pol II (polB), Pol IV (dinB), and Pol V (umuC) in addition to the constitutively expressed Pol I and Pol III. While Pol I, involved in repair and Okazaki fragment removal, and Pol III, the replicative DNA polymerase, can polymerize and incorporate the standard nucleotides opposite to their complementary bases, these inducible polymerases have the ability to incorporate and pair nucleotides opposite to specific forms of damaged DNA bases (ie, they promote translesion DNA synthesis). In vitro studies on single strand templates have shown that Pol III, the replicational polymerase of E coli, is arrested at DNA lesions such as those produced by UV irradiation [44, 45]. Several studies identified three DNA damage-inducible polymerases in E coli (as well as multiple homologs in eukaryotic cells, including XP-V) capable of incorporating nucleotides opposite DNA lesions with relatively high efficiency when Pol III had extended the template up to the blocking DNA lesion [9, 16, 44, 46, 47, 48, 49, 50]. Presumably, this allows lesions that would otherwise be blocks to DNA replication, a lethal event, to be bypassed and for replication to recover, a mutagenic event at worst. In fact, the first E coli translesion DNA polymerase, Pol V, encoded by umuC, was originally identified by its ability to produce mutations following UV irradiation, long before it was known to be a DNA polymerase [9]. In the absence of umuC (or umuD, see section: DNA damage replication checkpoint), the mutation rate drops dramatically following UV irradiation. Perhaps as a trade off for not mutating, these mutants are slightly more sensitive to UV irradiation than wild-type cells [9].
Following up on sequence similarities between the bypass DNA polymerase, REV1, identified by Nelson and colleagues [51] in yeast, both umuC and dinB were shown to be polymerases as well [47, 48]. Pol II, the product of the polB gene, was first identified as a polymerase and later shown to be inducible following damage, but its function has remained elusive since polB mutants are not sensitive to UV, which is sometimes interpreted to indicate a role in recovery of replication from DNA damage [13, 52, 53, 54]. Furthermore, no obvious replication defects are observed in polB mutants [55]. More recently, work from Rangarajan and colleagues [56] has shown that both the recovery of replication and the mutation spectra following UV irradiation is altered in dinB mutants, strongly suggesting that Pol II functions during the recovery process in vivo.
When and how these polymerases act at a DNA lesion in vivo remain unclear. Recent work from Napolitano and colleagues suggests that sometimes a polymerase might be specific for a specific DNA lesion but other times multiple polymerases may compete for the same DNA lesion, depending upon the nature of the DNA lesion and its sequence context [57]. Using a mutation assay based upon a plasmid construct that contain either an N-2-acetylaminofluorene (AAF) adduct, a benzo-a-pyrene adduct (BaP), or a 6-4 photoproduct, they demonstrated that the mutation frequency and spectra were altered in the absence of a specific bypass polymerase for each lesion. For instance, by measuring AAF induced −2 frameshifts, they found that mutagenesis was significantly reduced in polB mutants, but not in dinB nor umuC mutants. Conversely, when they measured BaP induced −1 frameshifts, they found that mutagenesis was significantly reduced in dinB and umuC mutants but not in polB mutants. This implies that, in vivo, different polymerases must be acting on different DNA lesions [57]. If and how the cell knows which polymerase to use at different DNA lesions remains an interesting but unknown question.
DNA DAMAGE REPLICATION CHECKPOINT
One of the bypass polymerases, Pol V (umuC) has an accomplice, umuD, which comprise an operon that is up-regulated rapidly following SOS induction. Since the initial discovery of these genes in screens for mutants that were not mutable by UV [9], extensive genetic and biochemical work has shown that they are required for error-prone translesion synthesis in E coli [48]. Following SOS induction, UmuD is proteolytically cleaved to form UmuD' by a mechanism similar to the LexA cleavage reaction [58, 59, 60, 61]. The proteins form a UmuD'2C complex that is believed to be the active form promoting lesion bypass during replication recovery after UV damage. The uncleaved form of UmuD can also form an UmuD2C complex that has been postulated to aid in the cessation of DNA replication following the detection of UV-induced lesions in the genome [61, 62]. The UV dose-dependent cleavage of UmuD, the checkpoint protein, to UmuD', the bypass protein, occurs with kinetics that parallel the rate at which nucleotide excision repair proteins remove the bulk of UV-induced lesions following irradiation [36, 63]. This processing time is proposed to act as a checkpoint that contributes to cell survival by allowing ample time to repair the genome prior to recovery of replication, avoiding the production of more deleterious forms of DNA damage which would result from further attempts at replicating a damaged DNA template (reviewed in Smith, 1998 and Sutton, 2001).
The checkpoint role of UmuD comes from the initial observation that constitutive over-expression of the uncleaved form of UmuD, but not the cleaved form UmuD', results in a cold sensitive phenotype and a modest inhibition of DNA synthesis [64]. Opperman and colleagues have since shown that expression of noncleavable Umu(S60A)D and UmuC from a plasmid partially complemented the UV sensitivity of a ΔumuDC strain and delayed DNA synthesis and cell growth following UV. As predicted, a ΔumuDC strain resumed DNA synthesis and cell growth within the first 10 minutes after UV, whereas deletion mutants complemented with umuC+umuD+ on a plasmid delayed resumption of replication for more than 45 minutes following 25 J/m2 UV. Complementation of a ΔumuDC strain with a umuD(S60A)C+ construct delayed DNA synthesis and cell growth over 60 minutes. Neither umuD'C+ nor umuD+C125 complemented the rapid resumption of DNA synthesis, despite the ability of both strains to perform translesion synthesis. It is important to note that resumption of replication was not observed in the umuD(S60A)C+ construct, suggesting that UmuD'-dependent translesion synthesis is required for proper resumption of replication after UV [63, 65, 66].
To perform their roles in replication inhibition and translesion synthesis in vivo, the UmuD2C and UmuD'2C complexes have been shown to interact with DNA polymerase III directly, but each in distinctive ways. Using affinity chromatography, Sutton et al demonstrated that UmuD2 has a greater affinity for the beta (processivity) subunit whereas UmuD'2 interacts most strongly with the alpha (catalytic) subunit of Pol III. Both the UmuD and UmuD' homodimers bound to the epsilon (proofreading) subunit of Pol III [67]. Consistent with these interactions are data showing that only over-expression of the epsilon and beta subunits, and not any of the other eight Pol III subunits, had an effect on the cold sensitive phenotype of a umuD+C+ over-expressing strain at the restrictive temperature [68]. Together, these data suggest that UmuD2C associates with the DNA replication complex directly and that these interactions serve to slow or block DNA synthesis, perhaps through sequestration of these subunits from the Pol III holoenzyme. It is interesting to note the similarity between this potential prokaryotic DNA damage checkpoint and the p21-dependent DNA damage inducible cell cycle checkpoint in eukaryotes. In eukaryotes, p21 targets PCNA, the eukaryotic analog of the beta subunit, to promote replication arrest after DNA damage [69].
DNA DAMAGE CELL DIVISION CHECKPOINT
E coli cells exposed to UV or other SOS-inducing treatments continue to elongate but fail to septate and thus grow as filaments. This inhibition of septation is an SOS-dependent process that involves the lexA+-regulated gene, sulA (sfiA). This gene was originally isolated as a suppressor of lon protease mutants, which are particularly prone to damage-induced filamentation [70]. The lon gene product is an ATP-dependent protease that degrades SulA in vivo. This degradation is rapid and, combined with LexA repression, serves to maintain SulA at low concentrations in the cell. Upon damage to the genome, sulA transcription is up-regulated dramatically, achieving 10-fold induction in the first five minutes after 25 J/m2 UV [21]. Therefore, SulA accumulates to inhibitory levels only when the SOS response is induced. The sulA gene product inhibits septation by interacting with FtsZ, a key cell division protein that forms the septation ring early in cell division [71]. Evidence exists that a second gene, sfiC, also works to inhibit cell septation after DNA damage through interaction with FtsZ. This protein is DNA damage inducible but its regulation is not dependent on lexA+recA+ [72, 73, 74].
Recently, two papers have suggested that a third mode of SOS-dependent septation inhibition exists in E coli. This mode of inhibition, is independent of sulA and sfiC, yet requires umuDC [65, 75]. In their studies of cold sensitivity induced by UmuDC overproduction, Opperman and colleagues noted the production of cell filaments at 30°C (the restrictive temperature) when umuDC were expressed constitutively from a single chromosomal copy of the operon. Over-expression of umuDC led to longer filaments and the production of filaments at 40°C. This role of UmuDC is apparently distinct from its role in cold sensitivity as physiological levels of the proteins induced cell division inhibition but not cold sensitivity. Unlike the cold sensitive phenotype, umuDC-dependent septation inhibition requires SOS induction, demonstrating the need for at least one other LexA-regulated gene in this process [65]. It is tempting to speculate that this gene might be ftsK, an essential cell division protein transcribed from dinH, an SOS-regulated promoter [76, 77]. It is unclear, however, how up-regulation of a gene required for septation would operate to inhibit this process after DNA damage. Perhaps high levels of FtsK prevent proper formation of the septation ring, perhaps by disrupting the stoichiometry of septation factors. Interestingly, over-expression of ftsQAZ from a plasmid suppressed umuDC-dependent filamentation [65]. This finding, combined with the recent demonstration that FtsK is required for recruitment of FtsQ and other factors to the septation ring [78], suggests that proper stoichiometry of septation proteins may indeed play a role in promoting the regulation of cell division in vivo. It is also possible that the UmuDC complex plays a direct inhibitory role in this process. Since it is the target of SulA and SfiC, FtsZ would be a logical target for inhibition by the UmuDC complex, but this interaction has yet to be demonstrated. There may also be a role for minCDE in regulating cell division and septation after DNA damage. These genes, which induce and regulate septum formation, are repressed following UV in a lexA-independent manner [21]. Clearly, further investigation of this division inhibition is warranted, especially in conjunction with the role of UmuDC in promoting the arrest of DNA replication after DNA damage.
RELIEVING STRESS: TURNING THE SOS RESPONSE OFF
Thus, the SOS response appears to upregulate genes that hold replication and cell division in place until the offending lesions are repaired (or polymerases that allow replication to occur over the lesions are expressed). Once this has occurred, the stress response must be turned off. Two mechanisms that appear to play a role in returning the cell to normalcy are the LexA repressor and the product of the dinI gene.
LexA represses its own gene, and following SOS induction, LexA repressor is heavily induced [79]. In the current model, as long as replication remains blocked and single stranded DNA is present at the replication fork to activate and bind RecA, the LexA produced will be cleaved and SOS genes will continue to be expressed (reviewed in [2]). Once replication has resumed and the single stranded regions have been filled in, RecA no longer remains in the activated state and the LexA protein accumulates, binds, and represses genes that contain LexA binding sites at their promoters. Eventually, enough LexA is produced to repress all genes under its control and the cell resumes replicating and dividing normally.
Once replication has resumed and repair has been completed, there still remains an excessive amount of RecA in the cell due to its large up-regulation following SOS induction. One additional gene induced during the SOS response, dinI, has recently been shown to help eliminate excess RecA and help terminate the SOS response through an unusual mechanism. DinI was discovered as an SOS inducible gene through the original screen by Kenyon and Walker [7]. The first hint that DinI may play a role in suppressing SOS response came from work by Ohmori and coworkers who demonstrated that it could suppress the cold-sensitive phenotype of an unusual dinD68 mutation. The dinD68 allele causes SOS induction without any DNA damaging agent at temperatures less than 20°C through a mechanism that is not yet understood [80]. However, subsequent work from Yasuda's group [81, 82] additionally showed that DinI, when over-expressed, conferred a UV sensitive phenotype to cells, inhibited induction of SulA, and impaired LexA and UmuD cleavage following DNA damage. These are all activities that are consistent with repression, rather than activation of SOS functions. Recently, Camerini-Otero and coworkers have shown that the down-regulation occurs directly through DinI binding to the RecA protein itself [83, 84]. This group demonstrated that the C-terminal alpha helix region of DinI has an unusually strong negatively charged surface that appears to mimic the structure and charge of single-stranded DNA, the substrate for RecA activation. However, unlike RecA bound to single stranded DNA, RecA bound to DinI is not an activated form and cannot induce the SOS response. Thus, these data strongly suggest that DinI functions to down-regulate the SOS response by titrating away the inducing “sensor” for SOS induction itself, the RecA protein.
SUMMARY
The picture emerging from these studies of E coli is one in which multiple events are choreographed spatially and temporally such that the entire process can restore and complete the duplication of the genome following a moderate dose of DNA damage without sacrificing viability or the integrity of the genetic information.
There are, however, some critical pieces to this picture which remain to be filled in. It will be interesting and important to now identify the specific structure of the replication fork when it arrests. We believe that single stranded DNA is produced (and activates RecA) to initiate the cascade, however, where it is formed and how the arrested replication fork is structured remain to be characterized. Furthermore, the timing and contribution that lesion repair, lesion bypass, or even recombination has on the recovery of replication is not at all clear and remains a critical question since each strategy has very different consequences for the integrity and fidelity of the genetic information as it is duplicated. Lastly and most importantly, when we look at the genes that are induced following DNA damage, fully half of these genes have not been well characterized as to their functions. While new information is constantly coming, it is apparent from recent studies reviewed here that our view to date is extremely DNA-centric and is limited by a small number of assays. It will be interesting to see which discoveries remain to be elucidated from the characterization of these unknown genes.
Acknowledgments
ACKNOWLEDGMENT
This work was supported in part by an award from the National Science Foundation MCB0130486 to J. Crowley.
References
- Radman M. Phenomenology of an inducible mutagenic DNA repair pathway in Escherichia coli: SOS repair hypothesis. In: Prakash L, Sherman F, Miller M, Lawrence C, Tabor H W, editors. Molecular and Environmental Aspects of Mutagenesis. Charles C Thomas; Springfield, Ill: 1999. pp. 128–142. [Google Scholar]
- Friedberg E C, Walker G C, Siede W. ASM Press; Washington, D.C.: 1995. DNA Repair and Mutagenesis. [Google Scholar]
- Koch W H, Woodgate R. The SOS response. In: Nickoloff J A, Hoekstra M F, editors. DNA Damage and Repair: DNA Repair in Prokaryotes and Lower Eukaryotes. Humana Press; Totowa, NJ: 1998. pp. 107–134. [Google Scholar]
- Sassanfar M, Roberts J W. Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J Mol Biol. 1990;212(1):79–96. doi: 10.1016/0022-2836(90)90306-7. [DOI] [PubMed] [Google Scholar]
- Brent R, Ptashne M. Mechanism of action of the lexA gene product. Proc Natl Acad Sci USA. 1981;78(7):4204–4208. doi: 10.1073/pnas.78.7.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little J W, Mount D W, Yanisch-Perron C R. Purified lexA protein is a repressor of the recA and lexA genes. Proc Natl Acad Sci USA. 1981;78(7):4199–4203. doi: 10.1073/pnas.78.7.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C J, Walker G C. DNA-damaging agents stimulate gene expression at specific loci in Escherichia coli. Proc Natl Acad Sci USA. 1980;77(5):2819–2823. doi: 10.1073/pnas.77.5.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G C. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev. 1984;48(1):60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagg A, Kenyon C J, Walker G C. Inducibility of a gene product required for UV and chemical mutagenesis in Escherichia coli. Proc Natl Acad Sci USA. 1981;78(9):5749–5753. doi: 10.1073/pnas.78.9.5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner C A, Hays S, McEntee K, Goodman M F. DNA polymerase II is encoded by the DNA damage-inducible dinA gene of Escherichia coli. Proc Natl Acad Sci USA. 1990;87(19):7663–7667. doi: 10.1073/pnas.87.19.7663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogliano M, Schendel P F. Evidence for the inducibility of the uvrB operon. Nature. 1981;289(5794):196–198. doi: 10.1038/289196a0. [DOI] [PubMed] [Google Scholar]
- Huisman O, D'Ari R. An inducible DNA replication-cell division coupling mechanism in E. coli. Nature. 1981;290(5809):797–799. doi: 10.1038/290797a0. [DOI] [PubMed] [Google Scholar]
- Iwasaki H, Nakata A, Walker G C, Shinagawa H. The Escherichia coli polB gene, which encodes DNA polymerase II, is regulated by the SOS system. J Bacteriol. 1990;172(11):6268–6273. doi: 10.1128/jb.172.11.6268-6273.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C J, Walker G C. Expression of the E. coli uvrA gene is inducible. Nature. 1981;289(5800):808–810. doi: 10.1038/289808a0. [DOI] [PubMed] [Google Scholar]
- Lloyd R G, Picksley S M, Prescott C. Inducible expression of a gene specific to the RecF pathway for recombination in Escherichia coli K12. Mol Gen Genet. 1983;190(1):162–167. doi: 10.1007/BF00330340. [DOI] [PubMed] [Google Scholar]
- Ohmori H, Hatada E, Qiao Y, Tsuji M, Fukuda R. dinP, a new gene in Escherichia coli, whose product shows similarities to UmuC and its homologues. Mutat Res. 1995;347(1):1–7. doi: 10.1016/0165-7992(95)90024-1. [DOI] [PubMed] [Google Scholar]
- Shurvinton C E, Lloyd R G. Damage to DNA induces expression of the ruv gene of Escherichia coli. Mol Gen Genet. 1982;185(2):352–355. doi: 10.1007/BF00330811. [DOI] [PubMed] [Google Scholar]
- Siegel E C. The Escherichia coli uvrD gene is inducible by DNA damage. Mol Gen Genet. 1983;191(3):397–400. doi: 10.1007/BF00425753. [DOI] [PubMed] [Google Scholar]
- Ohmori H, Saito M, Yasuda T, et al. The pcsA gene is identical to dinD in Escherichia coli. J Bacteriol. 1995;177(1):156–165. doi: 10.1128/jb.177.1.156-165.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis L K, Harlow G R, Gregg-Jolly L A, Mount D W. Identification of high affinity binding sites for LexA which define new DNA damage-inducible genes in Escherichia coli. J Mol Biol. 1994;241(4):507–523. doi: 10.1006/jmbi.1994.1528. [DOI] [PubMed] [Google Scholar]
- Courcelle J, Khodursky A, Peter B, Brown P O, Hanawalt P C. Comparative gene expression profiles following UV exposure in wild type and SOS deficient. Escherichia coli Genetics. 2001;158(1):41–64. doi: 10.1093/genetics/158.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satta G, Gudas L J, Pardee A B. Degradation of Escherichia coli DNA: evidence for limitation in vivo by protein X, the recA gene product. Mol Gen Genet. 1979;168(1):69–80. doi: 10.1007/BF00267935. [DOI] [PubMed] [Google Scholar]
- Horii Z, Suzuki K. Degradation of the DNA of Escherichia coli K12 rec- (JC1569b) after irradiation with ultraviolet light. Photochem photobiol. 1968;8(2):93–105. doi: 10.1111/j.1751-1097.1970.tb05976.x. [DOI] [PubMed] [Google Scholar]
- Clark A J, Margulies A D. Isolation and characterization of recombination-deficient mutants of Escherichia coli K12. Proc Natl Acad Sci USA. 1965;53(2):451–459. doi: 10.1073/pnas.53.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczykowski S C, Dixon D A, Eggleston A K, Lauder S D, Rehrauer W M. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58(3):401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roca A I, Cox M M. RecA protein: structure, function, and role in recombinationa repair. Prog Nucleic Acid Res Mol Biol. 1997;56:129–223. doi: 10.1016/s0079-6603(08)61005-3. [DOI] [PubMed] [Google Scholar]
- Smith G R. Homologous recombination in procaryotes. Microbiol Rev. 1988;52(1):1–28. doi: 10.1128/mr.52.1.1-28.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle J, Ganesan A K, Hanawalt P C. Therefore, what are recombination proteins there for? Bioessays. 2001;23(5):463–470. doi: 10.1002/bies.1065. [DOI] [PubMed] [Google Scholar]
- Courcelle J, Carswell-Crumpton C, Hanawalt P C. recF and recR are required for the resumption of replication at DNA replication forks in Escherichia coli. Proc Natl Acad Sci USA. 1997;94(8):3714–3719. doi: 10.1073/pnas.94.8.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle J, Crowley D J, Hanawalt P C. Recovery of DNA replication in UV-irradiated Escherichia coli requires both excision repair and recF protein function. J Bacteriol. 1999;181(3):916–922. doi: 10.1128/jb.181.3.916-922.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle J, Hanawalt P C. Participation of recombination proteins in rescue of arrested replication forks in UV-irradiated Escherichia coli need not involve recombination. Proc Natl Acad Sci USA. 2001;98(15):8196–8202. doi: 10.1073/pnas.121008898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlynn P, Mahdi A A, Lloyd R G. Characterisation of the catalytically active form of RecG helicase. Nucleic Acids Res. 2000;28(12):2324–2332. doi: 10.1093/nar/28.12.2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggleston A K, West S C. Cleavage of Holliday junctions by the E. coli RuvABC complex. J Biol Chem. 2000;275(34):26467–26476. doi: 10.1074/jbc.M001496200. [DOI] [PubMed] [Google Scholar]
- McGlynn P, Lloyd R G. RecG helicase activity at three- and four-strand DNA structures. Nucleic Acids Res. 1999;27(15):3049–3056. doi: 10.1093/nar/27.15.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlynn P, Lloyd R G. Modulation of RNA polymerase by (p)ppGpp reveals a RecG-dependent mechanism for replication fork progression. Cell. 2000;101(1):35–45. doi: 10.1016/S0092-8674(00)80621-2. [DOI] [PubMed] [Google Scholar]
- Crowley D J, Hanawalt P C. Induction of the SOS response increases the efficiency of global nucleotide excision repair of cyclobutane pyrimidine dimers, but not 6-4 photoproducts, in UV-irradiated Escherichia coli. J Bacteriol. 1998;180(13):3345–3352. doi: 10.1128/jb.180.13.3345-3352.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley D J, Hanawalt P C. The SOS-dependent upregulation of uvrD is not required for efficient nucleotide excision repair of ultraviolet light induced DNA photoproducts in Escherichia coli. Mutat Res. 2001;485(4):319–329. doi: 10.1016/s0921-8777(01)00068-4. [DOI] [PubMed] [Google Scholar]
- Siegel E C. Ultraviolet-sensitive mutator mutU4 of Escherichia coli inviable with polA. J Bacteriol. 1973;113(1):161–166. doi: 10.1128/jb.113.1.161-166.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn B K, Kushner S R. Construction and analysis of deletions in the structural gene (uvrD) for DNA helicase II of Escherichia coli. J Bacteriol. 1991;173(8):2569–2575. doi: 10.1128/jb.173.8.2569-2575.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruand C, Ehrlich S D. UvrD-dependent replication of rolling-circle plasmids in Escherichia coli. Mol Microbiol. 2000;35(1):204–210. doi: 10.1046/j.1365-2958.2000.01700.x. [DOI] [PubMed] [Google Scholar]
- George J W, Brosh R M. Jr, Matson S W. A dominant negative allele of the Escherichia coli uvrD gene encoding DNA helicase II. A biochemical and genetic characterization. J Mol Biol. 1994;235(2):424–435. doi: 10.1006/jmbi.1994.1003. [DOI] [PubMed] [Google Scholar]
- Moolenaar G F, Moorman C, Goosen N. Role of the Escherichia coli nucleotide excision repair proteins in DNA replication. J Bacteriol. 2000;182(20):5706–5714. doi: 10.1128/jb.182.20.5706-5714.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SaiSree L, Reddy M, Gowrishankar J. lon incompatibility associated with mutations causing SOS induction: null uvrD alleles induce an SOS response in Escherichia coli. J Bacteriol. 2000;182(11):3151–3157. doi: 10.1128/jb.182.11.3151-3157.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Pham P, Shen X, et al. Roles of E. coli DNA polymerases IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature. 2000;404(6781):1014–1018. doi: 10.1038/35010020. [DOI] [PubMed] [Google Scholar]
- Livneh Z. Replication of UV-irradiated single-stranded DNA by DNA polymerase III holoenzyme of Escherichia coli: evidence for bypass of pyrimidine photodimers. Proc Natl Acad Sci USA. 1986;83(13):4599–4603. doi: 10.1073/pnas.83.13.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner J, Nohmi T. Escherichia coli DNA polymerase IV mutator activity: genetic requirements and mutational specificity. J Bacteriol. 2000;182(16):4587–4595. doi: 10.1128/jb.182.16.4587-4595.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner J, Gruz P, Kim S R, et al. The dinB gene encodes a novel E. coli DNA polymerase, DNA pol IV, involved in mutagenesis. Mol Cell. 1999;4(2):281–286. doi: 10.1016/s1097-2765(00)80376-7. [DOI] [PubMed] [Google Scholar]
- Tang M, Shen X, Frank E G, O'Donnell M, Woodgate R, Goodman M F. UmuD'(2)C is an error-prone DNA polymerase, Escherichia coli pol V. Proc Natl Acad Sci USA. 1999;96(16):8919–8924. doi: 10.1073/pnas.96.16.8919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuven N B, Arad G, Maor-Shoshani A, Livneh Z. The mutagenesis protein UmuC is a DNA polymerase activated by UmuD', RecA, and SSB and is specialized for translesion replication. J Biol Chem. 1999;274(45):31763–31766. doi: 10.1074/jbc.274.45.31763. [DOI] [PubMed] [Google Scholar]
- Perry K L, Elledge S J, Mitchell B B, Marsh L, Walker G C. umuDC and mucAB operons whose products are required for UV light- and chemical-induced mutagenesis: UmuD, MucA, and LexA proteins share homology. Proc Natl Acad Sci USA. 1985;82(13):4331–4335. doi: 10.1073/pnas.82.13.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson J R, Lawrence C W, Hinkle D C. Deoxycytidyl transferase activity of yeast REV1 protein. Nature. 1996;382(6593):729–731. doi: 10.1038/382729a0. [DOI] [PubMed] [Google Scholar]
- Chen H, Bryan S K, Moses R E. Cloning the polB gene of Escherichia coli and identification of its product. J Biol Chem. 1989;264(34):20591–20595. [PubMed] [Google Scholar]
- Kornberg T, Gefter M L. DNA synthesis in cell-free extracts of a DNA polymerase-defective mutant. Biochem Biophys Res Commun. 1970;40(6):1348–1355. doi: 10.1016/0006-291x(70)90014-8. [DOI] [PubMed] [Google Scholar]
- Kornberg T, Gefter M L. Purification and DNA synthesis in cell-free extracts: properties of DNA polymerase II. Proc Natl Acad Sci USA. 1971;68(4):761–764. doi: 10.1073/pnas.68.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Z, Goodman M F. The Escherichia coli polB locus is identical to dinA, the structural gene for DNA polymerase II. Characterization of Pol II purified from a polB mutant. J Biol Chem. 1997;272(13):8611–8617. doi: 10.1074/jbc.272.13.8611. [DOI] [PubMed] [Google Scholar]
- Rangarajan S, Woodgate R, Goodman M F. A phenotype for enigmatic DNA polymerase II: a pivotal role for pol II in replication restart in UV-irradiated Escherichia coli. Proc Natl Acad Sci USA. 1999;96(16):9224–9229. doi: 10.1073/pnas.96.16.9224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napolitano R, Janel-Bintz R, Wagner J, Fuchs R P. All three SOS-inducible DNA polymerases (Pol II, Pol IV and Pol V) are involved in induced mutagenesis. EMBO J. 2000;19(22):6259–6265. doi: 10.1093/emboj/19.22.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh L, Walker G C. New phenotypes associated with mucAB: alteration of a MucA sequence homologous to the LexA cleavage site. J Bacteriol. 1987;169(5):1818–1823. doi: 10.1128/jb.169.5.1818-1823.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinagawa H, Iwasaki H, Kato T, Nakata A. RecA protein-dependent cleavage of UmuD protein and SOS mutagenesis. Proc Natl Acad Sci USA. 1988;85(6):1806–1810. doi: 10.1073/pnas.85.6.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burckhardt S E, Woodgate R, Scheuermann R H, Echols H. UmuD mutagenesis protein of Escherichia coli: overproduction, purification, and cleavage by RecA. Proc Natl Acad Sci USA. 1988;85(6):1811–1815. doi: 10.1073/pnas.85.6.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohmi T, Battista J R, Dodson L A, Walker G C. RecA-mediated cleavage activates UmuD for mutagenesis: mechanistic relationship between transcriptional derepression and posttranslational activation. Proc Natl Acad Sci USA. 1988;85(6):1816–1820. doi: 10.1073/pnas.85.6.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferentz A E, Walker G C, Wagner G. Converting a DNA damage checkpoint effector (UmuD2C) into a lesion bypass polymerase (UmuD'2C) EMBO J. 2001;20(15):4287–4298. doi: 10.1093/emboj/20.15.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opperman T, Murli S, Smith B T, Walker G C. A model for a umuDC-dependent prokaryotic DNA damage checkpoint. Proc Natl Acad Sci USA. 1999;96(16):9218–9223. doi: 10.1073/pnas.96.16.9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh L, Walker G C. Cold sensitivity induced by overproduction of UmuDC in Escherichia coli. J Bacteriol. 1985;162(1):155–161. doi: 10.1128/jb.162.1.155-161.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opperman T, Murli S, Walker G C. The genetic requirements for UmuDC-mediated cold sensitivity are distinct from those for SOS mutagenesis. J Bacteriol. 1996;178(15):4400–4411. doi: 10.1128/jb.178.15.4400-4411.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton M D, Walker G C. umuDC-mediated cold sensitivity is a manifestation of functions of the UmuD(2)C complex involved in a DNA damage checkpoint control. J Bacteriol. 2001;183(4):1215–1224. doi: 10.1128/JB.183.4.1215-1224.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton M D, Opperman T, Walker G C. The Escherichia coli SOS mutagenesis proteins UmuD and UmuD' interact physically with the replicative DNA polymerase. Proc Natl Acad Sci USA. 1999;96(22):12373–12378. doi: 10.1073/pnas.96.22.12373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton M D, Murli S, Opperman T, Klein C, Walker G C. umuDC-dnaQ Interaction and its implications for cell cycle regulation and SOS mutagenesis in Escherichia coli. J Bacteriol. 2001;183(3):1085–1089. doi: 10.1128/JB.183.3.1085-1089.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Waga S, Hannon G J, Beach D, Stillman B. Differential effects by the p21 CDK inhibitor on PCNA-dependent DNA replication and repair. Nature. 1994;371(6497):534–537. doi: 10.1038/371534a0. [DOI] [PubMed] [Google Scholar]
- George J, Castellazzi M, Buttin G. Prophage induction and cell division in E. coli. III. Mutations sfiA and sfiB restore division in tif and lon strains and permit the expression of mutator properties of tif. Mol Gen Genet. 1975;140(4):309–332. [PubMed] [Google Scholar]
- Bi E F, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354(6349):161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J F. Coupling of DNA replication and cell division: sulB is an allele of ftsZ. J Bacteriol. 1983;154(3):1339–1346. doi: 10.1128/jb.154.3.1339-1346.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoemaker J M, Gayda R C, Markovitz A. Regulation of cell division in Escherichia coli: SOS induction and cellular location of the sulA protein, a key to lon-associated filamentation and death. J Bacteriol. 1984;158(2):551–561. doi: 10.1128/jb.158.2.551-561.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C, Holland I B. Role of the SulB (FtsZ) protein in division inhibition during the SOS response in Escherichia coli: FtsZ stabilizes the inhibitor SulA in maxicells. Proc Natl Acad Sci USA. 1985;82(18):6045–6049. doi: 10.1073/pnas.82.18.6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill T M, Sharma B, Valjavec-Gratian M, Smith J. sfi-independent filamentation in Escherichia coli is lexA dependent and requires DNA damage for induction. J Bacteriol. 1997;179(6):1931–1939. doi: 10.1128/jb.179.6.1931-1939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorazi R, Dewar S J. The SOS promoter dinH is essential for ftsK transcription during cell division. Microbiology. 2000;146(11):2891–2899. doi: 10.1099/00221287-146-11-2891. [DOI] [PubMed] [Google Scholar]
- Wang L, Lutkenhaus J. FtsK is an essential cell division protein that is localized to the septum and induced as part of the SOS response. Mol Microbiol. 1998;29(3):731–740. doi: 10.1046/j.1365-2958.1998.00958.x. [DOI] [PubMed] [Google Scholar]
- Chen J C, Beckwith J. FtsQ, FtsL and FtsI require FtsK, but not FtsN, for co-localization with FtsZ during Escherichia coli cell division. Mol Microbiol. 2001;42(2):395–413. doi: 10.1046/j.1365-2958.2001.02640.x. [DOI] [PubMed] [Google Scholar]
- Brent R, Ptashne M. The lexA gene product represses its own promoter. Proc Natl Acad Sci USA. 1980;77(4):1932–1936. doi: 10.1073/pnas.77.4.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda T, Nagata T, Ohmori H. Multicopy suppressors of the cold-sensitive phenotype of the pcsA68 (dinD68) mutation in Escherichia coli. J Bacteriol. 1996;178(13):3854–3859. doi: 10.1128/jb.178.13.3854-3859.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda T, Morimatsu K, Kato R, Usukura J, Takahashi M, Ohmori H. Physical interactions between DinI and RecA nucleoprotein filament for the regulation of SOS mutagenesis. EMBO J. 2001;20(5):1192–1202. doi: 10.1093/emboj/20.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda T, Morimatsu K, Horii T, Nagata T, Ohmori H. Inhibition of Escherichia coli RecA coprotease activities by DinI. EMBO J. 1998;17(11):3207–3216. doi: 10.1093/emboj/17.11.3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez B E, Voloshin O N, Camerini-Otero R D, Bax A. Solution structure of DinI provides insight into its mode of RecA inactivation. Protein Sci. 2000;9(11):2161–2169. doi: 10.1110/ps.9.11.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voloshin O N, Ramirez B E, Bax A, Camerini-Otero R D. A model for the abrogation of the SOS response by an SOS protein: a negatively charged helix in DinI mimics DNA in its interaction with RecA. Genes Dev. 2001;15(4):415–427. doi: 10.1101/gad.862901. [DOI] [PMC free article] [PubMed] [Google Scholar]