Abstract
To identify the location of a domain of the β-subunit of Escherichia coli RNA polymerase (RNAP) on the three-dimensional structure, we developed a method to tag a nonessential surface of the multisubunit enzyme with a protein density easily detectable by electron microscopy and image processing. Four repeats of the IgG-binding domain of Staphylococcus aureus protein A were inserted at position 998 of the E. coli RNAP β-subunit. The mutant RNAP supported E. coli growth and showed no apparent functional defects in vitro. The structure of the mutant RNAP was determined by cryoelectron microscopy and image processing of frozen-hydrated helical crystals. Comparison of the mutant RNAP structure with the previously determined wild-type RNAP structure by Fourier difference analysis at 20-Å resolution directly revealed the location of the inserted protein domain, thereby locating the region around position 998 of the β-subunit within the RNAP three-dimensional structure and refining a model for the subunit locations within the enzyme.
Transcription in all cellular organisms is orchestrated by the multisubunit DNA-dependent RNA polymerase (RNAP), a multifunctional enzyme that synthesizes RNA and is a major target for the regulation of gene expression. The Escherichia coli RNAP, which is the best characterized, comprises an essential catalytic core of two α-subunits (each 36.5 kDa), one β-subunit (150.6 kDa), and one β′-subunit (155.2 kDa) that is responsible for transcript elongation and termination. The holoenzyme contains an additional regulatory subunit, normally σ70 (70.2 kDa), and is capable of promoter-specific recognition and transcription initiation. To identify the location of subunits within the RNAP, we developed a strategy to target insertions of a protein domain into surface-exposed, nonessential regions that can be visualized by electron microscopy (EM) and image processing.
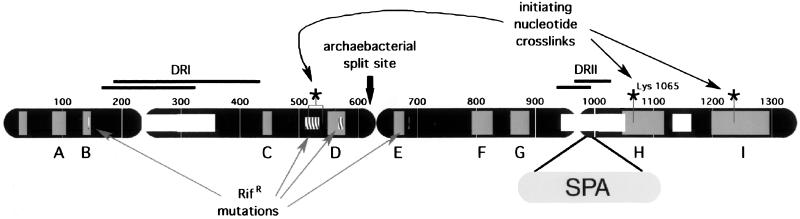
This strategy was readily applicable to either of the two largest subunits, β and β′, which have colinearly arranged regions of strong amino acid sequence similarity from bacteria to humans (1–3). The highly conserved regions are separated by relatively nonconserved spacer regions. In some organisms, the nonconserved regions contain large gaps or insertions compared with E. coli. In the β-subunit, nine conserved regions, labeled A through I, have been identified (ref. 3; Fig. 1). In addition, the β-subunit of E. coli RNAP contains two regions of poor sequence conservation, centered about residues 300 and 1,000, which are often missing in β-homologs from other organisms (Fig. 1).
Figure 1.
Staphylococcus aureus protein A (SPA) insertion in the β-subunit of E. coli RNAP. The primary sequence of the β-subunit is represented as a black bar. Evolutionarily conserved regions are shaded grey and labeled A–I (3). Evolutionarily hypervariable sequences are denoted by open, white boxes. Dispensable regions (DRI and DRII) are labeled, along with regions that have been deleted with little apparent consequence for basic RNAP assembly and transcription activity (black lines above sequence). The SPA insertion in DRII is shown. The sites of known rifampicin (rifampicin-resistant or RifR) mutations (6, 28) as well as split sites in archaebacterial genes and sites of cross-links to initiating nucleotide analogs (refs. 5 and 29; denoted as *) are also indicated.
As expected, mutations affecting the conserved regions frequently cause severe defects in RNAP function (ref. 4 and references therein). β-Residues 1,065 and 1,237, in conserved regions H and I, respectively, participate in the formation of the initiating site of the enzyme (5). Mutations in the β-subunit render the enzyme resistant to the antibiotic inhibitors rifampicin and streptolydigin (refs. 6–9; Fig. 1).
Alternatively, mutations in poorly conserved regions can have little or no apparent effect on RNAP function. In fact, large deletions in the two poorly conserved regions in β do not affect RNAP assembly and basic function in vitro (10, 11). These regions have thus been termed DRI (situated between conserved regions B and C, extending approximately from residues 170 to 430) and DRII (situated between conserved regions G and H, extending approximately from residues 930 to 1,030). Both of these regions are poorly conserved or absent in β-subunit homologs from other organisms; both can tolerate numerous mutations without fatal consequences in vivo; and both must be structurally autonomous to accommodate large deletions and insertions without disturbing the critical functions of the enzyme. Thus, both DRs are likely to comprise separate and relatively isolated domains on the RNAP structure.
Although neither DR of the β-subunit plays an important role in RNAP assembly or basic transcription activity, it is assumed that their presence points to a role in regulatory functions specific to E. coli that have not yet been identified. Indeed, DRI is targeted by the bacteriophage T4-Alc protein, which selectively induces premature termination of E. coli RNAP transcription on E. coli DNA (11), indicating that regulatory factors can impact transcription through these DRs.
Herein, we report a structural analysis of a mutant E. coli RNAP harboring an insertion of a protein domain in β-DRII. The mutant RNAP was used to locate directly the inserted domain on the three-dimensional structure of RNAP by using cryo-EM and difference analysis with the previously determined native RNAP structure, thereby locating its site of insertion. The approach used for this study is similar to the peptide-based difference mapping reported by Conway et al. (12), except that a larger protein domain was used to ensure visualization at the lower resolution of our analysis (20-Å compared with 11-Å resolution), and an internal rather than a terminal region of the polypeptide was located.
Materials and Methods
Construction of Strain RL1120.
Strain RL1120 was constructed from strain RL654 by recombination between the rpoB5142(HY526, SR643, RifR, SPA) gene in RL654 and the rpoB5201(SPA) gene in plasmid pRL715. RL654 (E. coli W3110 trpR tnaA2 rpoB5142) is the recA+ parent of the strain RL656 (4). pRL715 was constructed from the wild-type rpoB plasmid pRW408 (13) by replacement of a 3.6-kilobase BspEI to ScaI fragment with the corresponding fragment from pRW237rpoB5142. This replacement transferred the SPA insertion, but not the HY526 nor SR643 mutations, to pRL715. pRL715 expresses β∷SPA from a trp–lac fusion promoter (trc) but expresses little subunit in the absence of isopropyl β-d-thiogalactoside (IPTG), because the plasmid also carries a lacI gene. Thus, RL654 carrying pRL715 remains resistant to Rif (RifR) in medium lacking IPTG but will become Rif sensitive (Rifs) if the chromosomal rpoB5142 gene recombines with rpoB5201 on the plasmid. To obtain a Rifs recombinant, RL654 was transformed with pRL715 and subjected to two rounds of rifampicin-cycloserine selection (6) to enrich for Rifs recombinants. The Rifs recombinants were grown overnight in minimal glucose liquid medium without ampicillin, spread on LB plates, and screened for loss of the pRL715 plasmid by replica plating to LB-ampicillin plates.
Purification and Crystallization.
E. coli core SPA RNAP from strain RL1120 was prepared and crystallized as described (14, 15).
Sample Preparation and EM.
Samples of tubular crystals of E. coli SPA RNAP were applied to EM grids coated with holey carbon film. After blotting the solution with a filter paper, the grid was then immediately plunged into a slush of liquid ethane by using a guillotine device that allowed reproducible and rapid transfer into the cryogen. The high freezing rate ensured vitrification of the ice and minimized contamination by hexagonal and cubic ice that would disrupt morphological structure. The frozen specimen suspended over the holes and embedded in vitreous ice could then be stored indefinitely under liquid nitrogen.
Transmission EM was performed by using a CM12 microscope (Philips Electronic Instruments, Mahwah, NJ) operating at 120 kV. Low-dose procedures (<10 electrons per Å2) were used to minimize radiation damage. Images were recorded at 800–2,500 nm underfocus.
Image Processing.
Optical diffraction was used to select images that showed minimal astigmatism and drift based on the shape of the Thon rings in the contrast transfer function and strong symmetrical diffraction. The selected images were digitized at a 10-μm interval (corresponding to 2.8 Å on the specimen) by using a Perkin–Elmer flat-bed microdensitometer.
The analysis of the helical SPA RNAP crystals was performed as described (15) with a set of helical processing programs obtained from N. Unwin (Medical Research Council Laboratory of Molecular Biology, Cambridge, U.K.; refs. 16–18). For each image, defocus and astigmatism values were determined by dividing the amplitudes in the Fourier transforms of each tube into five sectors, averaging the sectors, and then fitting them to theoretical contrast transfer functions (19). After contrast transfer function correction, the tubes from different helical families were combined by using the method described by DeRosier et al. (20) involving alignment and averaging of the Fourier–Bessel coefficients.
To generate the three-dimensional difference map, the averaged map of SPA RNAP was aligned with the wild-type RNAP map (truncated to 20-Å resolution) by applying phase shifts to the Fourier–Bessel coefficients and minimizing the phase residual (21). The difference map was then calculated by using the command dr diff in spider (22).
Results
A Mutant RNAP for Structural Studies.
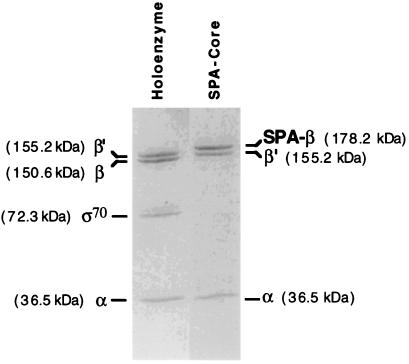
To obtain tubular, helical crystals of RNAP in which a site on one of the subunits could be identified by EM and image processing, we sought an insertion of a sufficient mass of protein in one of the subunits that fulfilled two criteria. First, the mutant subunit harboring the insertion should support normal E. coli growth when it was the only source of that subunit, ensuring that the mutant RNAP retained the essential functions of RNAP and thus did not have a disrupted structure. Second, the inserted protein mass should fold into a stable, well ordered structure to allow visualization of the extra density on the surface of the RNAP structure. A derivative of the E. coli RNAP β-subunit constructed in our lab was an excellent candidate for this study. A 234-amino acid segment of SPA, which encompassed four repeats of the protein A IgG-binding domain (repeats D–C), was inserted in DRII of the β-subunit on an expression plasmid (ref. 4; Fig. 1). E. coli RNAP is known to tolerate insertions and deletions in β-DRII (23, 24). Each SPA IgG-binding domain folds independently into a stable, 60-amino acid three-helix bundle (25, 26). An rpoB carrying the four-domain insertion between Leu-998 and Glu-999 together with an amino acid substitution conferring resistance to the antibiotic rifampicin (rpoB6; HY526) was transferred to the E. coli chromosome by recombination and found to yield viable bacteria. RNAP purified from this strain retained normal transcriptional activity (data not shown) and was bound specifically by IgG agarose (4), indicating that the SPA IgG-binding domains folded into their native structures on the surface of RNAP. However, this RifR RNAP was suboptimal for structural studies, because RNAP bound to rifampicin forms more ordered helical crystals (27). A Rifs version of SPA RNAP was obtained by constructing a strain carrying only the SPA insertion, but not the RifR mutation, in the chromosomal rpoB gene (see Materials and Methods). This strain had viability identical to an isogenic near wild-type strain from 17 to 42°C on minimal or rich media, verifying that the SPA insertion in DRII had little, if any, effect on RNAP function. This strain yielded core RNAP containing a mutant β-subunit harboring the SPA insertion (SPA RNAP) on purification (ref. 14; Fig. 2).
Figure 2.
Pure SPA core RNAP (Right) analyzed by SDS/PAGE on an 8–25% gradient PhastGel (Amersham Pharmacia). E. coli holoenzyme (Left) is shown as a standard. The SPA insertion in the β-subunit of SPA core RNAP increases the mass of that subunit compared with the wild-type enzyme.
Crystallization, EM, and Image Processing.
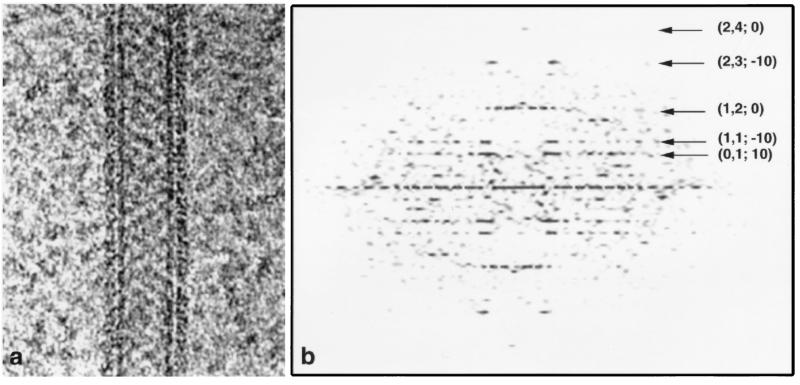
The purified SPA RNAP, complexed with rifampicin, formed tubular, helical crystals (Fig. 3a) in the presence of positively charged lipids as described for the wild-type core RNAP (14, 15). Cryo-EM images of three tubes embedded in amorphous ice were selected for further processing, and the raw diffraction patterns (Fig. 3b) were indexed. Each tube had a distinct helical symmetry (Table 1). The unprocessed diffraction patterns show layer lines visible to about 25-Å resolution, similar to the wild-type crystals (15).
Figure 3.
Electron micrograph (a) and raw Fourier transform (b) from a tube of the (−20, 10) helical family. (a) The tube, embedded in ice, is suspended over a hole in a carbon support film. The RNAP molecules are visible as a crystalline array surrounding the cylindrical lipid bilayer tube. The edge views of the head groups of the phospholipid bilayer forming the tube give rise to the dark vertical lines. (b) In the Fourier transform calculated from the raw image (uncorrected for distortions), the indices (h, k, and n) for the major layer lines are shown, where h and k are indices of corresponding surface lattice lines and n is the Bessel order of the layer lines (16).
Table 1.
Crystallographic data
| Film no. | Defocus, nm | Repeat length, Å | No. repeats | Indexing* (n;1;1;0), (n;1;0;1) | No. of molecules | Average phase residual vs. reference† degrees | Average phase residual vs. average‡ degrees |
|---|---|---|---|---|---|---|---|
| k1806 | 1,793 | 2,082 | 4 | (−20, 5), (10, 14) | 1,068 | 39.6 | 38.7 |
| k1833 | 1,940 | 1,715 | 2 | (−22, 5), (10, 19) | 594 | 47.9 | 46.8 |
| k1834 | 1,558 | 2,437 | 2 | (−21, 5), (10, 17) | 536 | 56.7 | 35.8 |
| Wild type§ | — | 1,715 | 6 | (−20, 4), (10, 11) | 2,134 | — | — |
According to the nomenclature of Toyoshima and Unwin (16).
† Average phase residual between the individual repeats and the alignment reference.
‡ Average phase residual of the Fourier–Bessel coefficients for all layer lines from an individual tube vs. the averaged Fourier–Bessel coefficients.
§ Average structure (15).
The three SPA RNAP tubes were analyzed separately with the set of helical processing programs from N. Unwin (16–18) as described (15). In this procedure, the repeat distance along the helical axis was determined, and then further calculations were performed on individual repeats. These calculations included refinement and correction for out-of-plane tilt, in-plane tube position and rotation, in-plane tilt, and repeat distance. Finally, the aligned repeats for each tube were averaged and corrected for the contrast transfer function by using the methods described in detail by Mimori et al. (19).
The three independent SPA RNAP structures obtained above were then averaged together by using the method described by DeRosier et al. (20). The final set of Gn,l(R,Z) from each tube was truncated to 20-Å resolution. The Fourier–Bessel coefficients, gn,l (r,Z), were then calculated in 2-Å radial steps. The tubes were aligned with the reference tube (chosen to be the −20, 10 tube) by applying phase and radial shifts to the Fourier–Bessel coefficients (21). Finally, the aligned and scaled sets of gn,l(r,Z) were averaged (20) to obtain the final map.
The Three-Dimensional Map.
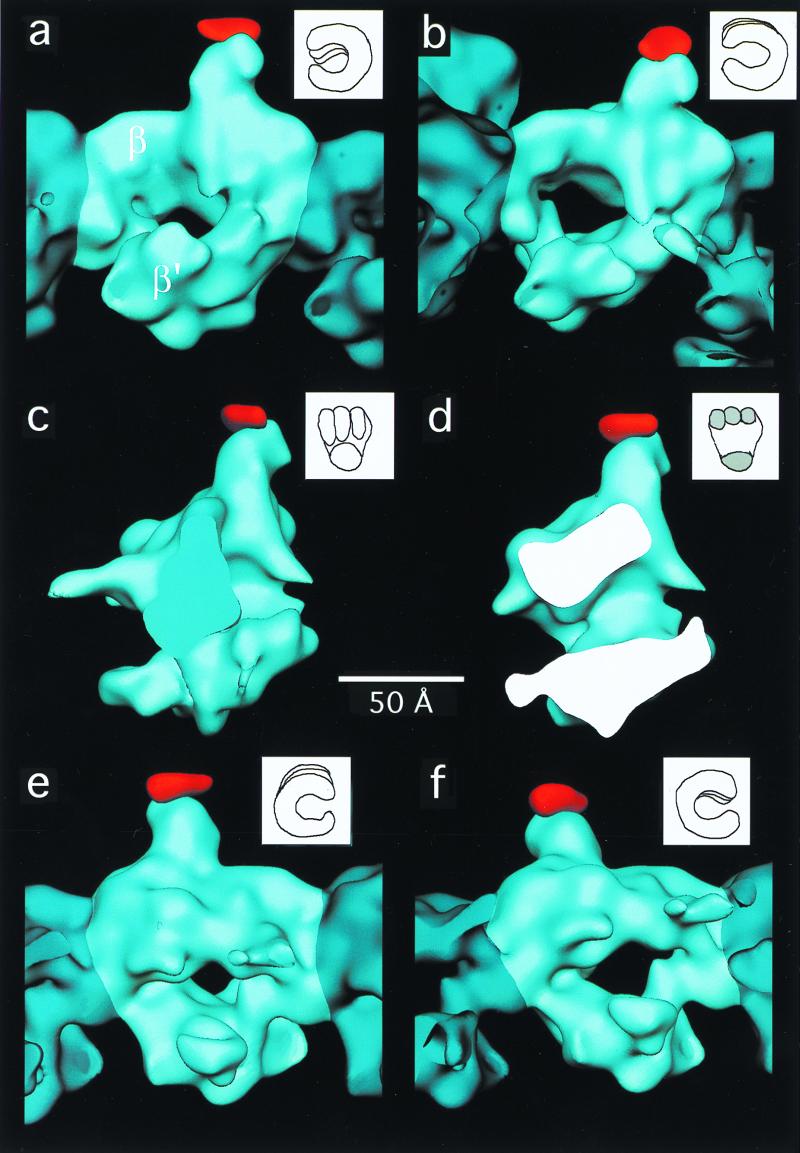
On inspection, the 20-Å structure of SPA RNAP core was very similar to the wild-type RNAP core structure (15), which had been truncated to 20-Å resolution. Differences between the wild-type RNAP and SPA RNAP maps were quantitated by a difference Fourier analysis. The averaged SPA RNAP data were aligned against the wild-type data (truncated to 20-Å resolution) previously obtained (15). The program package spider (22) was then used to generate the difference map illustrated in Fig. 4 (see Materials and Methods). A single 10σ (1σ corresponds to 1 SD above the mean) positive difference peak (shown in red in Fig. 4) indicates the position of the inserted SPA domain with respect to the wild-type core RNAP structure. No other positive or negative difference peaks of this magnitude were observed. The volume of the difference peak corresponds to a protein mass of about 15 kDa (assuming a protein density of 1.3 g/cm3), a little more than half the added mass of the SPA insertion (about 27 kDa).
Figure 4.
Views of the three-dimensional reconstruction of wild-type E. coli core RNAP (blue) with an extra density (red) corresponding to the SPA insertion in the β-subunit. In each view, a single core RNAP molecule is highlighted (light blue). For each view, a drawing of a “hand” is shown (Upper Right) in an orientation similar to that of the RNAP molecule. The difference map (red density) was calculated in real space. (a) View parallel to the helix axis. (b) View showing the wide end of the channel down its axis. (c) View perpendicular to the channel. (d) Cross section of the molecule in an orientation similar to the one shown in c. (e) View corresponding to the opposite side shown in a. (f) View showing the narrow end of the channel down its axis.
Discussion
We have described a strategy combining genetic and structural approaches to locate directly a specific site on the three-dimensional structure of E. coli RNAP. We generated and purified a mutant E. coli RNAP core harboring a large insertion at position 998 (within DRII) of the β-subunit. We demonstrated that the mutant RNAP was normal in its function both in vivo and in vitro, at least under the range of conditions that were investigated, indicating that the essential features of the molecular structure were retained. Finally, EM difference analysis unequivocally showed that the structure of the mutant RNAP contained a significant mass of extra density at the distal end of the domain protruding from the top of the structure (Fig. 4). Thus, we conclude that the extra density present in the mutant RNAP is the inserted SPA domain and that the point of attachment of this domain with the wild-type RNAP structure is position 998 of the β-subunit. This finding has important implications for the likely role of DRs in RNAP function and for the disposition of the subunits within the three-dimensional structure of the enzyme.
Several lines of evidence suggest that DRII of the β-subunit comprises a structurally autonomous domain that protrudes from the surface of the RNAP. First, DRII is poorly conserved or absent in β-subunit homologs from other organisms. Second, DRII can tolerate numerous mutations without fatal consequences in vivo. Third, and most importantly, DRII can be completely deleted from the E. coli RNAP β-subunit with little effect on the essential functions (and therefore structure) of the enzyme (10). For this reason, we propose that the domain protruding from the top of the RNAP structure (attached to the red difference peak in Fig. 4) comprises β-DRII.
Previously, along with the cryo-EM structure of the wild-type E. coli core RNAP, a preliminary model for the locations of the α-, β-, and β′-subunits within the structure was proposed (15, 27). In this model, the domain that we have shown herein to comprise β-DRII was assigned to a domain of the α-subunit. Our results are inconsistent with this previously proposed model and suggest instead that the upper half of the structure shown in Fig. 4 corresponds principally to the β-subunit, and the lower arm corresponds principally to β′, as shown in Fig. 4a (rather than the other way around, as proposed in the earlier model). This revised assignment is also consistent with the x-ray crystal structure of Thermus aquaticus core RNAP (30), which aligns extremely well with the low-resolution structure of E. coli core RNAP (S.A.D., N.O., A. Polyakov, and C.R., unpublished work).
Although deletion of β-DRII from E. coli RNAP had no apparent consequences in vivo or in vitro, its presence suggests a nonessential, regulatory role that has yet to be revealed. Interestingly, β-DRI has been shown to be capable of functioning as a target for a transcriptional regulatory factor (11). The direct localization of β-DRII reported herein contributes to our understanding of the structure–function relationship of the multisubunit and cellular RNAPs, provides an important constraint for models of the RNAP structure and function, and should aid in the design and interpretation of studies to elucidate the functional role of DRII.
Acknowledgments
N.O. was supported by funds from the Norman and Rosita Winston Foundation and from the Philippe Foundation. K.S. was supported by a Jane Coffin Childs Postdoctoral Fellowship and a Burroughs Wellcome Career Development Award. This work was supported in part by National Institutes of Health Grants GM38660 (to R.L.) and GM58020 (to S.A.D.) and a grant from the March of Dimes (to S.A.D.).
Abbreviations
- DR
dispensable region
- EM
electron microscopy
- Rifr/s
rifampicin resistant/sensitive
- RNAP
RNA polymerase
- SPA
Staphylococcus aureus protein A
References
- 1.Allison L A, Moyle M, Shales M, Ingles C J. Cell. 1985;42:599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- 2.Falkenburg D, Dworniczak B, Faust D M, Bautz E K F. J Mol Biol. 1987;195:929–937. doi: 10.1016/0022-2836(87)90496-7. [DOI] [PubMed] [Google Scholar]
- 3.Sweetser D, Nonet M, Young R A. Proc Natl Acad Sci USA. 1987;84:1192–1196. doi: 10.1073/pnas.84.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tavormina P L, Landick R, Gross C A. J Bacteriol. 1996;178:5263–5371. doi: 10.1128/jb.178.17.5263-5271.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mustaev A, Kashlev M, Lee J, Polyakov A, Lebedev A, Zalenskaya K, Grachev M, Goldfarb A, Nikiforov V. J Biol Chem. 1991;266:23927–23931. [PubMed] [Google Scholar]
- 6.Jin D J, Gross C A. J Mol Biol. 1988;202:45–58. doi: 10.1016/0022-2836(88)90517-7. [DOI] [PubMed] [Google Scholar]
- 7.Severinov K, Soushko M, Goldfarb A, Nikiforov V. Mol Gen Genet. 1994;244:120–126. doi: 10.1007/BF00283512. [DOI] [PubMed] [Google Scholar]
- 8.Heisler L M, Suzuki H, Landick R, Gross C A. J Biol Chem. 1993;268:25369–25375. [PubMed] [Google Scholar]
- 9.Severinov K, Markov D, Severinova E, Nikiforov V, Landick R, Darst S A, Goldfarb A. J Biol Chem. 1995;270:23926–23929. doi: 10.1074/jbc.270.41.23926. [DOI] [PubMed] [Google Scholar]
- 10.Borukhov S, Severinov K, Kashlev M, Lebedev A, Bass I, Rowland G C, Lim P-P, Glass R E, Nikiforov V, Goldfarb A. J Biol Chem. 1991;266:23921–23926. [PubMed] [Google Scholar]
- 11.Severinov K, Kashlev M, Severinova E, Bass I, McWilliams K, Kutter E, Nikiforov V, Snyder L, Goldfarb A. J Biol Chem. 1994;269:14254–14259. [PubMed] [Google Scholar]
- 12.Conway J F, Cheng N, Zlotnick A, Stahl S J, Wingfield P T, Steven A C. Proc Natl Acad Sci USA. 1998;95:14622–14627. doi: 10.1073/pnas.95.25.14622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Severinov K, Mooney R, Darst S A, Landick R. J Biol Chem. 1997;272:24137–24140. doi: 10.1074/jbc.272.39.24137. [DOI] [PubMed] [Google Scholar]
- 14.Polyakov A, Severinova E, Darst S A. Cell. 1995;83:365–373. doi: 10.1016/0092-8674(95)90114-0. [DOI] [PubMed] [Google Scholar]
- 15.Darst S A, Polyakov A, Richter C, Zhang G. J Struct Biol. 1998;124:115–122. doi: 10.1006/jsbi.1998.4057. [DOI] [PubMed] [Google Scholar]
- 16.Toyoshima C, Unwin N. J Cell Biol. 1990;111:2623–2635. doi: 10.1083/jcb.111.6.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Unwin N. J Mol Biol. 1993;229:1101–1124. doi: 10.1006/jmbi.1993.1107. [DOI] [PubMed] [Google Scholar]
- 18.Beroukhim R, Unwin N. Neuron. 1995;15:323–331. doi: 10.1016/0896-6273(95)90037-3. [DOI] [PubMed] [Google Scholar]
- 19.Mimori Y, Yamashita I, Murata K, Fujiyoshi Y, Yonekura K, Toyoshima C, Namba K. J Mol Biol. 1995;249:69–87. doi: 10.1006/jmbi.1995.0281. [DOI] [PubMed] [Google Scholar]
- 20.DeRosier D, Stokes D L, Darst S A. J Mol Biol. 1998;289:159–165. doi: 10.1006/jmbi.1999.2677. [DOI] [PubMed] [Google Scholar]
- 21.Hanein D, DeRosier D J. Ultramicroscopy. 1999;76:233–238. doi: 10.1016/s0304-3991(98)00086-2. [DOI] [PubMed] [Google Scholar]
- 22.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 23.Nene V, Glass R. Mol Gen Genet. 1984;196:64–67. doi: 10.1007/BF00334093. [DOI] [PubMed] [Google Scholar]
- 24.Severinov K, Mustaev A, Kashlev M, Borukhov S, Nikiforov V, Goldfarb A. J Biol Chem. 1992;267:12813–12819. [PubMed] [Google Scholar]
- 25.Deisenhofer J. Biochemistry. 1981;20:2361–2370. [PubMed] [Google Scholar]
- 26.Gouda H, Torigoe H, Saito A, Sato M, Arata Y, Shimada I. Biochemistry. 1992;31:9665–9672. doi: 10.1021/bi00155a020. [DOI] [PubMed] [Google Scholar]
- 27.Darst S A, Polyakov A, Richter C, Zhang G. Cold Spring Harbor Symp Quant Biol. 1998;63:269–276. doi: 10.1101/sqb.1998.63.269. [DOI] [PubMed] [Google Scholar]
- 28.Severinov K, Soushko M, Goldfarb A, Nikiforov A. J Biol Chem. 1993;268:14820–14825. [PubMed] [Google Scholar]
- 29.Severinov K, Mustaev A, Severinova E, Kozlov M, Darst S A, Goldfarb A. J Biol Chem. 1995;270:29428–29432. doi: 10.1074/jbc.270.49.29428. [DOI] [PubMed] [Google Scholar]
- 30.Zhang G, Campbell E A, Minakhin L, Richter C, Severinov K, Darst S A. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]