Abstract
Chlamydomonas reinhardtii is a unicellular eukaryotic alga possessing a single chloroplast that is widely used as a model system for the study of photosynthetic processes. This report analyzes the surprising structural and evolutionary features of the completely sequenced 203,395-bp plastid chromosome. The genome is divided by 21.2-kb inverted repeats into two single-copy regions of ∼80 kb and contains only 99 genes, including a full complement of tRNAs and atypical genes encoding the RNA polymerase. A remarkable feature is that >20% of the genome is repetitive DNA: the majority of intergenic regions consist of numerous classes of short dispersed repeats (SDRs), which may have structural or evolutionary significance. Among other sequenced chlorophyte plastid genomes, only that of the green alga Chlorella vulgaris appears to share this feature. The program MultiPipMaker was used to compare the genic complement of Chlamydomonas with those of other chloroplast genomes and to scan the genomes for sequence similarities and repetitive DNAs. Among the results was evidence that the SDRs were not derived from extant coding sequences, although some SDRs may have arisen from other genomic fragments. Phylogenetic reconstruction of changes in plastid genome content revealed that an accelerated rate of gene loss also characterized the Chlamydomonas/Chlorella lineage, a phenomenon that might be independent of the proliferation of SDRs. Together, our results reveal a dynamic and unusual plastid genome whose existence in a model organism will allow its features to be tested functionally.
INTRODUCTION
Plastids are a family of specialized organelles derived from an ancient endosymbiosis (Kowallik, 1994). Chloroplasts are studied primarily as the site of photosynthesis; however, they and other plastid types perform multiple and sometimes mysterious roles in organisms ranging from crop plants to the malarial parasites of mosquitoes (Köhler et al., 1997). Plastids, like mitochondria, possess remnants of their ancestral genomes, which vary considerably in size and gene content in spite of a probable single primary endosymbiosis (reviewed by Douglas, 1998). Complete plastid genome sequences were first obtained for tobacco (Shinozaki et al., 1986) and a liverwort (Ohyama et al., 1986); at present, there are 24 complete sequences representing all of the major lineages of the plant kingdom. With these sequences and the genome sequence of the cyanobacterium Synechocystis sp PCC6803 (Kaneko et al., 1995), which is related to the plastid ancestor (Palmer and Delwiche, 1996; Douglas, 1998), our ability to explore the evolution of plastid genes and functions, and chloroplast biogenesis, is at an unprecedented stage.
Sequence information has led in turn to phylogenetic comparisons, with the goal of understanding how this organelle has diverged since the primary endosymbiosis event (Martin and Herrmann, 1998). Martin and co-workers (1998) identified 45 protein-coding regions common to nine plastid genomes and Synechocystis (as an outgroup). This data set resulted in the discovery of multiple occurrences of parallel gene losses versus individual gene losses, and the analysis was extended recently to include predicted gene function and genome comparisons for 19 chloroplast DNAs (cpDNAs) (De Las Rivas et al., 2002). This approach yielded two main conclusions: first, regulatory units appear to be added to the energy-generating complexes (e.g., a group of genes encoding NADH dehydrogenase subunits); and second, a hierarchical structure develops within the plastid, resulting in a eukaryote-like genome organization (e.g., intron invasion and the presence of maturases). It has been hypothesized that organellar genomes persist to support certain regulatory mechanisms, allowing the structural proteins that balance redox potential to neutralize rapidly the side effects of altered electron transport (Race et al., 1999). Apart from strictly evolutionary implications, plastid genome sequencing has revealed new components of photosynthetic complexes (Hager et al., 1999; Swiatek et al., 2001), potential RNA editing sites (Maier et al., 1995), and coregulation of genes or gene clusters.
Although the plastid genomes of land plants are highly conserved in both sequence and structure (Wakasugi et al., 2001), algal plastid chromosomes exhibit tremendous variation that can be used to gain new insights into their evolution (Simpson and Stern, 2002). A good example is how the basal phylogeny of the Viridiplantae has been restructured based on whole chloroplast genomes. Data from the green algae Nephroselmis olivacea and Mesostigma viride revealed an early branch in the evolution of land plants (Turmel et al., 1999; Lemieux et al., 2000), placing Mesostigma sister to a clade containing both the chlorophytes (noncharophytic green algae) and streptophytes (land plants) and suggesting that Mesostigma represents a basal lineage that predates the separation of the chlorophytes and streptophytes. However, a more recent phylogeny, including many charophytic green algae but shorter sequences and different analytical approaches, placed Mesostigma basal to the streptophyte but not the chlorophyte lineage (Karol et al., 2001). Clearly, these evolutionary hypotheses would benefit from additional sequence information.
Verification of evolutionary and functional data ultimately requires, in many cases, manipulation of the plastid genome. Plastid transformation has been performed largely in tobacco and Chlamydomonas reinhardtii, enabling mechanistic insights to be gained, the expression of foreign proteins, and reverse genetic studies to be performed within the plastid. Chlamydomonas also is a key model for the study of photosynthesis, cell motility, and stress responses (Dent et al., 2001; Harris, 2001). Until now, the sequence of the Chlamydomonas chloroplast genome has not been fully assembled, in spite of numerous fragments deposited into the databases during the past 20 years. As part of a major Chlamydomonas genomics effort (Grossman, 2000), we have completed its sequence and used genome-wide analytical tools to reveal several uncharacteristic features, among them a preponderance of short dispersed repeats, a relatively low number of coding regions, and an atypical organization of the genes encoding the RNA polymerase. In the accompanying article (Lilly et al., 2002), we used this information to complete a genome-wide analysis of the expressed regions and to examine how the transcriptome responds to abiotic stimuli. The complete cpDNA sequence and its analysis, together with genome-wide expression studies, present challenging evolutionary enigmas and open new doors for functional analyses.
RESULTS
Genome Sequence, Content, and Organization
When this project was initiated, ∼120 kb of sequence was deposited in the databases, and 30 kb was known to exist as unpublished data (see supplemental data online). We elected to take a three-pronged strategy to complete the assembly. First, sequences published in peer-reviewed articles were taken at face value, except where we had indications from colleagues that they might not be fully accurate or where ambiguous positions had been deposited as such (particularly at the ends of sequence reads). Where multiple versions of genes or regions existed, these were aligned, and primer walking was used to resolve any disagreements. Second, we accepted unpublished sequences but performed single-pass sequencing to assess their overall quality. Third, we identified 14 gaps and used plasmid subcloning or PCR to generate the required fragments. All clones were obtained from the Chlamydomonas Genetics Center at Duke University, so that our sequence, of which 60 kb was newly obtained, would be congruent with previous submissions and with data to be obtained in the future by individual investigators (see supplemental data online). The complete sequence was made available on the Chlamydomonas genome project World Wide Web site (http://www.biology.duke.edu/chlamy_genome/chloro.html) in August 2001; a final assembly has been released to NCBI, in addition to the newly sequenced regions (see Methods), and we have created an interactive chloroplast genome/gene expression World Wide Web site (http://bti.cornell.edu/bti2/chlamyweb).
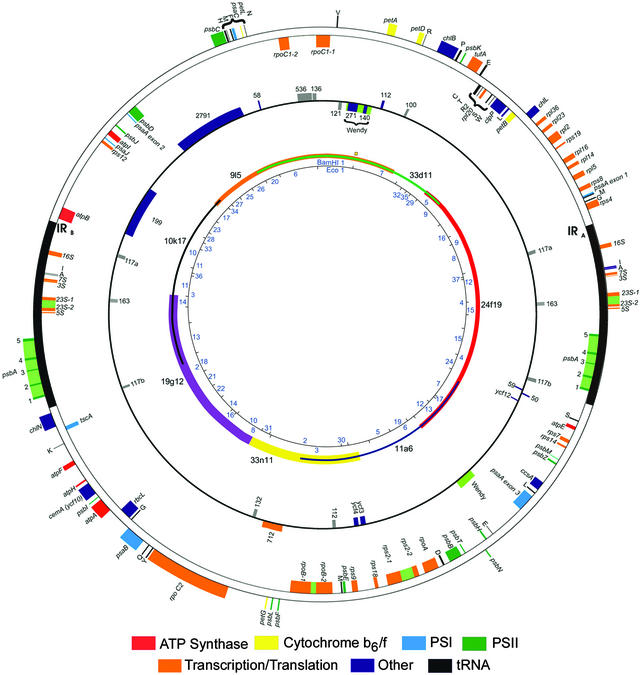
Our final assembly revealed a circular map of 203,395 bp (Figure 1), slightly larger than earlier estimates based on restriction fragment mapping (Rochaix, 1980). Position 1 was defined as the BamHI restriction site located within ORF271 of the “Wendy” element at the boundary between the plasmid clones Bam1 and Bam5 (Figure 1, orange square). The genome is 34.6% G+C with an average intergenic spacer of 0.7 kb, based on the gene definitions given below. The G+C content is comparable to that of Chlorella (31.6%) and Arabidopsis (36.3%). The genome possesses two copies of an inverted repeat sequence (22,211 bp; Figure 1, outer circle), which are separated by two nearly equally sized unique regions of 80,873 and 78,100 bp. The gene arrangement within the inverted repeat is typical of land plants, except that the 23S rRNA gene is divided by an intron (Rochaix and Malnoe, 1978) and the psbA gene is divided by four introns (Erickson et al., 1984). Some other Chlamydomonas species exhibit minor differences; for example, rbcL is found within the inverted repeat in C. moewusii (Boudreau et al., 1994), and atpB is found in the inverted repeat in C. gelatinosa (Boudreau and Turmel, 1996). Large insertions into the inverted repeat have expanded it substantially in C. eugametos and C. moewusii, without affecting gene content.
Figure 1.
The Plastid Chromosome of C. reinhardtii.
The inner circle shows BamHI and EcoRI restriction fragments mapped according to Rochaix (1980) and numbered according to Grant et al. (1980). Position 0 is shown by an orange square near the 12 o'clock position. The second concentric circle indicates seven overlapping BAC clones that span the genome. The third circle shows genes and ORFs of unknown function, including those for which disruption experiments were unsuccessful. Gray boxes represent newly identified ORFs. The outer circle shows genes of known or presumed function, with sequenced or hypothesized (rps2; see text) introns shown in olive green. Genes are color coded by function, as shown at bottom.
Genes contained in C. reinhardtii cpDNA are listed in Table 1. Although open reading frame (ORF) classifications are inevitably somewhat arbitrary, under the criteria used here, the number in C. reinhardtii is smaller than that in most other green algae or land plants (Simpson and Stern, 2002). Using a threshold of 225 bp for previously unidentified reading frames (see Methods for reasoning), 18 new ORFs were found (Table 1), but none was found to be expressed, based on RNA gel blot analysis (Lilly et al., 2002). The genome contains 72 bona fide protein-coding genes, of which only rpl36 had not been found previously, as well as 4 genes reported previously based on heterologous hybridization, for which we verified the locations. Three encode RNA polymerase components and one appears to encode a portion of rps2; these are discussed in more detail below because they differ from previous annotations. Thirty tRNA genes were identified (see Methods), giving a complete set for translation and an ultimate total of 99 expressed sequences (not including genes duplicated in the inverted repeat). This is the smallest plastid gene repertoire reported in the photosynthetic Viridiplantae and contrasts with 127 chloroplast genes in the green alga N. olivacea (Turmel et al., 1999) and 251 in the most gene-rich plastid DNA, that of the red alga Porphyra purpurea (Reith and Munholland, 1995).
Table 1.
Gene List for the Chlamydomonas reinhardtii Plastid Chromosome
| RNA Genes | |||
|---|---|---|---|
| Ribosomal RNAs | 23S rDNAa,b | 16S rDNAb | 7S rDNAb |
| 5S rDNAb | 3S rDNAb | ||
| Transfer RNAs | trnA (UGC) | trnA (UGC) | trnG (UCC) |
| trnC (GCA) | trnD (GUC) | trnE (UUC) | |
| trnE1 (UUC) | trnE2 (UUC) | trnF (GAA) | |
| trnG (GCC) | trnH (GUG) | trnI (GAU) | |
| trnI (GAU) | trnK (UUU) | trnL (UAA) | |
| trnL (UAG) | trnM (CAU) | trnM (CAU) | |
| trnM (CAU) | trnN (GUU) | trnP (UGG) | |
| trnQ (UUG) | trnR (ACG) | trnR2 (AGA) | |
| trnS (UGA) | trnS (GCU) | trnT (UGU) | |
| trnV (UAC) | trnW (CCA) | trnY (GUA) | |
| Small RNAs | tscA | ||
|
Protein-Coding Genes
|
|||
| Photosynthesis | |||
| Photosystem I | psaAa | psaB | psaC |
| psaJ | |||
| Photosystem II | psbAa,b | psbB | psbC |
| psbD | psbE | psbF | |
| psbH | psbJ | psbK | |
| psbL | psbM | psbN | |
| psbT | psbZ | ||
| Cytochrome b6/f | petA | petB | petD |
| petG | petL | ||
| ATP synthase | atpA | atpB | atpE |
| atpF | atpH | atpI | |
| Chlorophyll biosynthesis | chlB | chlL | chlN |
| Rubiscoc | rbcL | ||
| Ribosomal Proteins | |||
| Large subunit | rpl2 | rpl5 | rpl14 |
| rpl16 | rpl20 | rpl23 | |
| rpl36 | |||
| Small subunit | rps2d | rps3d | rps4 |
| rps7 | rps8 | rps9 | |
| rps12 | rps14 | rps18 | |
| rps19d | |||
| Transcription/translation | |||
| RNA polymerase | rpoAd | rpoBa | rpoC1ad |
| rpoC1bd | rpoC2 | ||
| Translation | tufA | ||
| Miscellaneous proteins | cemA | clpP | ccsA |
| Conserved proteins | ycf3 | ycf4 | ycf12 |
|
C. reinhardtii–Specific ORFs
|
|||
| Previously identified | ORF50 | ORF58 | ORF59 |
| ORF112 | ORF140 | ORF271 | |
| ORF1995 | ORF2971 | ||
| Newly identifiede | URF1 (100) | URF3 (162) | URF4 (111) |
| URF5 (117) | URF6 (163) | URF7 (117) | |
| URF8 (192) | URF9 (314)
|
URF12 (112) | |
| URF13 (208) | URF14 (111) | URF15 (117) | |
| URF16 (111) | URF17 (163) | URF18 (171) | |
| URF19 (266) | URF21 (536) | URF22 (136) | |
| URF24 (121) | |||
Intron containing gene.
Two copies due to inverted repeat.
Ribulose-1,5-bisphosphate carboxylase/oxygenase.
Limited similarity based on BLAST analysis.
Unidentified open readings labeled by consecutive order in genome; not shown in Figure 1. The predicted number of amino acids is in parentheses.
Short Dispersed Repeats Saturate Intergenic Regions
Small repeated sequences were first identified in the C. reinhardtii chloroplast using renaturation kinetics and other methods and were estimated to constitute 4 to 7% of the genome (Gelvin and Howell, 1979). Several studies also revealed repeats that were present in sequences flanking genes of interest (Dron et al., 1982; Schneider et al., 1985; Boynton et al., 1992; Fong and Surzycki, 1992b; Leu, 1998). These elements, which average 30 bp in length, often are marked by AatII and/or KpnI restriction sites. Because of their characteristics, we refer to them as short dispersed repeats (SDRs).
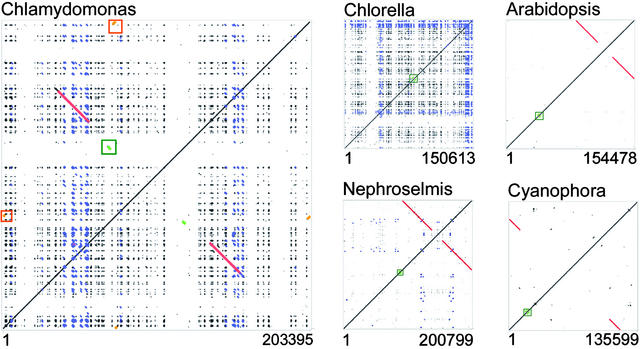
We used genome-wide dot-plot analysis to examine the extent of SDRs in Chlamydomonas and compared the results with those from several other sequenced chloroplast genomes, as shown in Figure 2. The dot plot is a visual representation of alignments generated by PipMaker, which uses a version of the BLAST (Basic Local Alignment Search Tool) algorithm (Schwartz et al., 2000). The self-comparisons in Figure 2 create diagonals of identity, with either side of the diagonal consisting of mirror images in which dots or short lines indicate regions of sequence similarity. The left panel shows results for Chlamydomonas, with the red lines perpendicular to the diagonal of identity representing the 22.1-kb inverted repeats. The orange boxes frame the duplicated Wendy-disabled transposon (Fan et al., 1995), and the green box highlights the related psaA and psaB genes encoding photosystem I apoproteins. Apart from these landmarks, a very large number of repeated DNA sequences are widely dispersed. The spaces between clusters of repeats are, in effect, the locations of genes. Dot plots for other selected plastid genomes are shown at right. In each of these cases (except Chlorella vulgaris), the rDNA-containing inverted repeat is shown as a red line, with its apparent location depending on where the genome was linearized for analysis. Chlorella, and to a lesser extent Nephroselmis, exhibit repetitive sequences that clearly are above the background level seen in Cyanophora paradoxa, Arabidopsis, and all other fully sequenced streptophytes (data not shown). The blue SDRs are those not within protein-coding sequences that are conserved between Chlamydomonas and the other species. For the most part, these homologies are restricted to a small number of SDRs in or near the inverted repeat (the remaining SDRs are shown in black). Surprisingly, the massive number of SDRs in C. vulgaris cpDNA was not reported along with the sequence (Wakasugi et al., 1997), although, as for Chlamydomonas, an earlier study had found some by chance in the inverted repeat region of a related Chlorella species (Yamada, 1991).
Figure 2.
Whole Chloroplast Genome Self-Comparison Dot Plots Generated by PipMaker.
Dots off the main diagonal represent alignments with >50% identity. Structural features off the main diagonal are color coded as follows: red, inverted repeats; blue, small dispersed repeats in C. reinhardtii that also are detected in C. vulgaris and N. olivacea; green, psaA/psaB genes; orange, Wendy elements. Accession numbers for the cpDNA sequences are given at the end of Methods.
We estimate that 19,500 SDRs are present in C. reinhardtii cpDNA, depending on the parameters used (J.E. Maul, unpublished data). BLAST analyses gave an approximate grouping of the repeats into ∼1000 classes, which were dispersed throughout the genome; this can be seen by following a vertical column in the dot plot (Figure 2). To further explore the SDRs, we compiled all noncoding repetitive sequences of >20 bp extracted from multiple BLAST alignments. These short elements were aligned and the resulting consensus sequences derived. Table 2 lists the consensus sequences and statistics for the 10 most prevalent SDRs, which together account for >10 kb (∼5%) of the plastid chromosome. An example of the limited amount of SDR sequence divergence is found in SDR9. Its exact consensus sequence is present in 36 locations; however, when the stringency was decreased to 90% (allowing three mismatches), an additional 15 copies were identified. The high level of sequence conservation suggests some type of functional role, a recent evolutionary origin, and/or frequent copy correction.
Table 2.
Most Highly Conserved Simple Dispersed Repeats Present in the C. reinhardtii Plastid Chromosome
| SDR No. | Size (bp) | Sequence a | Similarity b | Occurrence c | G+C (%) | Pyrimidines (%) |
|---|---|---|---|---|---|---|
| SDR1 | 26 | CTGCCTCCTTCCCCTTCCCATTCGGG | 90 | 48 | 65 | 80 |
| SDR2 | 27 | ATATAAATATTGGGCAAGTAAACTTAG | 90 | 28 | 26 | 37 |
| SDR3 | 33 | ATAAACTTTAGTTGCCCGAAGGGGTTTACATAC | 90 | 56 | 39 | 48 |
| SDR4 | 25 | AGGACAAATTTATTTATTGTGGTAC | 90 | 19 | 28 | 48 |
| SDR6.1 | 31 | GGACGTC AGTGGCAGTGGTACCGCCACTGCC | 90 | 37 | 68 | 48 |
| SDR6.2 | 23 | GTGGCAGTGGTACCGCCACTGCC | 90 | 68 | 70 | 52 |
| SDR7 | 33 | TCCACTAAAATTTATTTGCCGAAGGGGACGTCC | 90 | 51 | 45 | 51 |
| SDR8 | 32 | TAGGCAGTTGGCAGGCAACTGCACTGACGTCC | 90 | 34 | 59 | 46 |
| SDR9.1 | 24 | CTGCCAACTGCCGATATTTATATA | 100 | 36 | 37 | 58 |
| SDR9.2 d | 24 | CTGCCAACTGCCGATATTTATATA | 90 | 51 | 37 | 58 |
Underlined bases are AatII (GACGTC) and KpnI (GGTACC) restriction sites.
Percentage divergence among those sequences that share similarity based on BLAST analyses.
Number of copies in the full genome sequence.
If stringency is decreased to 90%, allowing three mismatches, an additional 15 copies of this repeat are detected.
We also performed BLASTn analyses to explore the similarity of SDR sequences to coding regions of Chlamydomonas cpDNA and those of 13 other species. One member of each of the 1000 repeat classes was used in this analysis. Many short (10- to 15-bp) identities were found between SDR sequences and various coding regions, but given the large number of sequence comparisons, none of these exceeded even a lenient significance cutoff of e−4. This approach was extended to search the nucleotide sequence of each genome and the entire GenBank database. One SDR (class 10) was found to share two palindromic 17-bp identities with a noncoding portion of the rice chloroplast genome (positions 55764 to 55780). The region in the rice plastid genome has been shown to be a mutational “hot spot” and gene conversion site (Morton and Clegg, 1993).
SDRs Are Polymorphic between C. reinhardtii Strains
During sequence assembly, we encountered minor differences between new data and previously published versions, which were derived from various laboratory strains and not the cpDNA plasmid collection at Duke University. This caused us to question whether SDR regions might exhibit polymorphisms, particularly if replication slippage, which is known in chloroplasts (Chang et al., 1996), was a common phenomenon in Chlamydomonas, which is typically maintained as a haploid vegetative culture. We analyzed five intergenic regions and the rbcL coding region from three representative strains in the C. reinhardtii 137c background (CC-125, CC-406, and CC-620), which share a common ancestor probably in the 1960s (and ultimately trace back to an isolate from ), and strain S1D2 (CC-2290; Gross et al., 1988), which was isolated in Minnesota in the 1980s. Two regions (ORF140-petA and ORF112-petD) failed to amplify in CC-2290 and CC-125, suggesting altered sequences at the priming sites, and were eliminated from the analysis. For those that did amplify, at least three independently cloned PCR products were sequenced. Single nucleotide polymorphisms (SNPs) were detected in all four sequenced regions, with a total fraction of polymorphic sites ranging from 0.4% (chlL-rpl23) to 0.9% (petD-chlB and psbZ-ccsA; Table 3). The intergenic regions all were polymorphic among the three 137c strains, implying the accumulation of SNPs within the 30 to 40 years since they shared a common ancestor. Additional polymorphisms distinguishing 137c from S1D2 could have accumulated under culture conditions or might be a reflection of existing variation between these independently isolated strains. By contrast, the rbcL coding region was uniform among the three 137c strains, with six SNPs separating the 137c and S1D2 strains. Thus, there appears to be a lower observable rate of SNP accumulation for this constrained gene sequence than for the repeat-rich intergenic regions. These results confirm the occurrence of plastid SNPs among C. reinhardtii strains and suggest that differences in cpDNA restriction fragment patterns might be anticipated. Our limited analysis also suggests that the SDR regions are not particularly unstable (e.g., prone to indels), at least over a short period.
Table 3.
Sequence Variation across Four C. reinhardtii Strains
| Percentage Polymorphic Sites
|
|
|||||
|---|---|---|---|---|---|---|
| Region | Size | Number of SNPs | Among 137c Isolates | (%) | 137c versus CC-2290 | (%) |
| rbcL | 932 | 6 | 0.0 | 0.7 | ||
| petD-chlB | 220 | 2 | 0.4 | 0.4 | ||
| chlL-rpl23 | 1205 | 6 | 0.2 | 0.4 | ||
| psbZ-ccsA | 755 | 7 | 0.7 | 0.2 | ||
The Chloroplast Genome Exists as Both Circular and Linear Molecules
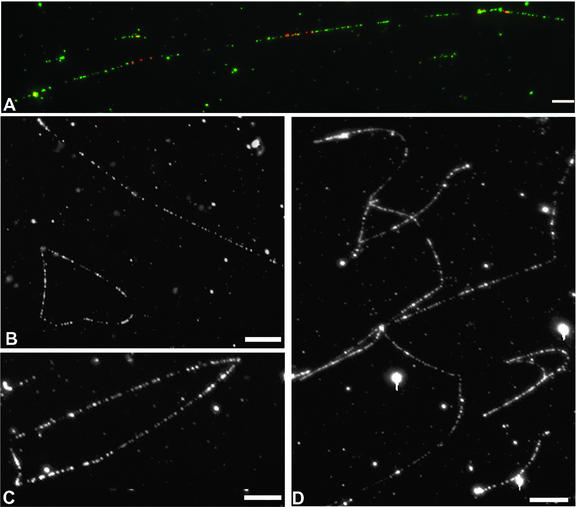
To gain insight into the structure of the Chlamydomonas plastid chromosome, a cytogenomic approach was used. It was already known that Chlamydomonas cpDNA, like other chloroplast genomes, can undergo “flip-flop” recombination between the large inverted repeats, leading to equimolar accumulation of these isomers (Palmer et al., 1985). We wished to determine whether the genome existed as monomers or higher order molecules and whether the DNA was primarily circular or linear in vivo. Previous analyses have used gel-based methods and/or microscopy of isolated cpDNAs (Bendich and Smith, 1990; Bendich, 1991). Pulsed-field gel electrophoresis of intact Chlamydomonas chloroplasts or whole (cell wall–deficient; cw15) cells embedded in agarose, followed by filter hybridization (data not shown), did not reveal the higher order organization that was observed previously for higher plant cpDNAs (Deng et al., 1989; Backert et al., 1995; Lilly et al., 2001), suggesting a lack of discrete genome conformations. In a second approach, shown in Figure 3, intact chloroplasts were spread on slides to obtain cpDNA fibers, which then were hybridized with BACs spanning the chloroplast genome (Figure 3A, green). To give landmarks, a second labeling procedure (Figure 3A, red) was performed with plasmids specific to the inverted repeat. Numerous observations revealed a predominance of small linear DNA fibers, many of which were <100 kb in size (Figures 3B to 3D). However, circular genomes also were identified in many different plastid isolations, and these included clearly monomeric (Figure 3B) and dimeric (Figure 3C) forms. Unlike higher plant cpDNAs, few molecules larger than trimer size were present, and all of these were linear. Genomes greater than monomer size could result directly from replication or could be diagnostic of intergenomic recombination. Because of the methods used here, breakage cannot be excluded as a source of some linear fibers. However, based on pulsed-field gel electrophoresis data, it seems reasonable to conclude that the Chlamydomonas plastid genome exists principally as a population of monomeric and dimeric linear and circular genomes.
Figure 3.
Cytogenomic Analyses of C. reinhardtii cpDNA.
(A) Linear cpDNA fiber with two genome copies. The green signal represents the entire plastid genome, and the three red signals result from the hybridization of clones P-14 and P-16, which span a 10-kb portion of the inverted repeat.
(B) Two monomeric cpDNA fibers visible in the same field of view.
(C) Chloroplast DNA dimer of ∼180 μm.
(D) A field showing various linear cpDNA fibers.
Bars = 10 μm.
Chlamydomonas cpDNA-Specific Features Visualized via Multiple Genome Analysis
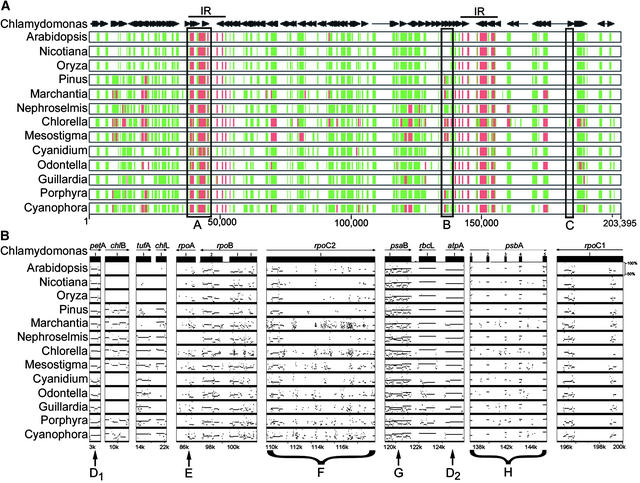
MultiPipMaker (http://bio.cse.psu.edu) is a new, World Wide Web–based tool for multiple genome analysis based on the PipMaker program for comparing two genome sequences (Schwartz et al., 2000). We applied it to simplify and enhance the arduous task of verifying and correcting initial gene annotations and to compare gene content and overall sequence similarity among 14 complete plastid genomes representing all of the major Viridiplantae lineages. The MultiPip overview is shown in Figure 4A, and selected features are detailed in Figure 4B (for complete results, see supplemental data online). MultiPipMaker works by keeping a single, reference genome in its actual linear order (Chlamydomonas genes are shown by arrows at the top of Figure 4A) and then aligns the additional genome sequences to the reference, irrespective of gene order. A genome-wide output shows global similarity, with red representing the highest similarity and green representing lower similarity. As an example, the highly conserved and ubiquitous rRNA genes are depicted in Figure 4A, box A. MultiPip also allows one to examine the presence or absence of a gene in different species (although not their relative order). As examples, boxes B and C show genes that are conserved in approximately half and only one of the selected genomes, respectively, and in the case of box B, with different amounts of similarity. Note that if a sequence is not present in the reference genome, it is not seen in the lower lines. Although Chlamydomonas is “gene poor,” ORFs specific to the chlorophyte lineage clearly are identified by this approach.
Figure 4.
MultiPipMaker Analysis of 14 Sequenced Plastid Genomes, Illustrating Similarities and Differences between Chlamydomonas and Other Photosynthetic Plastid Genomes.
(A) The reference genome, C. reinhardtii (top), was analyzed against 13 other species. Genes and their orientations are indicated by horizontal arrows, and the large inverted repeat (IR) is shown as a line. Positions on the linearized genome are shown across the bottom. The species for aligned genomes are shown at left, with accession numbers given at the end of Methods. Where alignments with the Chlamydomonas sequence were made, they are shown as 50 to 75% identity (green) or 75 to 100% identity (red). The boxes labeled A, B, and C denote features discussed in the text.
(B) Selected regions were extracted from the analysis shown in (A), with aligned regions displayed graphically as horizontal lines drawn to show average percentage identity between 50 and 100% (at right of Arabidopsis plot). Positions of the selected sequences in the Chlamydomonas chloroplast genome are shown at bottom. The genes or braced areas labeled D to H denote features discussed in the text.
Figure 4B illustrates features that can be examined by “zooming in” on parts of Figure 4A. In this case, gap-free sequence alignments are plotted as a series of lines with sequence identity between 50 and 100% (right side of the plot). Duplicated segments are displayed as multiple lines or dots at the appropriate levels of similarity, and completely identical sequences are displayed as superimposed lines.
Arrows D1 and D2 represent genes that are highly conserved along their entire lengths, although the percentage of identity is not constant. This type of discontinuity can be indicative of functional domains. Arrow E (rpoA) illustrates the opposite case, in which similarity is poor and intermittent. This can be interpreted as a gene with relatively few constrained domains, the presence of unknown introns, or other nonhomologous sequences and/or pseudogenes. Brace F highlights rpoC2, in which evidence of homology is intermittent and ranges from poor to good. With this type of alignment, one might suspect the presence of introns, a much clearer case of which is seen with psbA (brace H), in which the coding region is highly conserved. Finally, simple gene duplication is seen for psaB (arrow G), with which the psaB genes of other species give a high-similarity alignment and the related psaA gene aligns intermittently and at a lower similarity. The reciprocal plot is not seen (psaB aligning to psaA), because psaA is split into three distantly located exons in Chlamydomonas (Kück et al., 1987) (Figure 1). This feature is quite interesting because it is the only clear case for gene duplication other than the inverted repeat, and the psaA-psaB tandem repeat feature is more conserved across plastid genomes than the often-lost inverted repeat. Among sequenced plastid genomes, only Chlamydomonas has experienced mutations that have separated the copies of psaA and psaB to distant locations. Limited similarities among intergenic regions were identified between Chlamydomonas, Chlorella, and Nephroselmis (see supplemental data online).
Accelerated Protein Evolution and Gene Loss among the Chlorophyceae
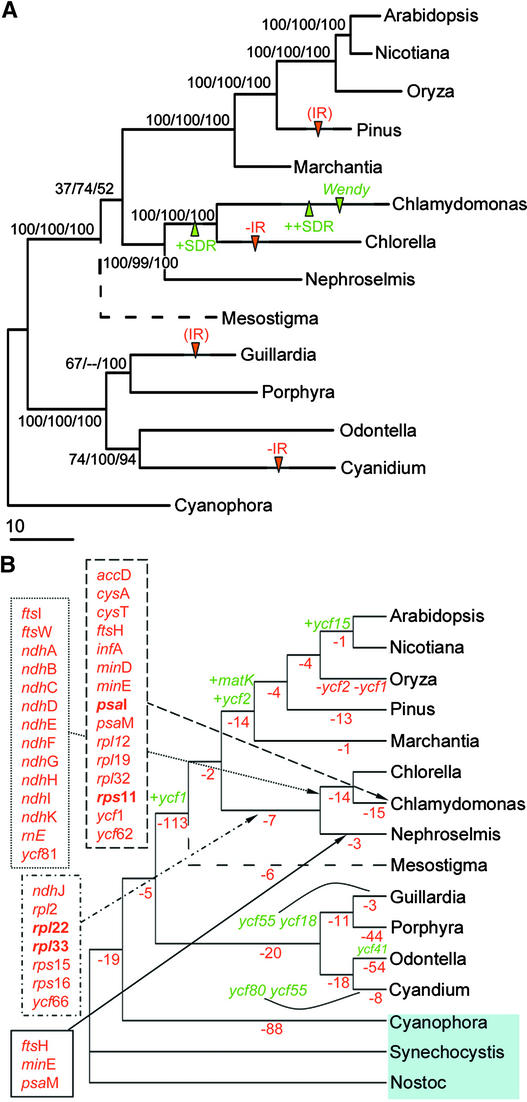
Using a set of 8856 concatenated amino acid characters and various phylogenetic analyses, we placed Chlamydomonas reinhardtii into a phylogeny based on 39 coding regions common to 14 photosynthetic plastid genomes, using the gene-rich C. paradoxa cyanelle genome as the outgroup (Figure 5; see also supplemental data online). The topology generated is congruent with the results of phylo-genetic analysis of proteome data sets for fewer species (Martin and Herrmann, 1998) and similar to those from multivariate statistical analyses of gene presence/absence data (De Las Rivas et al., 2002). Chlorophytes (C. reinhardtii, N. olivacea, and C. vulgaris) are strongly resolved as a monophyletic group, as are the land plants and a clade containing the nongreen algae (rhodophytes [Cyanidium caldarium and P. purpurea]) plus algae with plastids from secondary symbiosis (Odontella sinensis [diatom, a chrysophyte] and Guillardia theta [a cryptophyte]) (McFadden, 2001). All branches in this phylogeny are supported strongly with the exception of the placement of Mesostigma (Figure 5, dashed line) and the arrangements among the nongreen algae.
Figure 5.
Plastid Genome Phylogeny with Changes in Gene Content and Structural Features for 14 Fully Sequenced Plastid Genomes and 2 Cyanobacterial Genomes.
(A) Maximum likelihood phylogeny for 39 concatenated proteins totaling 8856 amino acids after removal of gaps and regions of ambiguous alignment. The tree is drawn with branch lengths reconstructed under maximum likelihood with the JTT model. Bootstrap support values are shown for nodes with 50% or greater support in maximum parsimony/neighbor joining/maximum likelihood RELL BP analyses. The dotted branch leading to Mesostigma indicates the uncertainty of the branching position for this taxon (see text). Arrows highlight noteworthy structural changes, including loss (−IR) or near loss (IR) of one copy of the inverted repeat, invasion by SDRs (+SDR) and their further proliferation (++SDR), and invasion by the Wendy element in Chlamydomonas.
(B) Unambiguous gene losses (in red) and gains (in green) on each branch, assuming that a gene with a detected homolog in cyanobacterial outgroups (Synechocystis or Nostoc; shaded) was present in the ancestral plastid genome. Each gene gain is indicated; specific gene losses are shown for green algal branches only. Losses inferred to have occurred only once throughout the plastome phylogeny are shown in boldface; all other losses are inferred in multiple plastome lineages (see supplemental data online for mapping of all gene changes on every branch).
The Chlamydomonas genome appears on a long branch in the chlorophytes, suggesting increased substitution rates for amino acids (Figure 5A). We first compared the likelihood of the tree with no molecular clock (ln L = 113,742.2) with that obtained with an enforced molecular clock (ln L = 124,509.4). The likelihood ratio was significant (P << 0.001), and global rate homogeneity was rejected. As a follow-up, we performed a relative rate test for the three green algal taxa (Chlorella versus Chlamydomonas with outgroup Nephroselmis) using HY-PHY. Homogeneity of rates was rejected (P = 4e-6). Thus, the amino acid substitution rates are heterogeneous for different lineages in this data set, especially within the Chlorophyceae, in which the rate for the Chlamydomonas branch is significantly higher than that for Chlorella.
The phylogram shown in Figure 5A also shows the gain of SDRs, the Wendy putative transposon, and the evolution of the large inverted repeat. The inferred loss (or major reduction) of the inverted repeat has occurred four times under this scenario, including independent complete losses from the green alga Chlorella and loss or near loss from the nongreen algae Cyanidium and Porphyra. The current phylogeny suggests that the loss of the inverted repeat from Cyanidium and the nearly complete loss from Porphyra were independent events. Alternatively, shared inverted repeat reductions in Cyanidium and Porphyra would be consistent with an alternative branching pattern, as reported by De Las Rivas et al. (2002). The Chlorophyceae clade is supported not only by protein-coding sequences (bootstrap values of 99 or greater) but also by the invasion and increasing amount of repetitive sequences (Figure 5A, SDRs).
We used parsimony to determine the number of unambiguous gene gain (green) and gene loss (red) events on the phylogeny using the extant chloroplast ancestors Synechocystis sp PCC6803 and Nostoc sp PCC7120 as outgroups (Figure 5B, blue box). Gene gains and losses were treated as equally probable except for genes also present in cyanobacteria, which were assumed to be ancestrally present in plastid genomes (Martin and Herrmann, 1998). Complete mapping of gene gains and multiple loss events is provided in the supplemental data online. Figure 5B shows massive gene loss across the plant lineage but highlights the fact that occasional functional gene gains also have occurred, presumably as a result of gene duplication and/or horizontal transfer (Palmer and Delwiche, 1996; Palmer, 1997). The frequency of these 9 putative gene gain events is extremely low compared with the overall genome history-wide 427 inferred loss events. Given this extraordinary bias toward gene loss, correct reconstruction of the phylogeny based on gene content is unlikely. However, both gene content and inverted repeat distributions would be consistent with monophyly of Cyanidium + Porphyra and Odontella + Guillardia, a hypothesis not supported by the concatenated protein data set.
Gene losses are observed from a wide range of functional categories, including photosystem I (psaI and psaM), small (rps11, rps15, and rps16) and large (rpl2, rpl12, rpl19, rpl22, rpl32, and rpl33) ribosomal subunit proteins and translation initiation factor 1 (infA), the full set of ndh genes, and numerous miscellaneous proteins and conserved ORFs. The lineage leading to C. reinhardtii is notable for an apparent acceleration of gene loss, with 29 losses relative to genomes more distant than Chlorella. This includes 10 of 11 ndh genes as well as diverse other proteins. Surprisingly, no additional gene losses were inferred in Chlorella, whereas gene loss has continued in Chlamydomonas, with 15 additional genes lost since the separation of the Chlamydomonas and Chlorella lineages.
This concatenated protein alignment generated additional information about the expressed regions of Chlamydomonas. When the 39 coding regions were aligned, 15 genes in Chlamydomonas were shown to possess differences in com-pletely conserved residues among the 15 additional species (see supplemental data online), some of which could be accounted for, in principle, by C-to-U RNA editing, which is well known in a range of species (Bock, 2000). Four of these instances (in rbcL, atpB, psbB, and psbD) were investigated, but there were no differences between the cDNA and genomic sequences, which supports the general belief that, unlike those of higher plants, Chlamydomonas cpRNAs do not undergo editing (Barkan and Goldschmidt-Clermont, 2000).
Evidence for Unusual RNA Polymerase Gene Organization
The plastid-encoded RNA polymerase (PEP) resembles that of Escherichia coli, whose core RNA polymerase is encoded by rpoA, rpoB, and rpoC. In chloroplast genomes and cyanobacteria, rpoC is divided into two separate genes/proteins, rpoC1 and rpoC2, with rpoC1 sometimes containing an intron (Downie et al., 1996). In Chlamydomonas, a location for rpoA had been reported based on heterologous filter hybridization (Watson and Surzycki, 1983); however, we found no evidence of homology with other rpoA genes in that region. On the other hand, an ORF with high similarity to rpoA was identified in a previously uncharacterized region of the genome (between psbB and rps2; Figure 1). The rpoB gene is a single ORF in other chloroplast genomes; however, it was reported as two closely linked ORFs in C. reinhardtii (Fong and Surzycki, 1992a). For the normally divided rpoC gene, rpoC2 but not rpoC1 had been found in Chlamydomonas (Fong and Surzycki, 1992a). In addition, the rpoC2 homology was embedded in a much longer ORF that otherwise lacked similarity to any other gene.
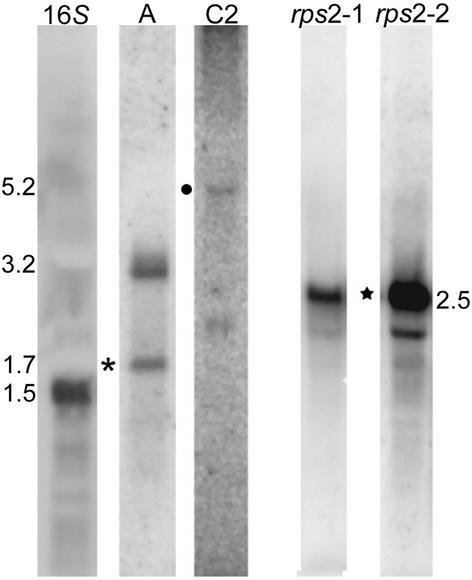
Because transcriptional activity would be evidence that bona fide PEP-encoding genes had been found, RNA filter hybridization (Figure 6) and reverse transcriptase–PCR (RT-PCR) (Figure 7) were performed. In the case of rpoA, both revealed expression, with the gel blots identifying 1.7- and 3.2-kb transcripts in wild-type cells grown under optimum conditions. The shorter of these transcripts (Figure 6, asterisk) could encode the entire predicted protein of 553 amino acids. The N terminus of rpoA aligns fairly well with other rpoA sequences and has 43% identity to the most closely related sequence (Figure 8A); however, substantial divergence occurs toward the C terminus. In addition, homology is interrupted throughout the ORF by insertions in the Chlamydomonas sequence relative to all other organisms except Nephroselmis and to some extent Chlorella. Interestingly, these are the two other plastid genomes with significant numbers of SDRs, and this may be another indication of a closer relationship within the green algal chloroplasts.
Figure 6.
RNA Gel Blot Analysis for Selected PEP Genes and rps2.
Total RNA (40 μg) from CC-125 cells grown in continuous light was analyzed by filter hybridization using 32P-labeled probes derived by PCR from rpoA and rpoC2 (A and C2, respectively). Probes corresponding to the rps2 ORFs were used in the lanes marked rps2-1 (previously ORF570) and rps2-2 (ORF208). Individual transcript sizes are noted and were estimated by comparison with RNA markers (Invitrogen) and the 16S rRNA hybridization control (16S). The symbols mark transcripts that are discussed in the text, generally those considered most likely to encode the protein product based on size or on hybridization with two probes from the same gene.
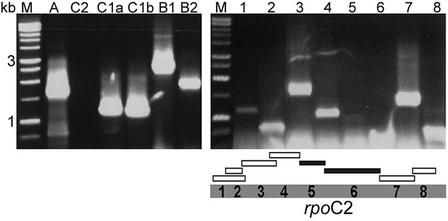
Figure 7.
RT-PCR for Genes That Encode PEP Subunits.
Approximately 1 μg of total RNA from light-grown CC-125 cells was subjected to a 1-h reverse transcription reaction, followed by 30 cycles of PCR using gene-specific primers (Lilly et al., 2002). At left, RT-PCR products for an internal region of each PEP gene. Lanes are as follows: M, molecular size markers; A, rpoA; C2, rpoC2; C1a, rpoC1a; C1b, rpoC1b; B1, rpoB1; and B2, rpoB2. At right, RT-PCR products spanning the rpoC2 gene. Lanes correspond to regions of the 3119–amino acid rpoC2 ORF, as indicated by the diagram below the gel. Open bars represent regions that were amplified by RT-PCR, and closed bars represent regions not amplified by RT-PCR.
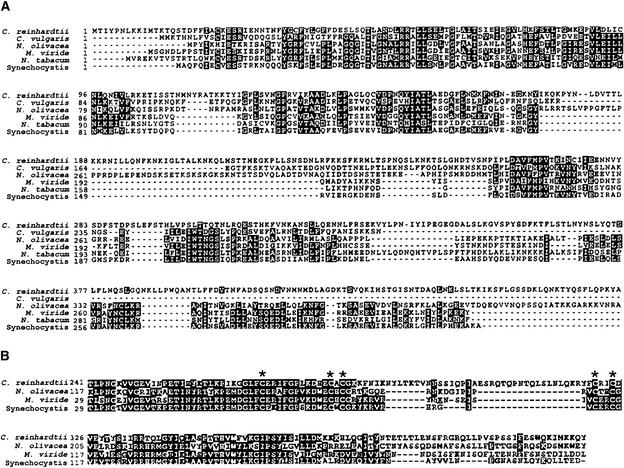
Figure 8.
BOXSHADE Alignments of Newly Identified PEP Coding Regions.
(A) Multiple sequence alignment of the translated rpoA sequence of C. reinhardtii with the five most similar peptide sequences, as determined by tBLASTX analysis.
(B) A section of the multiple sequence alignment of the translated rpoC1 sequence from C. reinhardtii with those of other related species. This excerpt highlights the zinc binding domain (conserved Cys residues are denoted by asterisks), which is found in most prokaryote-like rpoC1 proteins and implicated in transcription termination.
Full names and accession numbers are given at the end of Methods.
As mentioned above, rpoB was suggested to have an unusual structure with two separately transcribed exons (B1 and B2), yet it was predicted not to have an intron based on the lack of consensus secondary structures in the RNA. Corroboration of the transcription of both exons came from RT-PCR (Figure 7, left); however, a cDNA spanning the entire gene could not be amplified. A large transcript was revealed on RNA gel blots with probes derived from PCR products of each exon, but many cross-reacting bands confounded the analysis (data not shown).
The rpoC1 gene was identified in a previously unsequenced region, just upstream of the disabled Wendy transposon (Figure 1, ORF271 and ORF140). A predicted protein of 607 amino acids aligned with the first part of other rpoC1 proteins, albeit with a long N-terminal extension, whereas a downstream ORF of 507 amino acids predicted a protein similar to the C-terminal portion of rpoC1. A computer-based fusion of the two ORFs yielded a predicted peptide of 1113 amino acids with 49% identity to the most closely related rpoC1 sequence, that of Nephroselmis. The central portion of this alignment is shown in Figure 8B. Even within this highly conserved region, Chlamydomonas contains an insertion relative to all other sequences, as does Chlorella somewhat farther downstream. Although no discrete transcript of a length sufficient to contain both ORFs was detected using RNA gel blot hybridization (data not shown), RT-PCR demonstrated transcription of both coding regions (Figure 7, left), but attempts to span the 2.1-kb junction between the two ORFs using RT-PCR failed to generate any products. Based on the lack of evidence for a single transcript, and the fact that this arrangement was predicted by hybridization analysis of both C. reinhardtii and C. moewusii (Boudreau et al., 1994), we assigned the gene names rpoC1a and rpoC1b, suggesting that the division of rpoC1 into two parts occurred before the divergence of these Chlamydomonas species.
The C-terminal portion of the bacterial rpoC gene is represented by rpoC2 in chloroplast genomes. In C. reinhardtii, an ORF of 1175 amino acids was reported previously, of which only the N-terminal half appears to share homology with other predicted rpoC2 proteins (Fong and Surzycki, 1992a). Subsequently, however, a frameshift error was reported in this sequence, extending the ORF to 3119 amino acids, with additional C-terminal similarity to rpoC2. To determine if this longer ORF was transcribed, we subdivided the ORF into eight regions and used RT-PCR to search for transcription. As shown in Figure 7, transcription of six of the eight regions could be demonstrated, but two regions in the center could not be amplified from RNA, although cpDNA as a template yielded the expected products (data not shown). This finding is consistent with the presence of an intron; however, attempts to amplify across this putative ∼3-kb intron were unsuccessful. A discrete ∼5.2-kb transcript was detected by RNA filter hybridization with the N-terminal portion (Figure 6, circle), a size consistent with a spliced version, whereas an unspliced version would be >9 kb. These results leave open two interpretations. One is that rpoC2, like rpoC1, has now been divided into distinct genes and that the ORF remains complete by chance. In this case, C. reinhardtii would contain rpoC2a and rpoC2b, as proposed previously (Boudreau et al., 1994). A second interpretation is that a contiguous large mRNA has some portion spliced out; therefore, the gene should be termed rpoC2. Because of this ambiguity, we have retained the simpler rpoC2 nomenclature.
The last ORF investigated in detail was ORF208, which is adjacent to ORF570 (Figure 1, rps2-2). The predicted translation product of ORF570 was noted previously to have limited similarity to the N-terminal part of rps2 (Leu, 1998). Although ORF570 did not encode the C-terminal portion, another study (Boudreau et al., 1994) had shown that a C. eugametos rps2 probe hybridized to the downstream Bam6 fragment, where ORF208 is located. When ORF208 was translated and aligned with other rps2 sequences, it showed similarity to the predicted proteins (data not shown). RNA filter hybridization with ORF570- and ORF208-specific probes identified apparently identical transcripts of ∼2.5 kb (Figure 6, star). We were unable to amplify across the ORF570-ORF208 region using RT-PCR (data not shown). One of several interpretations of these findings is that the two 2.5-kb RNAs are not identical. Purification and sequencing of the expressed protein may clarify whether both ORFs actually are functional; indeed, proteomic analysis of the small ribosomal subunit identified an ORF570 but not an ORF208 translation product, confirming that at least ORF570 is an active gene (Yamaguchi et al., 2002).
DISCUSSION
A Gene-Poor Chloroplast Genome
The sequence analysis reported here indicates that the green algal lineage is a “hotbed” for chloroplast genome evolution, including invasion by repeat elements, gene loss, ORF gain, and loss of the inverted repeat. The completion of the C. reinhardtii plastid genome culminates a 20-year effort that began with the sequencing of rbcL (Dron et al., 1982). It might appear surprising that no other group completed what appeared to be a straightforward sequencing endeavor, especially considering the impact of a completed plastid sequence. We quickly discovered that the reason for the abundance of gaps in the genome was the presence of SDRs. The SDR-rich regions cause problems in primer design for sequence walking and additional problems in the sequencing reactions themselves. This, together with the lack of novel expressed ORFs, resulted in a nearly complete set of coding sequences being deposited over time but the notable absence of accurately sequenced intergenic regions.
The coding regions of the C. reinhardtii chloroplast genome are typical and encode highly conserved proteins with well-defined functions, with the exception of the PEP genes, certain ORFs, and ycfs (Figure 1, Table 1). Interestingly, many genes present in either related green algal or higher plant cpDNAs appear to have been lost from Chlamydomonas; however, five of these “lost” genes have been identified in the large EST database (Grossman, 2000; Lilly et al., 2002), suggesting that they have been successfully functionally transferred to the nucleus. The nature of conservation between Chlamydomonas genes and their counterparts in other genomes is readily revealed by the MultiPip analysis shown in Figure 4 (complete information is available in the supplemental data online). However, the genome is unique in that a single small RNA species (tscA) is responsible for the trans-splicing of the tripartite psaA mRNA (Goldschmidt-Clermont et al., 1991), and it possesses two copies of the ancient transposon Wendy, which has been proposed to be responsible for rearrangements in this genome since the divergence of C. reinhardtii (Fan et al., 1995). These unique features, together with the abundance of rearranged and split genes, make further analysis compelling.
Interestingly, these two dramatic alterations to the chloroplast genome of Chlamydomonas—the accumulation of repetitive elements and the loss of numerous coding regions—have resulted in a genome with no net contraction in size relative to other chlorophyte plastid genomes. The observed phylogenetic correlation of increased noncoding repeats and an accelerated rate of gene losses could related if dispensable coding regions were allowed to accumulate repeat sequences, or they may reflect a common mechanism that simultaneously promotes massive gene transfer and repeat proliferation. The first possibility could be supported by the observation that repeats were derived from coding regions that have been lost from either the Chlamydomonas or an earlier green algal lineage.
Functional and Evolutionary Implications of Small Dispersed Repeats
Perhaps the most interesting feature of this genome is the abundance of repetitive DNAs, which account for >20% of the sequence. Highly repetitive sequences are rare in chloroplast genomes compared with nuclear DNA. Using dot-plot analysis as a foundation, we initially documented 10 SDRs that together account for >5% of the plastid genome. Repetitive DNAs had been reported previously, but their abundance throughout the genome had not been quantified. For example, the presence of SDRs was implicated by BLAST searches in a study describing the rps2 gene, and downstream regions of rpoB and rpoC2, as well as upstream regions of rbcL, were reported to contain repeat elements (Dron et al., 1982; Fong and Surzycki, 1992a). Interestingly, although these repetitive sequences were seen in all isolates examined that are interfertile with C. reinhardtii (Boynton et al., 1992), they are not ubiquitous within the Volvocales. DNA filter hybridizations have shown previously that cpDNA of C. gelatinosa, a close relative of C. reinhardtii, also harbors at least one repeat family present in at least 35 genomic locations; however C. moewusii and C. pitschmannii lack SDR-like sequences, and members of this clade possess fewer rearrangements relative to one another than do the C. reinhardtii/C. gelatinosa group (Turmel et al., 1987; Boudreau and Turmel, 1995, 1996). These findings suggest that the highly rearranged C. reinhardtii and C. gelatinosa plastid chromosomes are correlated with the high level of repetitive DNA dispersed throughout the genome (Palmer, 1985). Repeat-mediated cpDNA rearrangements also have been proposed to occur in land plants, but on a smaller scale (Palmer et al., 1987; Palmer, 1991).
It is well documented that different cpDNA regions may have different evolutionary rates. For example, introns and intergenic spacers have a significantly higher level of divergence and presence of indels than adjacent coding regions (Kelchner and Wendel, 1996). To determine whether SDRs are relatively unstable, we investigated three SDR-containing intergenic regions across three closely related C. reinhardtii laboratory strains compared with a natural isolate from a different location. There was little intrastrain sequence variation, although it would be interesting to extend this analysis to a broader range of sequences and isolates (Table 3). Our analysis, coupled with previously documented SDRs, illustrate four main features. First, SDRs appear to be dispersed randomly throughout the intergenic regions, with the exception of a repeat-poor region approximately equidistant from the two repeat-rich inverted repeats. Second, SDR-rich intergenic regions evolve rapidly; 30 to 40 years in culture has been sufficient to accumulate detectable SNPs in these regions, whereas the functionally constrained rbcL coding region has not evolved SNPs among strains derived from the same isolate. Third, there is no evidence that SDRs have or result from retrotransposon-like activity, because they lack footprints or conserved sequence elements flanking the repetitive DNAs. Fourth, different copies of each SDR sequence share high identity with other copies around the chloroplast genome, which could indicate evolutionary conservation, as might be expected of a regulatory element or other functionally constrained sequence, the existence of a copy correction function, or that the multiple copies have proliferated only recently. This highly complex and diverse repeat family will be described in detail elsewhere (J.E. Maul and D.B. Stern, unpublished results).
One hypothesis for the function of at least one SDR is transcriptional regulation. It was demonstrated in both Chlamydomonas cells and E. coli that transcription from the so-called “PA promoter,” which in fact appears to be an SDR cluster, was enhanced when cells were treated with novobiocin, a DNA gyrase inhibitor (Thompson and Mosig, 1987), and that this segment of cpDNA changed conformation when cells were transferred between dark and light (Thompson and Mosig, 1990). However, genome-wide analysis of transcription rates have failed to correlate novobiocin-induced modulation with the number or abundance of flanking SDRs (Lilly et al., 2002).
With its widespread repetitive DNA, we suspected that Chlamydomonas cpDNA might exist in conformations more typical of complex higher plant mitochondrial genomes (Bendich, 1996; Oldenburg and Bendich, 2001). On the other hand, it does contain a large inverted repeat, a feature that has been implicated in conferring stability not only to plastid genomes (Palmer and Thompson, 1982) but also to viral and bacterial sequences (Rayko, 1997). The Chlamydomonas plastid genome represents what appears to be a structural intermediate, by virtue of possessing both the inverted repeats and many small tandem and dispersed repeated DNAs. Our fiber–fluorescence in situ hybridization analysis yielded results atypical of those obtained earlier with higher plants (Lilly et al., 2001). In particular, few higher order DNA fibers were present, and no circular genomes greater than a tetrameric equivalent were identified. Additionally, there were many DNA fibers of less than genome equivalent sizes present on all slides. We hypothesize that the SDRs might facilitate intramolecular recombination in much the same way that direct repeats of mitochondrial genomes do, resulting in a diverse population of molecules. Analysis of Chlorella cpDNA would be interesting in this regard, because it also contains abundant SDRs.
Phylogenetic History: Gene Loss Dominates the Dynamics of Plastid Genomes
The phylogenetic analyses of plastid genomes give an overall well-supported phylogeny, with four major lineages: streptophytes (land plants), chlorophytes (green algae), rho-dophytes-heterokonts-cryptophytes (nongreen algae), and glaucocystophytes. The Chlamydomonas plastid genome resides in the chlorophyte lineage in all analyses, with Chlorella being the closest relative. However, the branch length is notably long compared with that in land plant chloroplasts, suggesting that the amino acid substitution rate increased in green algae (Figure 5A). Here, we have shown that the rates of amino acid substitution are heterogeneous across plastome history, with a modest but significant acceleration specifically in the Chlamydomonas lineage. The position of M. viride has been under debate, as evidence from different sources strongly supports either the Mesostigma + land plants topology or the Mesostigma + all green plants topology (Lemieux et al., 2000; Karol et al., 2001). We performed gene-by-gene and combined-data-set parsimony analyses to look for significant conflict among genes that might in some cases support alternative arrangements. The results show that most individual genes do not favor one topology over another significantly, with the exception of the RNA polymerase genes (which support Mesostigma + all green plants; results not shown). Because of the sparse taxon sampling, the relationship is barely resolved by maximum likelihood analysis. More plastid sequence data from Chlorophyceae will help increase the resolution.
In the red algal lineage, secondary endosymbiosis has led to cryptophytes (G. theta) and heterokonts (O. sinensis). It has been proposed that a cyanobacteria-like prokaryote was engulfed by a eukaryote phagotroph (McFadden, 2001). Guillardia still retains the remnant plastid nuclear genome, termed the nucleomorph. Although plastids from both species are derived from secondary endosymbiosis, multiprotein analyses generally support the independent origins of their plastids, suggesting that there might have been more than one secondary endosymbiosis event in this group.
Structural events inferred throughout plastome evolution include the surprisingly frequent loss or massive reduction of the otherwise characteristic inverted repeat, occasional gene gains, changes in gene order, and invasion by SDR elements (in green algae) and the Wendy element (in Chlamydomonas). The retention of ribosomal protein clusters and the ancient tandemly duplicated psaA/psaB gene pair are among the few reliably retained ancestral structural traits in plastid genomes. The fact that the psaA/psaB gene cluster is highly separated in Chlamydomonas alone is a noteworthy indicator of the degree to which this genome has been rearranged. However, one of the most consistent patterns of structural evolution observed here has been the continued loss of plastid genes throughout plastome history. We inferred 39 gene loss events to have occurred since the last common ancestor of the chlorophytic green algae Nephroselmis, Chlorella, and Chlamydomonas. Most of these gene losses are either shared by Chlamydomonas and Chlorella (14) or specific to the Chlamydomonas lineage (15). Few gene losses are unique to green algae. All but four of the genes lost (rpl22, rpl33, rps11, and psaI) in the chlorophytes also have been lost from other photosynthetic plastid genomes (see supplemental data online). This finding underscores the observation that many gene losses are repeated and highly homoplastic events (Martin et al., 1998). Attempts to reconstruct phylogenetic relationships based on the pattern of gene presence/absence alone may be difficult. The unexpected positions of Chlorella and Pinus on trees generated from multivariate statistical analyses of gene presence/absence data (De Las Rivas et al., 2002) may have been a reflection of shared losses of ndh genes by these lineages.
The accelerated loss of plastid genes is quite surprising given the simultaneous tendency for Chlamydomonas and Chlorella to rapidly accumulate noncoding repeat sequences. Because both Chlorella and Chlamydomonas have accumulated many SDRs (Figure 2), but only Chlamydomonas has continued to rapidly lose plastid genes (no gene losses are seen specific to Chlorella), there is no clear relationship between the accumulation of SDR sequences and gene loss in these plastomes. Therefore, we conclude that these two principal features of the plastid genome of Chlamydomonas could be attributable to independent underlying causes.
It is not yet clear which of these genes have been transferred successfully to the green algal nuclear genome and which may have been lost entirely. A limited number of these lost genes have been identified in the Chlamydomonas EST database. However, no EST sequences have been detected from the ndh genes, suggesting that these genes may have been lost entirely and not transferred functionally to the nucleus. The complete nuclear genome sequence of Chlamydomonas (expected in late 2002) should give a more complete picture of chloroplast gene transfer to the nucleus.
Gene Rearrangement: Unique Organization of the PEP Coding Regions
Plastid DNAs of photosynthetic organisms encode the PEP (reviewed by Weihe and Börner, 1999; Cahoon and Stern, 2001), with specificity added by sigma factors that in higher plants are encoded by nuclear gene families (Allison, 2000). Higher plants also possess a second, phage-like polymerase (nucleus-encoded polymerase) for both chloroplasts and mitochondria that is nucleus encoded. Transcription inhibitor analysis failed to demonstrate nucleus-encoded polymerase activity in the Chlamydomonas chloroplast, suggesting that PEP performs all plastid transcription (Lilly et al., 2002).
The PEP genes described here, with the exception of rpoA, are structurally divergent from even their phylogenetically closest relatives, and often they gave ambiguous results in transcript analyses and alignments. RT-PCR gave conclusive evidence for the expression of each of these genes at the RNA level. Conversely, the multiple and sometimes weak bands on filter blots, coupled with mostly unsuccessful attempts to identify potential intron splice sites, left us seeking additional molecular or proteomic data. The fact that multiple sequence alignments of the translated PEP genes exhibited regions of high similarity among related species and included conserved core domains (Figure 8) suggests that these genes encode functional peptides. Interestingly, even though the newly identified rpoC1 possesses five highly conserved Cys residues, which in many species are known to function in the termination of transcription (Clerget et al., 1995), there is a large spacer between the third and fourth Cys residues that may destroy the zinc finger domain. This feature could be one reason there has been little evidence for efficient transcription termination in Chlamydomonas chloroplasts (Rott et al., 1996).
The complete and annotated chloroplast genome represents an important resource for engineering the genome to address basic questions of gene expression and photosynthesis, and perhaps for biotechnology as well. Given our results, genome-wide expression studies (Grossman, 2000; Lilly et al., 2002), and a nearly completed nuclear genome sequence, this organism remains an important model for the plant biology community.
METHODS
Sequence Analysis
Plasmid clones spanning regions of the Chlamydomonas reinhardtii genome for which sequences had not been deposited previously (see supplemental data online) were obtained from the Chlamydomonas Genetics Center at Duke University. The collection of BamHI, EcoRI, and PstI clones for the chloroplast genome is derived entirely from strains in the 137c background. Most of these plasmids were constructed from the wild-type strain CC-125. A few, including those used by Rochaix and colleagues (Rochaix, 1980) for early sequencing studies, were derived from a cell wall–deficient strain (cw15) that shares ancestry with CC-125 dating back approximately to the 1960s. Comparative sequence studies were made with the 137c strains CC-125, CC-406 (cw15), and CC-620 and the natural isolate CC-2290 (Gross et al., 1988). Plasmids were sequenced using subcloning and vector primers combined with sequencing templates generated using an Escherichia coli transposon-based approach (Template Generation System; Finnzymes, Espoo, Finland). Final sequencing and confirmation of ambiguities were completed using individually designed primers. The completed and annotated nucleotide sequence is available in the Third Party Annotation Section of the DDBJ/EMBL/GenBank databases. Accession numbers are given at the end of Methods.
DNA sequence files were trimmed, aligned, and assembled using the Lasergene software suite (DNAstar, Madison, WI). Gene annotation and open reading frame (ORF) recognition were performed with the Gene Construction Kit version 2.5 (Textco, West Lebanon, NH). The tRNA complement was identified using the tRNAscan program (www.genetics.wustl.edu/eddy/tRNAscan-SE/). New ORFs were defined by an initial AUG codon and 75 amino acids as minimal criteria. Allowing for shorter ORFs increased the total number, but in no case did it lead to the identification of putative functional genes, based on BLAST (Basic Local Alignment Search Tool) analyses. Initial sequence homologies were identified using tBLASTX (Altschul et al., 1997). Open reading frames that had no clear homologs in the database were designated ORFX, with X representing the number of amino acid codons. Multiple amino acid sequence alignments were generated with CLUSTAL W using the Blosum scoring matrix with equally weighted gap penalties. BOXSHADE was used to generate alignments of homologous genes.
Individual classes of short dispersed repeats (SDRs) were compiled by extracting and comparing sequences that were prevalent repeatedly in BLASTN output. These individual sequences then were aligned using a 90% similarity index across a 20-bp window. The resulting classes were combined further into core SDR units defined as short noncoding DNA sequences 20 to 35 bp in size and present at least 25 times with 90% identity. The proportion of the genome occupied by a given SDR was estimated by multiplying the size of the repeat by the number of occurrences and dividing the total base pairs by the full genome size of 203,395 bp. After performing this calculation for the remaining SDR classes, a figure for the overall proportion of repetitive DNAs was obtained.
Informatics Analysis
PipMaker and MultiPipMaker are World Wide Web–based genome analysis tools (Schwartz et al., 2000) that allow users to compare large DNA sequences, currently up to 2 Mb, and identify regions of high sequence similarity. The World Wide Web interface can be found at http://bio.cse.psu.edu/. PipMaker and MultiPipMaker compute alignments and similarity scores for two or more DNA sequences over the length of the “reference” sequence. Sequence alignments are determined by the modified BLAST algorithm BLASTZ (Altschul et al., 1997), with scoring parameters given by Chiaromonte et al. (2002), where a gap of k nucleotides is penalized 400 + 30k. Similarity scores are calculated as the percentage identity across a contiguous aligned region. The output from the PipMaker server can be visualized in four forms: a percentage identity plot (PIP), a dot plot, a typical BLAST-like alignment, and a concise list of the coordinates of the aligned segments.
In this study, the MultiPipMaker percentage identity plot output (Figure 4) was used to compare the reference sequence of C. reinhardtii chloroplast DNA with the plastid genomes of 13 other species: Arabidopsis thaliana, Chlorella vulgaris, Cyanidium caldarium, Cyanophora paradoxa cyanelle, Guillardia theta, Marchantia polymorpha, Mesostigma viride, Nephroselmis olivacea, Nicotiana tabacum, Odontella sinensis, Oryza sativa, Pinus thunbergii, Porphyra purpurea, Spinacia oleracea, and Zea mays. Accession numbers for these sequences are given at the end of Methods.
Phylogenetic Reconstruction and Rate Analysis
Thirty-nine proteins (see supplemental data online) shared by 18 selected plastid genomes were aligned independently and concatenated to a data set of 14,372 amino acids. We later removed four angiosperm taxa (Lotus japonica, Oenothera elata hookeri, Spinacia oleracea, and Zea mays) and deleted all gap characters. Thus, 8856 amino acid characters for 14 plastid genomes were used in the following analyses. Maximum parsimony was performed with PAUP* 4.0b8 (Swofford, 1993) with a heuristic search including 100 random addition sequences and branch swapping. Neighbor-joining analysis was performed with NEIGHBOR in PHYLIP 3.573 (Felsenstein, 1993) using the JTT model. In both cases, 250 bootstrap replications were performed. Maximum likelihood and minimum evolution analyses were performed through protml in MOLPHY2.3 (Adachi and Hasegawa, 1996) using the JTT-F model and RELL BP. All bootstrap values >50% are presented in Figure 5.
The gene content for each genome was determined by extracting all coding sequences and performing cross-comparisons using BLASTP, with the E-value threshold for a positive match set at 1e-6. Two cyanobacterial genomes were used as a reference for the ancestral plastid genome. A matrix consisting of 245 protein-coding genes and 14 taxa was built to summarize the gene content information (see supplemental data online). We used MacClade 4.0 (Maddison and Maddison, 1989) to map the gene presence/absence data onto the plastid genome phylogeny and to infer the unambiguous gene gains and losses along each branch by the principle of maximum parsimony.
The global heterogeneity of evolutionary rates was determined using likelihood ratio tests. The likelihood was obtained using PHYLIP version 3.6a2 programs proml and promlk with the JTT model (Felsenstein, 1993). The heterogeneity of substitution rates in specific lineages also was determined by the three taxa relative rates test with the Jones model using HY-PHY version 0.9b (http://peppercat.stat.ncsu.edu/~hyphy/).
Expression Analyses
Total RNA from CC-125 liquid cultures was isolated as described previously (Drager et al., 1999). RNA gels, transfer to nylon membrane, and filter hybridizations were as described previously (Higgs et al., 1999). DNA probes specific to the coding regions of rpoA, rpoB1, rpoB2, rpoC1, rpoC2, and rps2 were amplified by PCR using standard conditions with total DNA as a template and gene-specific primers (Lilly et al., 2002).
Total CC-125 RNA was used for single-tube reverse transcriptase–PCR (RT-PCR) with the Access RT-PCR system (Promega, Madison, WI) with gene-specific oligonucleotides (Lilly et al., 2002). Cycling conditions were 48°C for 1 h, 94°C for 2 min, followed by 30 cycles of 94°C for 45 s, 55°C for 1 min, and 72°C for 5 min. PCR products were resolved on 1.0% agarose gels. Fragment sizes were estimated by comparison with a 1-kb DNA ladder (Promega).
Sequencing was performed to ensure that PCR products were in fact derived from rpo cDNAs. DNA fragments were excised from gels, purified using Qiaex II resin (Qiagen, Valencia, CA), and cloned using the TOPO-TA cloning vector (Invitrogen, Carlsbad, CA), and DNA from positive colonies was end sequenced with vector primers. Similar RT-PCR and sequencing methods were used for the analysis of cDNAs investigated for the possibility of RNA editing.
Cytogenomic Methods
Mid-log-phase cultures from light-grown cell wall–deficient CC-406 cells were used for chloroplast isolations. Approximately 500 mL of a culture of 106 cells/mL was collected by centrifugation at 1000g. Cells were washed in nebulization medium (0.45 M sorbitol, 50 mM Tris, pH 7.6, 5 mM EDTA, 0.2% [w/v] BSA, 1.0% PVP-360, 0.025% spermine, 0.025% spermidine, and 1 mM β-mercaptoethanol), collected by centrifugation, and resuspended to a concentration of 1 × 108 cells/mL. This solution was passed through the BioNeb Cell Disruption System (Glascol Systems, Terra Haute, IN) under N2 gas (18 p.s.i.). The centrifugation was repeated, and the crude chloroplast pellet was resuspended in 36 mL of nebulization medium minus BSA. Chloroplasts were loaded onto Percoll step gradients (45%/75%) in 15-mL Corex centrifuge tubes. The gradients were centrifuged at 12,000g for 10 min in a swinging-bucket rotor. The chloroplast band at the 45%/75% interface was removed and diluted with 3 volumes of wash buffer plus 20 mM EDTA. Chloroplasts were pelleted at 3500g and resuspended in 2 mL of wash buffer plus 20 mM EDTA. Probe labeling, chloroplast fiber–fluorescence in situ hybridization, detection, and image capture were as described previously (Lilly et al., 2001). Each experiment was replicated a minimum of three times. Images of chloroplast DNAs for size estimations were collected randomly across four slides. Determination of the absence of nuclear DNA contamination was based on the lack of signal from a nuclear clone (rDNA clone P-92; Nikaido et al., 1994) when used as a probe on chloroplast DNA fibers. DNA fiber sizes were estimated based on the conversion 1.0 kb = 0.3 μM ± 0.04 kb. Final image adjustments (brightness/contrast) were made with Photoshop 6 (Adobe Systems, Mountain View, CA).
Upon request, all novel materials described in this article will be made available in a timely manner for noncommercial research purposes. No restrictions or conditions will be placed on the use of any materials described in this article that would limit their use for noncommercial research purposes.
Accession Numbers
Newly generated sequences were deposited in GenBank. The positions of these regions and their associated accession numbers are as follows: AF541860 (11142 to 11973), AF541861 (34012 to 36702), AF541862 (45836 to 48765), AF541863 (55547 to 57420), AF541864 (62012 to 63985), AF541865 (69176 to 69282), AF541866 (72027 to 73333), AF541867 (82275 to 88072), AF541868 (158441 to 159819), AF541869 (167594 to 169060), and AF541870 (189956 to 203333). Accession numbers for the 13 species used to compare the reference sequence of C. reinhardtii cpDNA are as follows: Arabidopsis thaliana, NC_000932; Chlorella vulgaris, NC_001865; Cyanidium caldarium, NC_001840; Cyanophora paradoxa cyanelle, NC_001675; Guil-lardia theta, NC_000926; Marchantia polymorpha, NC_001319; Mesostigma viride, NC_002186; Nephroselmis olivacea, NC_000927; Nicotiana tabacum, NC_001879; Odontella sinensis, NC_001713; Oryza sativa, NC_001320; Pinus thunbergii, NC_001631; Porphyra purpurea, NC_000925; Spinacia oleracea, NC_002202; and Zea mays, NC_001666. Accession numbers for sequences shown in other figures are NC_000911 (Synechocystis sp PCC6803) and NC_003272 (Nostoc sp PCC7120).
Supplementary Material
Acknowledgments
We thank P. Kerr Wall of Pennsylvania State University for assembling the gene database from sequenced chloroplast genomes, two anonymous reviewers for valuable comments, and members of the Chlamydomonas community for communicating unpublished results, including Paul Liu, Saul Purton, David Herrin, and Steve Surzycki. This work was funded by National Science Foundation awards MCB 9975765 (J.E.M., J.W.L., E.H.H., and D.B.S.) and DBI-0115684 (L.C. and C.W.d.), award HG02238 from the National Human Genome Research Institute to W.M., and a National Institutes of Health–National Research Service Award postdoctoral fellowship to J.W.L.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.006155.
Footnotes
Online version contains Web-only data.
References
- Adachi, J., and Hasegawa, M. (1996). MOLPHY version 2.3: Programs for molecular phylogeny based on maximum likelihood. In Computer Science Monographs 28. (Tokyo: Institute of Statistical Mathematics).
- Allison, L.A. (2000). The role of sigma factors in plastid transcription. Biochimie 82, 537–548. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backert, S., Dorfel, P., and Börner, T. (1995). Investigation of plant organellar DNAs by pulsed-field gel electrophoresis. Curr. Genet. 28, 390–399. [DOI] [PubMed] [Google Scholar]
- Barkan, A., and Goldschmidt-Clermont, M. (2000). Participation of nuclear genes in chloroplast gene expression. Biochimie 82, 559–572. [DOI] [PubMed] [Google Scholar]
- Bendich, A.J. (1991). Moving pictures of DNA released upon lysis from bacteria chloroplasts and mitochondria. Protoplasma 160, 121–130. [Google Scholar]
- Bendich, A.J. (1996). Structural analysis of mitochondrial DNA molecules from fungi and plants using moving pictures and pulsed-field gel electrophoresis. J. Mol. Biol. 255, 564–588. [DOI] [PubMed] [Google Scholar]
- Bendich, A.J., and Smith, S.B. (1990). Moving pictures and pulsed-field gel electrophoresis show linear DNA molecules from chloroplasts and mitochondria. Curr. Genet. 17, 421–425. [DOI] [PubMed] [Google Scholar]
- Bock, R. (2000). Sense from nonsense: How the genetic information of chloroplasts is altered by RNA editing. Biochimie 82, 549–557. [DOI] [PubMed] [Google Scholar]
- Boudreau, E., Otis, C., and Turmel, M. (1994). Conserved gene clusters in the highly rearranged chloroplast genomes of Chlamydomonas moewusii and Chlamydomonas reinhardtii. Plant Mol. Biol. 24, 585–602. [DOI] [PubMed] [Google Scholar]
- Boudreau, E., and Turmel, M. (1995). Gene rearrangements in Chlamydomonas chloroplast DNAs are accounted for by inversions and by the expansion/contraction of the inverted repeat. Plant Mol. Biol. 27, 351–364. [DOI] [PubMed] [Google Scholar]
- Boudreau, E., and Turmel, M. (1996). Extensive gene rearrangements in the chloroplast DNAs of Chlamydomonas species featuring multiple dispersed repeats. Mol. Biol. Evol. 13, 233–243. [DOI] [PubMed] [Google Scholar]
- Boynton, J.E., Gillham, N.W., Newman, S.M., and Harris, E.H. (1992). Organelle genetics and transformation in Chlamydomonas. In Cell Organelles, T. Hohn, ed (Vienna: Springer-Verlag), pp. 364–389.
- Cahoon, A.B., and Stern, D.B. (2001). Plastid transcription: A ménage à trois? Trends Plant Sci. 6, 45–46. [DOI] [PubMed] [Google Scholar]
- Chang, T.L., Stoike, L.L., Zarka, D., Schewe, G., Chiu, W.L., Jarrell, D.C., and Sears, B.B. (1996). Characterization of primary lesions caused by the plastome mutator of Oenothera. Curr. Genet. 30, 522–530. [DOI] [PubMed] [Google Scholar]
- Chiaromonte, F., Yap, V.B., and Miller, W. (2002). Scoring pairwise genomic sequence alignments. Pac. Symp. Biocomputing 7, 115–126. [DOI] [PubMed] [Google Scholar]
- Clerget, M., Ding, J.J., and Weisberg, R.A. (1995). A zinc-binding region in the β′ subunit of RNA polymerase is involved in anti termination of early transcription of phage HK022. J. Mol. Biol. 248, 768–780. [DOI] [PubMed] [Google Scholar]
- De Las Rivas, J., Lozano, J.J., and Ortiz, A.R. (2002). Comparative analysis of chloroplast genomes: Functional annotation, genome-based phylogeny, and deduced evolutionary patterns. Genome Res. 12, 567–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, X.W., Wing, R.A., and Gruissem, W. (1989). The chloroplast genome exists in multimeric forms. Proc. Natl. Acad. Sci. USA 86, 4156–4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent, R.M., Han, M., and Niyogi, K.K. (2001). Functional genomics of plant photosynthesis in the fast lane using Chlamydomonas reinhardtii. Trends Plant Sci. 6, 364–371. [DOI] [PubMed] [Google Scholar]
- Douglas, S.E. (1998). Plastid evolution: Origins, diversity, trends. Curr. Opin. Genet. Dev. 8, 655–661. [DOI] [PubMed] [Google Scholar]
- Downie, S.R., Llanas, E., and KatzDownie, D.S. (1996). Multiple independent losses of the rpoC1 intron in angiosperm chloroplast DNA's. Syst. Bot. 21, 135–151. [Google Scholar]
- Drager, R.G., Higgs, D.C., Kindle, K.L., and Stern, D.B. (1999). 5′ to 3′ exoribonucleolytic activity is a normal component of chloroplast mRNA decay pathways. Plant J. 19, 521–532. [DOI] [PubMed] [Google Scholar]
- Dron, M., Rahire, M., and Rochaix, J.D. (1982). Sequence of the chloroplast DNA region of Chlamydomonas reinhardtii containing the gene of the large subunit of ribulose bisphosphate carboxylase and parts of its flanking genes. J. Mol. Biol. 162, 775–793. [DOI] [PubMed] [Google Scholar]
- Erickson, J.M., Rahire, M., and Rochaix, J.D. (1984). Chlamydomonas reinhardtii gene for the 32 000 mol. wt. protein of photosystem II contains four large introns and is located entirely within the chloroplast inverted repeat. EMBO J. 3, 2753–2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, W.H., Woelfle, M.A., and Mosig, G. (1995). Two copies of a DNA element, ‘Wendy’, in the chloroplast chromosome of Chlamydomonas reinhardtii between rearranged gene clusters. Plant Mol. Biol. 29, 63–80. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J. (1993). PHYLIP (Phylogeny Inference Package). (Seattle: Department of Genetics, University of Washington).
- Fong, S.E., and Surzycki, S.J. (1992. a). Chloroplast RNA polymerase genes of Chlamydomonas reinhardtii exhibit an unusual structure and arrangement. Curr. Genet. 21, 485–497. [DOI] [PubMed] [Google Scholar]
- Fong, S.E., and Surzycki, S.J. (1992. b). Organization and structure of plastome psbF, psbL, petG and ORF712 genes in Chlamydomonas reinhardtii. Curr. Genet. 21, 527–530. [DOI] [PubMed] [Google Scholar]
- Gelvin, S.B., and Howell, S.H. (1979). Small repeated sequences in the chloroplast genome of Chlamydomonas reinhardtii. Mol. Gen. Genet. 173, 315–322. [Google Scholar]
- Goldschmidt-Clermont, M., Choquet, Y., Girard-Bascou, J., Michel, F., Schirmer-Rahire, M., and Rochaix, J.D. (1991). A small chloroplast RNA may be required for trans-splicing in Chlamydomonas reinhardtii. Cell 65, 135–144. [DOI] [PubMed] [Google Scholar]
- Grant, D.M., Gillham, N.W., and Boynton, J.E. (1980). Inheritance of chloroplast DNA in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 77, 6067–6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, C.H., Ranum, L.P.W., and Lefebvre, P.A. (1988). Extensive restriction fragment length polymorphisms in a new isolate of Chlamydomonas reinhardtii. Curr. Genet. 13, 503–508. [DOI] [PubMed] [Google Scholar]
- Grossman, A.R. (2000). Chlamydomonas reinhardtii and photosynthesis: Genetics to genomics. Curr. Opin. Plant Biol. 3, 132–137. [DOI] [PubMed] [Google Scholar]
- Hager, M., Biehler, K., Illerhaus, J., Ruf, S., and Bock, R. (1999). Targeted inactivation of the smallest plastid genome-encoded open reading frame reveals a novel and essential subunit of the cytochrome b6/f complex. EMBO J. 18, 5834–5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, E.H. (2001). Chlamydomonas as a model organism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 363–406. [DOI] [PubMed] [Google Scholar]
- Higgs, D.C., Shapiro, R.S., Kindle, K.L., and Stern, D.B. (1999). Small cis-acting sequences that specify secondary structures in a chloroplast mRNA are essential for RNA stability and translation. Mol. Cell. Biol. 19, 8479–8491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko, T., Tanaka, A., Sato, S., Kotani, H., Sazuka, T., Miyajima, N., Sugiura, M., and Tabata, S. (1995). Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. I. Sequence features in the 1 Mb region from map positions 64% to 92% of the genome. DNA Res. 2, 153–166. [DOI] [PubMed] [Google Scholar]
- Karol, K.G., McCourt, R.M., Cimino, M.T., and Delwiche, C.F. (2001). The closest living relatives of land plants. Science 294, 2351–2353. [DOI] [PubMed] [Google Scholar]
- Kelchner, S.A., and Wendel, J.F. (1996). Hairpins create minute inversions in non-coding regions of chloroplast DNA. Curr. Genet. 30, 259–262. [DOI] [PubMed] [Google Scholar]
- Köhler, S., Delwiche, C.F., Denny, P.W., Tilney, L.G., Webster, P., Wilson, R.J.M., Palmer, J.D., and Roos, D.S. (1997). A plastid of probable green algal origin in apicomplexan parasites. Science 275, 1485–1489. [DOI] [PubMed] [Google Scholar]
- Kowallik, K.V. (1994). From endosymbionts to chloroplasts: Evidence for a single prokaryotic eukaryotic endocytobiosis. Endocytobiosis Cell Res. 10, 137–149. [Google Scholar]
- Kück, U., Choquet, Y., Schneider, M., Dron, M., and Bennoun, P. (1987). Structural and transcription analysis of two homologous genes for the P700 chlorophyll a-proteins in Chlamydomonas reinhardtii: Evidence for in vivo trans-splicing. EMBO J. 6, 2185–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux, C., Otis, C., and Turmel, M. (2000). Ancestral chloroplast genome in Mesostigma viride reveals an early branch of green plant evolution. Nature 403, 649–652. [DOI] [PubMed] [Google Scholar]
- Leu, S. (1998). Extraordinary features in the Chlamydomonas reinhardtii chloroplast genome: (1) rps2 as part of a large open reading frame; (2) A C. reinhardtii specific repeat sequence. Biochim. Biophys. Acta 1365, 541–544. [DOI] [PubMed] [Google Scholar]
- Lilly, J.W., Havey, M.J., Jackson, S.A., and Jiang, J. (2001). Cytogenomic analyses reveal the structural plasticity of the chloroplast genome in higher plants. Plant Cell 13, 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly, J.W., Maul, J.E., and Stern, D.B. (2002). The Chlamydomonas reinhardtii organellar genomes respond transcriptionally and post-transcriptionally to abiotic stimuli. Plant Cell 14, 2681–2706. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Maddison, W.P., and Maddison, D.R. (1989). Interactive analysis of phylogeny and character evolution using the computer program Macclade. Folia Primatol. 53, 190–202. [DOI] [PubMed] [Google Scholar]
- Maier, R.M., Neckermann, K., Igloi, G.L., and Koessel, H. (1995). Complete sequence of the maize chloroplast genome: Gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J. Mol. Biol. 251, 614–628. [DOI] [PubMed] [Google Scholar]
- Martin, W., and Herrmann, R.G. (1998). Gene transfer from organelles to the nucleus: How much, what happens, and why? Plant Physiol. 118, 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, W., Stoebe, B., Goremykin, V., Hansmann, S., Hasegawa, M., and Kowallik, K.V. (1998). Gene transfer to the nucleus and the evolution of chloroplasts. Nature 393, 162–165. [DOI] [PubMed] [Google Scholar]
- McFadden, G.I. (2001). Primary and secondary endosymbiosis and the origin of plastids. J. Phycol. 37, 951–959. [Google Scholar]
- Morton, B.R., and Clegg, M.T. (1993). A chloroplast DNA mutational hotspot and gene conversion in a noncoding region near rbcL in the grass family (Poaceae). Curr. Genet. 24, 357–365. [DOI] [PubMed] [Google Scholar]
- Nikaido, S.S., Locke, C.R., and Weeks, D.P. (1994). Automated sampling and RNA isolation at room temperature for measurements of circadian rhythms in Chlamydomonas reinhardtii. Plant Mol. Biol. 26, 275–284. [DOI] [PubMed] [Google Scholar]
- Ohyama, K., et al. (1986). Chloroplast gene organization deduced from complete sequence of liverwort (Marchantia polymorpha) chloroplast DNA. Nature 322, 572–574. [Google Scholar]
- Oldenburg, D.J., and Bendich, A.J. (2001). Mitochondrial DNA from the liverwort Marchantia polymorpha: Circularly permuted linear molecules, head-to-tail concatamers, and a 5′ protein. J. Mol. Biol. 310, 549–562. [DOI] [PubMed] [Google Scholar]
- Palmer, J.D. (1985). Comparative organization of chloroplast genomes. Annu. Rev. Genet. 19, 325–354. [DOI] [PubMed] [Google Scholar]
- Palmer, J.D. (1991). Plastid chromosomes: Structure and evolution. In The Molecular Biology of Plastids, I.K. Vasil, ed (San Diego, CA: Academic Press).
- Palmer, J.D. (1997). Organelle genomes: Going, going, gone! Science 275, 790–791. [DOI] [PubMed] [Google Scholar]
- Palmer, J.D., Boynton, J.E., Gillham, N.W., and Harris, E.H. (1985). Evolution and recombination of the large inverted repeat in Chlamydomonas chloroplast DNA. In Molecular Biology of the Photosynthetic Apparatus, C. Arntzen, L. Bogorad, S. Bonitz, and K. Steinback, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 269–278.
- Palmer, J.D., and Delwiche, C.F. (1996). Second-hand chloroplasts and the case of the disappearing nucleus. Proc. Natl. Acad. Sci. USA 93, 7432–7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, J.D., Nugent, J.M., and Herbon, L.H. (1987). Unusual structure of Geranium chloroplast DNA: A triple-sized inverted repeat, extensive gene duplications, multiple inversions and two repeat families. Proc. Natl. Acad. Sci. USA 84, 769–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, J.D., and Thompson, W.F. (1982). Chloroplast DNA rearrangements are more frequent when a large inverted repeat sequence is lost. Cell 29, 537–550. [DOI] [PubMed] [Google Scholar]
- Race, H.L., Herrmann, R.G., and Martin, W. (1999). Why have organelles retained genomes? Trends Genet. 15, 364–370. [DOI] [PubMed] [Google Scholar]
- Rayko, E. (1997). Organization, generation and replication of amphimeric genomes: A review. Gene 199, 1–18. [DOI] [PubMed] [Google Scholar]
- Reith, M., and Munholland, J. (1995). Complete nucleotide sequence of the Porphyra purpurea chloroplast genome. Plant Mol. Biol. Rep. 13, 333–335. [Google Scholar]
- Rochaix, J.-D. (1980). Restriction fragments from Chlamydomonas chloroplast DNA. Methods Enzymol. 65, 785–795. [DOI] [PubMed] [Google Scholar]
- Rochaix, J.D., and Malnoe, P. (1978). Anatomy of the chloroplast ribosomal DNA of Chlamydomonas reinhardtii. Cell 15, 661–670. [DOI] [PubMed] [Google Scholar]
- Rott, R., Drager, R.G., Stern, D.B., and Schuster, G. (1996). The 3′ untranslated regions of chloroplast genes in Chlamydomonas reinhardtii do not serve as efficient transcriptional terminators. Mol. Gen. Genet. 252, 676–683. [DOI] [PubMed] [Google Scholar]
- Schneider, M., Darlix, J.-L., Erickson, J., and Rochaix, J.D. (1985). Sequence organization of repetitive elements in the flanking regions of the chloroplast ribosomal unit of Chlamydomonas reinhardtii. Nucleic Acids Res. 13, 8531–8541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, S., Zhang, Z., Frazer, K.A., Smit, A., Riemer, C., Bouck, J., Gibbs, R., Hardison, R., and Miller, W. (2000). PipMaker: A web server for aligning two genomic DNA sequences. Genome Res. 10, 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki, K., et al. (1986). The complete nucleotide sequence of the tobacco chloroplast genome: Its gene organization and expression. EMBO J. 5, 2043–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, C.L., and Stern, D.B. (2002). The treasure trove of algal chloroplast genomes: Surprises in architecture and gene content, and their functional implications. Plant Physiol. 129, 957–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiatek, M., Kuras, R., Sokolenko, A., Higgs, D., Olive, J., Cinque, G., Muller, B., Eichacker, L.A., Stern, D.B., Bassi, R., Herrmann, R.G., and Wollman, F.A. (2001). The chloroplast gene ycf9 encodes a photosystem II (PSII) core subunit, PsbZ, that participates in PSII supramolecular architecture. Plant Cell 13, 1347–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford, D.L. (1993). PAUP: Phylogenetic Analysis Using Parsimony. (Champaign, IL: Illinois Natural History Survey).
- Thompson, R.J., and Mosig, G. (1987). Stimulation of a Chlamydomonas chloroplast promoter by novobiocin in situ and in E. coli implies regulation by torsional stress in the chloroplast DNA. Cell 48, 281–287. [DOI] [PubMed] [Google Scholar]
- Thompson, R.J., and Mosig, G. (1990). Light affects the structure of Chlamydomonas chloroplast chromosomes. Nucleic Acids Res. 18, 2625–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turmel, M., Bellemare, G., and Lemieux, C. (1987). Physical mapping of differences between the chloroplast DNAs of the interfertile algae Chlamydomonas eugametos and Chlamydomonas moewusii. Curr. Genet. 11, 543–552. [Google Scholar]
- Turmel, M., Otis, C., and Lemieux, C. (1999). The complete chloroplast DNA sequence of the green alga Nephroselmis olivacea: Insights into the architecture of ancestral chloroplast genomes. Proc. Natl. Acad. Sci. USA 96, 10248–10253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasugi, T., et al. (1997). Complete nucleotide sequence of the chloroplast genome from the green alga Chlorella vulgaris: The existence of genes possibly involved in chloroplast division. Proc. Natl. Acad. Sci. USA 94, 5967–5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasugi, T., Tsudzuki, T., and Sugiura, M. (2001). The genomics of land plant chloroplasts: Gene content and alteration of genomic information by RNA editing. Photosynth. Res. 70, 107–118. [DOI] [PubMed] [Google Scholar]
- Watson, J.C., and Surzycki, S.J. (1983). Both the chloroplast and nuclear genomes of Chlamydomonas reinhardtii share homology with Escherichia coli genes for transcriptional and translational components. Curr. Genet. 7, 201–210. [DOI] [PubMed] [Google Scholar]
- Weihe, A., and Börner, T. (1999). Transcription and the architecture of promoters in chloroplasts. Trends Plant Sci. 4, 169–170. [DOI] [PubMed] [Google Scholar]
- Yamada, T. (1991). Repetitive sequence-mediated rearrangements in Chlorella ellipsoidea chloroplast DNA: Completion of nucleotide sequence of the large inverted repeat. Curr. Genet. 19, 139–147. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, K., Prieto, S., Beligni, M.V., Haynes, P.A., McDonald, W.H., Yates, J.R., III, and Mayfield, S.P. (2002). Proteomic characterization of the small subunit of the Chlamydomonas reinhardtii chloroplast ribosome: Identification of a novel S1 domain–containing protein and unusually large orthologs of bacterial S2, S3, and S5. Plant Cell 14, 2957–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.