Abstract
Human immunodeficiency virus type 1 (HIV-1) reverse transcription is primed by the cellular tRNA3Lys molecule, which binds, with its 3"-terminal 18 nucleotides (nt), to a complementary sequence in the viral genome, the primer-binding site (PBS). Besides PBS-anti-PBS pairing, additional interactions between viral RNA sequences and the tRNA primer are thought to regulate the process of reverse transcription. We previously identified a novel 8-nt sequence motif in the U5 region of the HIV-1 RNA genome that is critical for tRNA3Lys-mediated initiation of reverse transcription in vitro. This motif activates initiation from the natural tRNA3Lys primer but is not involved in tRNA placement and was therefore termed primer activation signal (PAS). It was proposed that the PAS interacts with the anti-PAS motif in the TΨC arm of tRNA3Lys. In this study, we analyzed several PAS-mutated viruses and performed reverse transcription assays with virion-extracted RNA-tRNA complexes. Mutation of the PAS reduced the efficiency of tRNA-primed reverse transcription. In contrast, mutations in the opposing leader sequence that trigger release of the PAS from base pairing stimulated reverse transcription. These results are similar to the reverse transcription effects observed in vitro. We also selected revertant viruses that partially overcome the reverse transcription defect of the PAS deletion mutant. Remarkably, all revertants acquired a single nucleotide substitution that does not restore the PAS sequence but that stimulates elongation of reverse transcription. These combined results indicate that the additional PAS-anti-PAS interaction is needed to assemble an initiation-competent and processive reverse transcription complex.
The replication cycles of human immunodeficiency virus type 1 (HIV-1) and other retroviruses are characterized by reverse transcription of the viral RNA (vRNA) genome into double-stranded DNA that integrates into the host cell genome (40). This process is mediated by the virion-associated reverse transcriptase (RT) enzyme, and a cellular tRNA3Lys molecule is used as a primer (33). The tRNA primer, with its 3"-terminal 18 nucleotides (nt), binds to a complementary sequence in the viral genome, the primer-binding site (PBS; Fig. 1A , green). The PBS is located in the 5"-untranslated leader region of the viral genome and is part of a highly structured RNA domain. The PBS is flanked by the upstream U5-top hairpin and the U5-leader stem, which is formed by the base pairing of sequences in the upstream U5 region and the downstream leader region (8, 10, 11, 15, 38). Besides the PBS-anti-PBS pairing, additional interactions between vRNA sequences and the tRNA primer are thought to regulate the process of reverse transcription. For instance, it was proposed that nt +168 to +172 in HIV-1 RNA (the A-rich loop) interact with the U-rich anticodon loop of tRNA3Lys and that nucleotides +142 to +145 interact with the 3" portion of the anticodon stem of the tRNA molecule (3, 5,19-21, 23, 31, 42, 43).
We recently identified an 8-nt motif in the U5 region of HIV-1 RNA (positions +123 to +130) that is critical for tRNA3Lys-mediated initiation of reverse transcription in vitro (8). This U5 motif is not important for tRNA annealing but rather for activation of the PBS-bound tRNA primer to initiate reverse transcription and is therefore referred to as primer activation signal (PAS). Deletion or mutation of the PAS element severely impairs reverse transcription initiated from the tRNA primer but not from DNA oligonucleotide primers. We proposed that the PAS interacts with the anti-PAS motif in the TΨC arm of tRNA3Lys (Fig. 1A; orange), similar to the interaction proposed for the Rous sarcoma virus (RSV) genome and the corresponding tRNATrp primer (1, 2, 13, 14, 29, 35, 36). Interestingly, the PAS is occluded by base pairing in the U5 leader stem of HIV-1 RNA (Fig. 1A). We demonstrated that reverse transcription is greatly upregulated by exposure of the PAS through mutation of the “opposing” leader sequence. The presence of the PAS enhancer element in a repressive RNA structure may provide a mechanism to regulate HIV-1 reverse transcription (8).
FIG. 1.
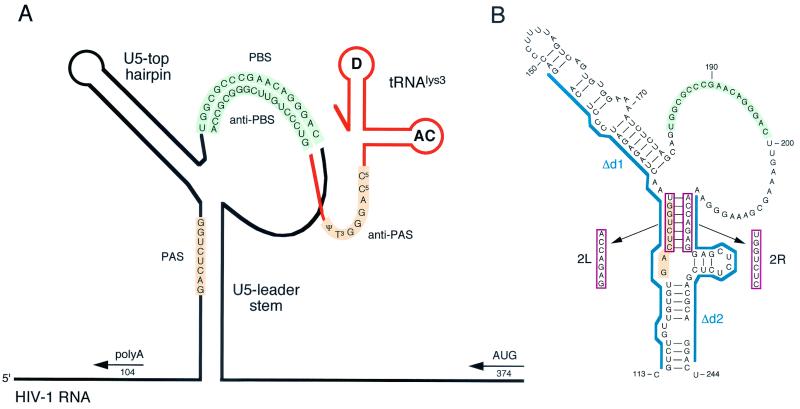
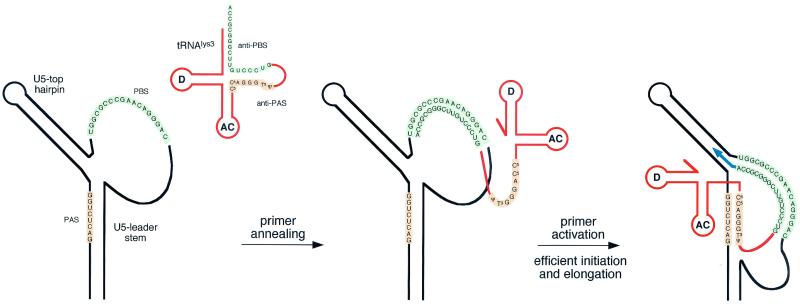
Secondary structure model for the U5-PBS leader region of HIV-1. (A) Schematic of HIV-1 RNA (black line) with an annealed tRNA3Lys primer (orange line; AC, anticodon loop; D, D loop). The tRNA3Lys molecule is used as a primer for reverse transcription by HIV-1 and binds to the PBS (green, PBS and anti-PBS). It was proposed that an additional interaction between the PAS element in the vRNA and the anti-PAS sequence in the tRNA primer (orange) activates the PBS-bound tRNA primer for initiation of reverse transcription (8). The HIV-1 RNA is folded into the U5 top hairpin upstream of the PBS and the U5 leader stem, which occludes the PAS in base pairing. The positions of the upstream poly(A) and downstream AUG DNA oligonucleotide primers are indicated. (B) Blue line, d1 and d2 deletions. Double mutant d1/2 combines both deletions. The 2L and 2R mutations (red box) are 7-nt substitutions on the left and right sides, respectively, of the U5-leader stem. Double mutant 2LR combines both mutations. The PAS element is inactivated in mutants 2L, d1, 2LR, and d1/2. The PAS is exposed by mutation of the opposite sequence in the U5-leader stem in mutants 2R and d2.
In this study, we analyzed several PAS-mutated viruses and performed reverse transcription assays with virion-extracted vRNA-tRNA complexes. Deletion of the PAS reduced the efficiency of tRNA-primed reverse transcription, whereas mutations in the opposing leader sequence stimulated reverse transcription. These results are similar to the previous in vitro results. We also selected revertant viruses that partially overcome the reverse transcription defect of the PAS deletion mutant. Remarkably, all revertants acquired a single nucleotide substitution that does not restore the PAS sequence but that stimulates elongation of reverse transcription. These results indicate that the additional PAS-anti-PAS interaction is needed to assemble an initiation-competent and processive reverse transcription complex.
MATERIALS AND METHODS
DNA constructs.
A derivative of full-length proviral HIV-1 clone pLAI was used to produce wild-type and U5-leader stem-mutated viruses. This construct, pLAI-R37, has been described previously and contains a unique U5 region in the 5" long terminal repeat (LTR) (16). Nucleotide numbers refer to positions on HIV-1 genomic RNA, with +1 being the capped G residue. For mutation of the U5-leader stem, we used construct Blue-5"LTR (25), which contains an XbaI-ClaI fragment of pLAI encompassing the 5" LTR, complete leader, and 5" end of the gag gene (positions −454 to +386) cloned into pBluescript KS(+) (Stratagene). Mutations were introduced as described previously (8). The mutated XbaI-ClaI fragments were introduced into proviral clone pLAI-R37. All mutant constructs were verified by sequence analysis of the complete leader region.
Cells, virus replication, and virus revertants.
SupT1 T cells were grown in RPMI 1640 medium supplemented with 10% fetal calf serum at 37°C and 5% CO2. SupT1 cells (5 × 106) were transfected with 1 μg of the HIV-1 proviral constructs by electroporation (250 V, 960 μF). Fresh SupT1 cells (0.5 × 106) were added after transfection to support virus replication. Cells were split 1 to 10 twice a week. CA-p24 levels in the culture medium were determined by enzyme-linked immunosorbent assay (6).
For the selection of revertant viruses, SupT1 cells were transfected with 1 μg of the 2L and 2R proviral constructs or 40 μg of the d1, d2, and d1/2 proviral constructs. After transfection, the cultures were split into several independent cultures, which were maintained for up to 3.5 months. At the peak of virus production, 0.1 to 100 μl of the culture supernatant was used to infect fresh SupT1 cells. At each passage, cells and supernatant samples were stored at −70°C. The replication capacities of virus samples were assayed by infection of fresh SupT1 cells with an equal amount of virus (10 ng of CA-p24). For the analysis of the revertant viruses present in the cultures, the cells were pelleted by centrifugation at 3,000 × g for 4 min and washed with phosphate-buffered saline. To isolate total cellular DNA, the cells were lysed in 10 mM Tris-HCl (pH 8.0)-1 mM EDTA-0.5% Tween 20 and incubated with 200 μg of proteinase K per ml at 56°C for 1 h. Proteinase K was subsequently inactivated by incubation at 95oC for 10 min. The 5" LTR leader region was amplified by PCR from the total cellular DNA with sense R region primer T7-1 (positions −54 to −34) and antisense primer AD-GAG (positions +442 to +463). The PCR products were directly sequenced, thus providing the average sequence of the viral population (population sequence). For insertion of revertant sequences into the wild-type proviral construct, the PCR product of the d1 revertant (culture A, day 72) was first cloned into pBlue-5"LTR as a HindIII/NarI fragment and then was cloned as an XbaI/ClaI fragment into proviral plasmid pLAI-R37. Introduction of the revertant sequences into pLAI-R37 was verified by sequence analysis.
Transient transfection, virus production, and isolation of viral RNA.
C33A cells were grown in Dulbecco's modified Eagle's medium with 10% fetal calf serum at 37°C and 5% CO2. For the production of virions, C33A cells were transiently transfected by the calcium phosphate method. Cells were grown to 60% confluence in 20 ml of culture medium in a 75-cm2 flask. Forty micrograms of the proviral construct in 880 μl of water was mixed with 1 ml of 50 mM HEPES (pH 7.1)-250 mM NaCl-1.5 mM Na2HPO4-120 μl of 2 M CaCl2, incubated at room temperature for 20 min, and added to the culture medium. The culture medium was changed after 16 h. Three days after transfection of C33A cells, the culture medium (20 ml) was centrifuged at 2,750 × g for 15 min to remove cells. The supernatant was subsequently filtered through a 0.45-μm-pore-size filter (Schleicher & Schuell), and the virions were pelleted by centrifugation at 25,000 rpm for 30 min in a Beckman SW28 rotor. Virions were resuspended in 400 μl of 10 mM Tris-HCl (pH 8.0)-100 mM NaCl-1 mM EDTA. To isolate vRNA, the viruses were incubated for 30 min at 37°C in the presence of 100 μg of proteinase K/ml-0.5% sodium dodecyl sulfate, followed by extraction with phenol-chloroform-isoamyl alcohol (25:24:1) and precipitation in 0.3 M sodium acetate (pH 5.2)-ethanol at −20°C. The vRNA was pelleted by centrifugation (16,000 × g, 20 min), washed with 70% ethanol, and dried. The pellet was dissolved in 20 μl of 10 mM Tris-HCl (pH 8.0)-1 mM EDTA and stored at −70°C.
Reverse transcription assays.
In the oligonucleotide and tRNA primer extension assays, vRNA corresponding to 50 ng of CA-p24 was incubated with or without an oligonucleotide primer (20 ng) in 12 μl of 83 mM Tris-HCl (pH 7.5)-125 mM KCl at 85°C for 2 min and at 65°C for 10 min, followed by cooling to room temperature for 1 h to allow the annealing of the primer. The natural tRNA primer or the annealed DNA primer or both were extended by addition of 6 μl of RT buffer (9 mM MgCl2; 30 mM dithiothreitol; 150 μg of actinomycin D/ml; 30 μM dATP, dGTP, and dTTP; 1.5 μM dCTP), 0.5 μl of [α-32P]dCTP, and 0.5 U of either HIV-1 RT (p66/p51) (Medical Research Council) or RNase H-negative RT (E478Q) (National Institutes of Health) (28, 39). The samples were incubated at 37°C for 30 min. The cDNA product was precipitated in 25 mM EDTA-0.3 M sodium acetate (pH 5.2)-80% ethanol at −20°C. The products were analyzed on a denaturing 6% polyacrylamide-urea sequencing gel. The antisense primers used are poly(A) (positions +88 to +104) and AUG (positions +348 to +368, with six additional nucleotides at its 5" end).
RESULTS
Mutation of the U5 leader stem.
We previously reported the construction of a set of HIV-1 leader RNA mutants that were tested in in vitro reverse transcription assays. This study focuses on in vivo experiments, for which we selected deletion mutants d1 and d2, double mutant d1/2, substitution mutants 2L and 2R, and double mutant 2LR (Fig. 1B) (8). A large deletion that includes the PAS element (positions +112 to +148) was introduced in mutant d1, and mutant d2 contains a large deletion in the downstream leader region, which is base paired to the PAS (positions +216 to +242). Double mutant d1/2 combines both deletions. In mutant 2L, a seven-nucleotide sequence overlapping the PAS element on the left side of the U5-leader stem was replaced. In mutant 2R, a seven-nucleotide sequence on the right side of the U5-leader stem was replaced. Double mutant 2LR combines both mutations. Thus, the PAS element is inactivated in mutants 2L, 2LR, d1, and d1/2. Another putative tRNA interaction site was removed in mutant d1 (positions +142 to +145; see the introduction). The downstream sequence that is base paired to the PAS is mutated in 2R and d2, thus making the PAS more accessible for base pairing with the anti-PAS motif in tRNA3Lys.
Replication studies and selection of revertant viruses.
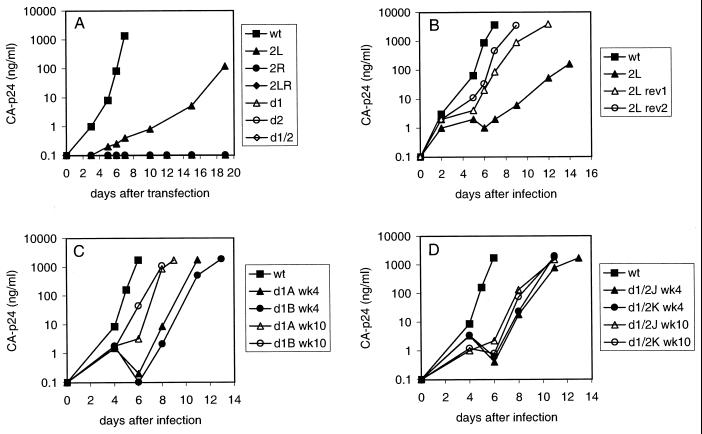
To study the replication potential of the mutant viruses, we transfected wild-type and mutant proviral genomes into the SupT1 T-cell line. These cells express the CD4-CXCR4 receptors and are fully susceptible to replication of the LAI strain. Virus replication was monitored by measuring CA-p24 in the culture medium at several days posttransfection. Transfection of proviral constructs d1, d2, and d1/2 demonstrated that the large deletions abolish virus replication (Fig. 2A) . The 2L mutation significantly delayed virus replication. Mutants 2R and 2LR are replication defective, but a low level of virus replication was observed when more DNA was transfected (5 μg instead of 1 μg; results not shown).
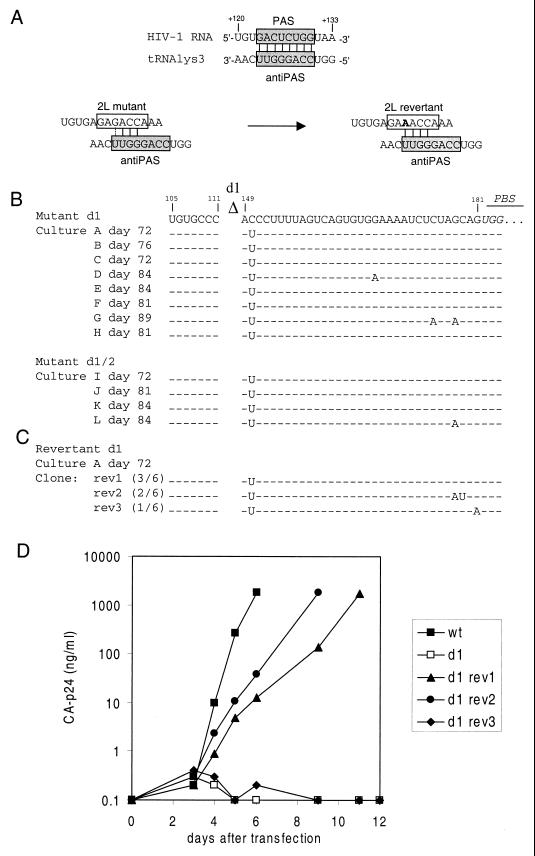
Prolonged culturing of mutant viruses may result in the selection of faster-replicating revertant viruses. Therefore, we transfected SupT1 cells with proviral constructs 2L, 2R, and 2LR and maintained the cultures for up to 3.5 months. No revertants were obtained for mutants 2R and 2LR. Faster-replicating revertants were observed in five out of six independent 2L cultures, and virus and cell samples were taken at several times. The replication capacities of two of these putative virus revertants were assayed by infection of fresh SupT1 cells (Fig. 2B). The replication of the revertant viruses was significantly increased compared with that of the original mutant, 2L. To determine the sequence of the leader region of the phenotypic 2L revertants, total DNA was isolated from the infected cells after 3.5 months. The complete 5" LTR leader region of the integrated HIV-1 proviral DNA was amplified by PCR and sequenced. All five revertants acquired the same G-to-A mutation at position 127 within the mutated PAS element (Fig. 3A) . Realignment of the tRNA-vRNA sequences allows the mutant PAS to form a 4-bp interaction. The mutation at position 127 optimizes this realigned interaction by replacing a G-U base pair with a more stable A-U base pair. This initial result suggests that the PAS-anti-PAS interaction is important for virus replication.
FIG. 2.
Replication of wild-type (wt), mutant, and revertant HIV-1 viruses. (A) SupT1 cells were transfected with 1 μg of the proviral constructs. CA-p24 production was measured in the culture medium at several days posttransfection. (B) Increased replication of the 2L virus after 3.5 months of culturing. Virus samples obtained from cultures 1 and 2 (corresponding to rev1 and rev2, respectively) were assayed for their replication capacities by infection of SupT1 cells with the same amount of virus (10 ng of CA-p24). (C) Increased replication capacity of the mutant d1 virus upon prolonged culturing. Virus samples obtained from d1 cultures A and B after 4 and 10 weeks were assayed for their replication capacities by infection of fresh SupT1 cells with the same amount of virus (10 ng of CA-p24). (D) Increased replication capacity of the mutant d1/2 virus in cultures J and K after 4 and 10 weeks of culturing.
To select for revertant viruses of the severely defective mutants d1, d2, and d1/2, we performed a SupT1 transfection with a large amount of DNA (40 μg). The transfected cells were split into multiple independent cultures (nine d1 cultures, six d2 cultures, and six d1/2 cultures). The cultures were maintained for up to 3 months, and replicating virus was noticed in several of the d1 and d1/2 cultures after a variable lag phase. No revertants were obtained for mutant d2. The replication capacities of several virus samples were assayed by infection of fresh SupT1 cells. The mutant d1 virus present at week 4 in cultures A and B showed improved replication kinetics, and replication was further optimized for the week 10 sample (Fig. 2C). The replication capacities of virus present in d1/2 cultures J and K also increased after 4 and 10 weeks of culturing (Fig. 2D). We isolated cellular DNA of the end point samples and amplified by PCR the complete HIV-1 leader region for each putative revertant. The PCR products were directly sequenced, thus providing the average sequence of the viral population (population sequence). Strikingly, the same C150U change was present in all revertants (Fig. 3B). We again sequenced the d1 proviral construct to exclude the possibility that this mutation was present in the original plasmid. Several revertants had acquired additional mutations in the U5-top hairpin (cultures D, G, and L). In particular, G-to-A changes were frequently observed in this region, consistent with previous results (9).
FIG. 3.
Sequence analysis and replication of revertant viruses. (A) The proposed PAS-anti-PAS interaction is shown on top (gray boxes). The PAS was mutated in mutant 2L (open box). This mutant reverted by means of the G127A mutation within the mutated PAS motif (altered nucleotide in boldface). This mutation is proposed to optimize the realigned interaction with the tRNA primer. (B) Sequences of the 5" leader regions of the viruses in d1 cultures A to H and d1/2 cultures I to L were determined by population sequencing. Mutations were found exclusively in the U5 region. The position of the d1 deletion is indicated, and the PBS is in italics. Dashes, nucleotides that are identical to those of the input proviral clone. The C150U mutation is present in all d1 and d1/2 revertants, and additional mutations are present in cultures D, G, and L. (C) The d1 revertant sequence of culture A was introduced into the wild-type proviral construct. The sequences of six individual clones were determined (frequencies in parentheses). (D) Replication of d1 revertant clones. SupT1 cells were transfected with 2 μg of the proviral constructs. CA-p24 production was measured in the culture medium at several days posttransfection. wt, wild type.
Because the C150U mutation was observed in all d1 and d1/2 revertants, we focused on this mutation. To verify its role in the phenotypic reversion of mutants d1 and d1/2, the PCR fragment of culture A was introduced in the d1 proviral genome. The sequences of six individual clones were determined (Fig. 3C). The C150U mutation is present in all clones, two clones contain additional mutations at positions 178 and 179, and one clone contains a G181A mutation. Mutation C150U is indeed sufficient to restore replication of mutant d1 (Fig. 3D, rev1). The additional mutations at positions 178 and 179 in rev2 slightly improved replication. The additional mutation G181A in rev3 completely abolished replication, and this mutation may have been introduced during PCR amplification.
Analysis of mutant HIV-1 virions and reverse transcription.
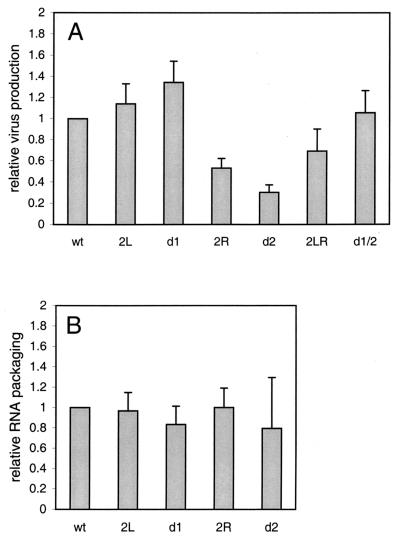
We transfected C33A cells (human cervix carcinoma cells not expressing CD4) with the wild-type and mutant proviral constructs to obtain virion particles for subsequent analyses. The amount of virions produced was determined by CA-p24 enzyme-linked immunosorbent assay (Fig. 4A) . Virus production was not affected or even slightly enhanced by the 2L and d1 mutations, respectively. Production of virions was reduced up to fourfold for mutants 2R and d2, and this defect was partially repaired for the double mutants, 2LR and d1/2. These results suggest that the leader region mutated in 2R and d2 encodes a motif involved in viral gene expression or virus assembly.
vRNA was isolated from C33A-produced virions, and the amount of vRNA per virion was determined by primer extension analysis with the poly(A) primer (Fig. 5, lanes 1 to 6). The CA-p24 values were used to control for the amount of virions used per sample. Because of the severe virus production defect of mutant d2, we were unable to use the standard amount of this virus. Packaging of vRNA was not determined for the double mutants, 2LR and d1/2. The primer extension analysis indicated that all mutants package a wild-type level of vRNA (Fig. 4B).
FIG. 5.
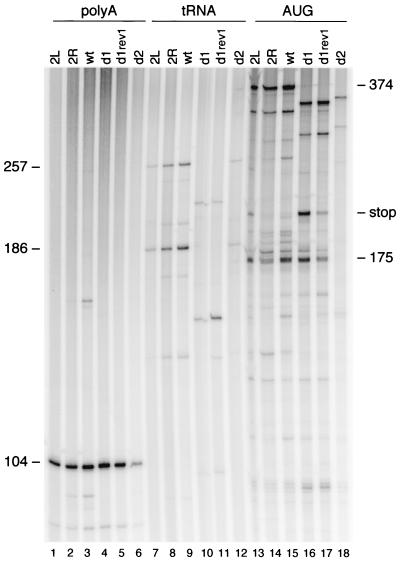
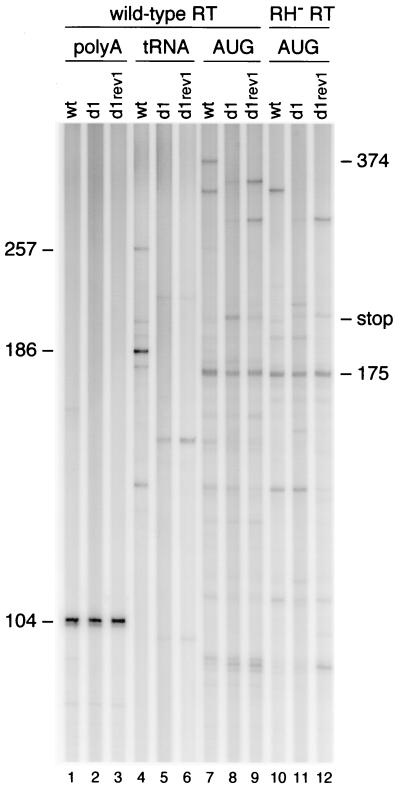
Reverse transcription assays with vRNA-tRNA complexes isolated from wild-type and mutant virions. The amount of vRNA was quantitated by DNA primer extension with the poly(A) primer, which produced a 104-nt product (lanes 1 to 6). Extension of the PBS-bound tRNA primer resulted in a 257-nt tRNA-cDNA product on the wild-type (wt) template (lanes 7 to 12). The products are shorter on the d1 and d1/2 template due to the d1 deletion in the U5 region. A major RT pause product with a length of 186 nt is visible on the wild-type template. The PBS-tRNA occupancy was determined by extension of the downstream AUG primer (lanes 13 to 18). When reverse transcription is blocked by the annealed tRNA primer, a 175-nt cDNA is produced. Free RNA templates will produce a full-length cDNA product of 374 nt. An additional stop product is observed on the 2L and d1 templates (lanes 13 and 16; stop). This stop is partially resolved on the d1 rev1 template (lane 17).
FIG. 4.
Analysis of wild-type and mutant HIV-1 virions. (A) Virus production in the culture medium after transfection of C33A cells with the proviral constructs. The results of three independent transfection experiments are summarized. Virus production for the wild-type (wt) pLAI construct was arbitrarily set at 1. (B) Packaging of HIV-1 RNA in virus particles. The packaging efficiency was measured by extension of the poly(A) primer on virion-isolated vRNA (Fig. 5, lanes 1 to 6). The results of three independent assays were quantitated, and the CA-p24 values were used to control for the amount of virions used per sample. Packaging was arbitrarily set at 1 for the wild-type virus.
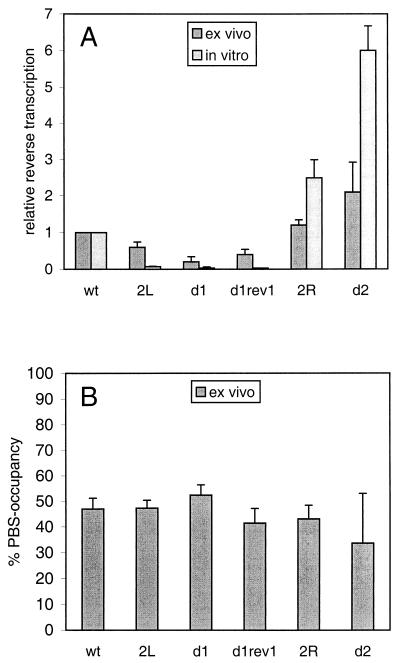
To study the role of the PAS during reverse transcription in vivo, one can either use a PCR-based method to quantitate cDNA products in the infected cell or perform an ex vivo analysis with vRNA-tRNA complexes isolated from virions. The first method is complicated by the fact that it is difficult to discriminate between defects in tRNA annealing, initiation, and elongation of reverse transcription. In addition, defects in virus production and/or assembly indirectly affect the level of reverse transcription. We therefore studied reverse transcription by the ex vivo method. Viral particles were produced in C33A cells, and the vRNA-tRNA complex was isolated. Equal amounts of wild-type and mutant virions (based on CA-p24) were used in reverse transcription assays, except for mutant d2, for which we were unable to use the normal amount of virions due to the severe virus production defect. The tRNA primer was extended by the addition of deoxynucleoside triphosphates and the HIV-1 RT enzyme, which results in a 257-nt tRNA-cDNA product on the wild-type template (Fig. 5, lane 9). A major RT pause product with a length of 186 nt was observed on the wild-type template. The amounts of full-length and pause tRNA-cDNA products were quantitated for the different templates (lanes 7 to 12). These values were combined and corrected for the amount of input vRNA as determined by poly(A) primer extension (lanes 1 to 6). The reverse transcription efficiency was calculated and is plotted in Fig. 6A . The efficiency was reduced to 60 and 30% of the wild-type value on PAS-negative templates 2L and d1, respectively. The C150U mutation in rev1 only slightly increased the efficiency of full-length cDNA synthesis, but the overall reverse transcription activity was significantly improved as demonstrated by the more prominent pause product. The 2R and d2 mutations that make the PAS element more accessible stimulate reverse transcription to 125 and 210% of the wild-type value, respectively. We were unable to use the standard amount of the mutant d2 virus, which resulted in a significant variation in the reverse transcription activity measured for this mutant. In general, the results obtained with this reverse transcription assay are consistent with the previously published in vitro results (8), which are plotted in Fig. 6A for comparison. Mutation of the PAS causes a reverse transcription defect, and making the PAS more accessible by opening base pairs in the U5-leader stem results in higher-than-wild-type activity. Both up and down effects appear less pronounced in the assay with virion-extracted vRNA-tRNA complexes than in the in vitro reverse transcription assay (Fig. 6A).
FIG. 6.
Relative reverse transcription activities of the wild-type and mutant templates. (A) Reverse transcription activity of virion-isolated vRNA-tRNA complexes as measured in Fig. 5 (ex vivo method). The results of three independent experiments were quantitated. There is a significant standard deviation for the d2 sample, which is due to the severe virus production defect of this mutant. The activity of the wild-type (wt) template was arbitrarily set at 1. For comparison, the results obtained previously with in vitro-assembled vRNA-tRNA complexes are included (8). (B) The tRNA occupancy of the PBS is shown for the wild-type and mutant vRNA genomes. The 374- and 175-nt products for three independent experiments were quantitated. The total of these products was arbitrarily set at 100%, and the percentage of 175-nt product is plotted.
The observed differences in reverse transcription efficiency may be caused by differences in the amount of tRNA primer annealed onto the mutant templates. We therefore determined the tRNA occupancy of the PBS. In this assay, vRNA-tRNA complexes were used as a template for extension of downstream DNA primer AUG (Fig. 1A). vRNA templates lacking a tRNA yielded a full-length cDNA product of 374 nt for the wild-type template (Fig. 5, lane 15). When tRNA3Lys was annealed onto the PBS, both the DNA and tRNA primers were extended by the RT enzyme. Reverse transcription initiated from the downstream AUG primer was partially blocked by the annealed tRNA primer, yielding a cDNA stop product of approximately 175 nt (Fig. 5, lane 15). Extension of the AUG primer yielded an additional stop product on the d1 template and to a lesser extent also on the 2L template (Fig. 5, lanes 16 and 13, stop). This stop product is partially resolved on the d1 rev1 template (lane 17). Quantitation of the 374- and 175-nt products indicated that the wild-type and mutant viral genomes are equally occupied by tRNA (Fig. 5, lanes 13 to 18). The tRNA occupancy of all templates is approximately 50%. The results are summarized in Fig. 6B and demonstrate that the PAS is not involved in tRNA placement onto the PBS, which is consistent with the previous in vitro results (8).
RT pausing on PAS-negative templates.
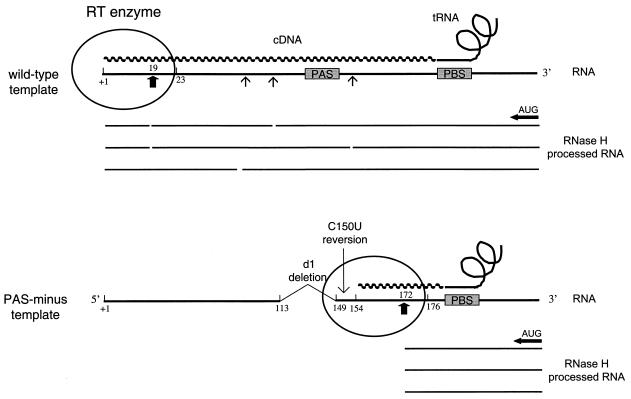
We became interested in the nature of the additional stop product observed in the PBS occupancy assay with the d1 and 2L templates (Fig. 5, lanes 16 and 13) because it seems specific for PAS-negative templates. This stop is most pronounced for the d1 template and is partially resolved on the d1 rev1 template (lane 17). Because this stop product is not observed upon tRNA extension (lanes 7 to 12), it must result from extension of the downstream AUG primer. A sequencing reaction with the AUG primer was analyzed in parallel and demonstrated that reverse transcription stopped at position +172. Intriguingly, this stop at position +172 on the d1 template is largely resolved on the d1 rev1 template, which contains the C-to-U mutation at position +150, far upstream of the actual stop position.
The following hypothesis may explain these seemingly paradoxical results. Enhanced pausing of the RT enzyme during tRNA-primed reverse transcription may result in enhanced RNase H-mediated degradation of the vRNA template, with a hot spot of cleavage at position +172. AUG-primed reverse transcription complexes that displace the PBS-bound tRNA primer stop at this position. This mechanistic explanation is illustrated in Fig. 7. Because the RNase H domain of HIV-1 RT is located 18 nt from the polymerase active site (4, 24, 26), RNase H cleavage at position +172 would mean that RT pausing actually occurred at position +154 during tRNA extension. The d1 reversion mutation at position +150 therefore occurs near the polymerase active site. In fact, this position is within the RT complex that covers approximately 8 nt upstream of the polymerization site (4, 34, 44). Thus, we propose that PAS-negative templates exhibit both initiation and elongation problems in tRNA-mediated reverse transcription. A nonprocessive reverse transcription complex that pauses predominantly around position +154 is assembled, yielding a major RNase H cleavage at position +172, which we detect in the AUG primer extension.
FIG. 7.
Model for RNase H-mediated degradation of the vRNA template during reverse transcription. In the PBS occupancy assay, both the downstream AUG DNA primer and the tRNA primer are extended by RT. Extension of the tRNA primer yields a full-length tRNA-cDNA on the wild-type template (top) but not on the PAS-negative template (bottom). Extension of the tRNA primer triggers RNase H-mediated degradation of the RNA template (arrows and breaks in the processed RNA). The RNase H domain of HIV-1 RT is located 18 nt from the polymerase active site (24, 26). Thus, enhanced pausing of the RT enzyme near template position +154 on the PAS-negative template results in pronounced RNase H cleavage at position +172 (thick arrow). AUG-primed reverse transcription complexes that displace the PBS-bound tRNA primer stop at this position are shown. The HIV-1 RT enzyme covers approximately 8 nt upstream and 22 nt downstream of the polymerization site (34, 44), such that the reversion-based C150U mutation is within the RT complex.
To test this hypothesis, we performed AUG primer extension with an RNase H-inactive RT mutant (Fig. 8, lanes 10 to 12). Indeed, the major stop observed with the wild-type RT enzyme on the d1 template (lane 8) is not observed for the RNase H-negative enzyme (lane 11). This indicates that the major stop product is caused by RNase H-mediated cleavage by a stalled RT enzyme during tRNA extension. We note that several other stop products are visible with the RNase H-negative RT enzyme on the d1 template. These stops most likely represent a collision of the AUG-initiated RT enzyme with stalled tRNA-initiated RT complexes. These signals are also specific for the PAS-negative d1 template (lane 11) compared with the wild-type template (lane 10), and the signals are partially resolved on the d1 rev1 template (lane 12). Thus, although the discrete RNase H-mediated stop is lost in assays with the RNaseH-negative RT enzyme, this result provides new evidence for poor processivity of reverse transcription on the PAS-negative d1 template. These combined results suggest that an RT initiation complex with reduced processivity is assembled on PAS-negative templates, resulting in more-frequent RT pausing (Fig. 9).
FIG. 8.
Reverse transcription with the wild-type and RNase H-negative RT enzyme. vRNA-tRNA complexes isolated from wild-type, d1, and d1 rev1 virions were used as templates for extension of the downstream AUG DNA primer (lanes 7 to 12). Reverse transcription was performed with the wild-type HIV-1 RT enzyme (lanes 7 to 9) and the RNase H-negative RT enzyme (RH− RT; lanes 10 to 12). The major stop product (stop) on the d1 template that is observed upon extension of the AUG primer with wild-type RT (lane 8) is not observed with RNase H-negative RT (lane 11). AUG-primed reverse transcription with the RNase H-negative RT did not yield the full-length 374-nt product but produced several stop products. This is probably caused by the reduced processivity of the mutant RT enzyme. Control reactions were performed with the poly(A) primer (lanes 1 to 3) and the tRNA primer (lanes 4 to 6).
FIG. 9.
Model for HIV-1 reverse transcription. Shown is a schematic of the secondary structure in the PBS region of the HIV-1 RNA genome (black line) and the secondary structure of the tRNA3Lys molecule (orange line; AC, anticodon loop; D, D loop) that is used as a primer for reverse transcription. The tRNA primer with its 3"-terminal 18 nt anneals to the PBS for reverse transcription (PBS and anti-PBS are green), but an additional interaction between the PAS in the viral RNA and the anti-PAS in the tRNA primer (orange) is necessary for efficient initiation and elongation of reverse transcription.
DISCUSSION
We previously identified an 8-nt sequence motif in the U5 region of the HIV-1 RNA genome that is critical for efficient reverse transcription (8). This motif is not involved in tRNA3Lys annealing but stimulates initiation from the natural tRNA3Lys primer. This motif is not required for initiation of DNA-primed reactions. We refer to this motif as the PAS. It is proposed that the PAS interacts with the anti-PAS motif in the TΨC arm of tRNA3Lys (Fig. 1A and 9), similar to the interaction that was proposed for the RSV genome and the corresponding tRNATrp primer (1, 2,13, 14, 29, 35, 36). In reverse transcription assays with virion-extracted vRNA-tRNA complexes, we now confirm that the efficiency of tRNA-primed reverse transcription is reduced on PAS-negative templates d1 and 2L. Furthermore, reverse transcription was enhanced on the 2R and d2 templates with mutations in the downstream leader region, which is base paired to the PAS in the extended U5-leader stem. No significant differences in the amount of tRNA primer annealed onto the wild-type and mutant templates were observed. These results are consistent with the results of reverse transcription assays with in vitro-synthesized RNA templates and heat-annealed tRNA (8). However, the reverse transcription effects are less pronounced in the assays with virion-derived vRNA-tRNA complexes. It is possible that cofactors in the virion particle, e.g., the NC protein, which acts as an RNA chaperone (reviewed in reference 37), facilitate the assembly of an initiation-competent vRNA-tRNA complex on PAS-negative templates.
The replication capacities and reverse transcription efficiencies of PAS-negative viruses are strongly reduced, the effect of the PAS deletion mutant d1 being more pronounced than that of substitution mutant 2L. Additional tRNA interaction sites may have been removed in mutant d1, for instance, the +142 to +145 motif, which, it has been proposed, interacts with the anticodon stem of the tRNA molecule (3, 5,19-21, 23, 31, 42, 43). Additional evidence for the importance of the PAS motif in virus replication was obtained from the analysis of revertant viruses. All revertants of substitution mutant 2L acquired a G-to-A mutation at position 127 within the mutated PAS motif. This mutation optimizes the potential interaction between the mutant vRNA template and the anti-PAS motif in the tRNA primer (Fig. 3A). Multiple virus revertants with improved replication capacity were obtained from PAS-negative deletion mutants d1 and d1/2. Surprisingly, all revertants acquired the same C150U mutation, which was subsequently demonstrated to partially restore virus replication. This C150U reversion did not repair the PAS sequence but profoundly enhanced the production of tRNA-primed cDNAs, although full-length cDNA synthesis was only slightly increased. In particular, the d1 reversion partially resolved a major stop product that was observed upon extension of the downstream AUG primer. The same stop product was observed for mutant 2L, suggesting that the stop is specific for PAS-negative templates. The stop was mapped to position +172 and was not observed with an RNase H-negative RT enzyme. We propose that tRNA-primed reverse transcription on the PAS-negative templates pauses near position +154, resulting in pronounced RNase H-mediated cleavage of the template at position +172, which is responsible for the stop product with the downstream AUG primer (Fig. 7). Thus, tRNA-primed reverse transcription that is initiated without the PAS-anti-PAS interaction is not processive, and most RT polymerases stop after incorporation of approximately 28 nt. At this point, the C150U reversion mutation can trigger prolonged elongation of tRNA-primed reverse transcription. This resulted in the accumulation of more-extended cDNA products in tRNA-primed reverse transcription, although most RT enzymes still did not produce full-length cDNAs. We currently do not understand the mechanism by which the C150U mutation enhances elongation. The combined results suggest that vRNA-tRNA complexes assembled on a PAS-negative template are not properly activated, leading to initiation and elongation defects (Fig. 9).
The 2R and d2 mutations, which make the PAS element more accessible, yield reverse transcription levels greater than that measured for the wild-type HIV-1 template. However, replication of these virus mutants was significantly reduced. This may be due to the virus production defect of these mutants (Fig. 4A). However, it is also possible that reverse transcription needs to be strictly modulated and that mutational enhancement of reverse transcription is not beneficial for virus replication. We previously described a reverse transcription defect for a mutant HIV-1 RNA template with a stabilized U5 leader stem (7). This mutant reverted by a deletion-insertion event that opened the stabilized U5-leader stem, making the PAS more accessible for base pairing with the tRNA primer. Thus, the presence of the PAS enhancer element in the repressive U5-leader stem structure may provide a mechanism to regulate initiation of reverse transcription during virus replication. We propose that the binding of tRNA3Lys to the PBS may occur relatively early in the virus-producing cell but that primer activation requires maturation of the vRNA-tRNA complex by means of the PAS-anti-PAS interaction and possibly other vRNA-tRNA contacts. The viral NC protein may be involved in this maturation, although it is not required for PAS stimulation in vitro (8). Because NC is released only from the Gag precursor protein during assembly of virion particles, this mechanism controls the precise timing of reverse transcription.
Other viral RNA-tRNA interactions were proposed previously for HIV-1, based primarily on biochemical probing studies (20-23, 27). For instance, it has been proposed that the A-rich loop (A168 to A171) of the U5-PBS hairpin interacts with the U-rich anticodon loop of the tRNA3Lys primer (U33 to U36). We recently presented an alternative folding for the PBS region based on extensive RNA secondary structure probing experiments with mutant transcripts (8). In this structure, the A-rich sequence is present in an internal loop of the U5-top hairpin (Fig. 1B). It has been reported that mutation of the A-rich loop affects initiation and elongation of reverse transcription (22, 27, 30-32, 45). Deletion of the A-rich loop affects virus replication, and the A-rich sequence is restored upon long-term culturing (31). However, the presence of overlapping sequence motifs for integration of the viral DNA genome complicates the interpretation of these results (12, 17, 41). Furthermore, other members of the lentivirus genus that utilize tRNA3Lys as a primer do not possess an A-rich loop in the region 5" to the PBS, indicating that this putative A-loop interaction is not a general property of this retrovirus genus.
In conclusion, we propose that the PAS in HIV-1 RNA interacts with the anti-PAS in the TΨC arm of tRNA3Lys. This interaction activates the PBS-bound tRNA primer for reverse transcription, resulting in efficient initiation and more-processive elongation of reverse transcription (Fig. 9). A similar interaction between a U5 motif in the RSV genome and tRNATrp stimulates initiation of reverse transcription in this avian retrovirus (1). The original RSV mechanism led us to propose a similar vRNA-tRNA interaction for HIV-2 (11), which was recently confirmed in a detailed RNA probing study (18). Specifically, a profound footprint was found in the TΨC arm of tRNA3Lys upon the binding of the HIV-2 RNA template (18). Phylogenetic analysis of the U5 sequence near the PBSs of several retroviruses that use different tRNA primers (including bovine leukemia virus, Moloney leukemia virus, mouse mammary tumor virus, simian sarcoma virus, simian retrovirus type 1, gibbon ape leukemia virus, baboon endogenous virus, Mason-Pfizer monkey virus, equine infectious anemia virus, feline leukemia virus, and feline immunodeficiency virus) suggests that the PAS-anti-PAS interaction, but obviously not the sequences themselves, is conserved in evolution (1). Thus, retrovirus reverse transcription seems to be regulated by a common mechanism. Additional viral RNA-tRNA interactions, including the previously discussed A-rich loop interaction, can also play a role in HIV-1 reverse transcription. During the initial phases of reverse transcription, there may be multiple structural transitions in the initiation complex, and several interactions may be formed and resolved. It is likely that retroviruses have evolved this complexity to increase the level of specificity of the reverse transcription reaction and to enable its regulation. For instance, the additional vRNA-tRNA contacts may restrict priming events by nonself tRNA primers and may preclude premature reverse transcription.
Acknowledgments
We thank Wim van Est for photography work, Truus Abbink and Atze Das for critical reading of the manuscript, D. Stammers for the gift of the purified HIV-1 RT enzyme (obtained through the MRC AIDS Reagent Project), and S. Le Grice for the donation of the RNase H-negative mutant HIV-1 RT enzyme (E478Q) (obtained through the NIH AIDS Research and Reference Program).
This work was supported in part by The Netherlands Foundation for Chemical Research with financial aid from The Netherlands Organization for Scientific Research (NWO-CW).
REFERENCES
- 1.Aiyar, A., D. Cobrinik, Z. Ge, H. J. Kung, and J. Leis. 1992. Interaction between retroviral U5 RNA and the TYC loop of the tRNATrp primer is required for efficient initiation of reverse transcription. J. Virol. 66:2464-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aiyar, A., Z. Ge, and J. Leis. 1994. A specific orientation of RNA secondary structures is required for initiation of reverse transcription. J. Virol. 68:611-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arts, E. J., M. Ghosh, P. S. Jacques, B. Ehresmann, and S. F. J. Le Grice. 1996. Restoration of tRNALys,3-primed (−)-strand DNA synthesis to an HIV-1 reverse transcriptase mutant with extended tRNAs. J. Biol. Chem. 271:9054-9061. [DOI] [PubMed] [Google Scholar]
- 4.Arts, E. J., and S. F. J. Le Grice. 1998. Interaction of retroviral reverse transcriptase with template-primer duplexes during replication. Prog. Nucleic Acid Res. Mol. Biol. 58:339-393. [DOI] [PubMed] [Google Scholar]
- 5.Arts, E. J., S. R. Stetor, Y. Li, J. W. Rausch, K. J. Howard, B. Ehresmann, T. W. North, B. M. Wohrl, R. S. Goody, M. A. Wainberg, and S. F. J. Le Grice. 1996. Initiation of (−) strand DNA synthesis from tRNALys3 on lentiviral RNAs: implications of specific HIV-1 RNA-tRNALys3 interactions inhibiting primer utilization by retroviral reverse trancriptases. Proc. Natl. Acad. Sci. USA 93:10063-10068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Back, N. K. T., M. Nijhuis, W. Keulen, C. A. B. Boucher, B. B. Oude Essink, A. B. P. van Kuilenburg, A. H. Van Gennip, and B. Berkhout. 1996. Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J. 15:4040-4049. [PMC free article] [PubMed] [Google Scholar]
- 7.Beerens, N., F. Groot, and B. Berkhout. 2000. Stabilization of the U5-leader stem in the HIV-1 RNA genome affects initiation and elongation of reverse transcription. Nucleic Acids Res. 28:4130-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beerens, N., F. Groot, and B. Berkhout. 2001. Initiation of HIV-1 reverse transcription is regulated by a primer activation signal. J. Biol. Chem. 276:31247-31256. [DOI] [PubMed] [Google Scholar]
- 9.Berkhout, B., A. T. Das, and N. Beerens. 2001. HIV-1 RNA editing, hypermutation and error-prone reverse transcription. Science 292:7.. [DOI] [PubMed] [Google Scholar]
- 10.Berkhout, B. 1996. Structure and function of the human immunodeficiency virus leader RNA. Prog. Nucleic Acid Res. Mol. Biol. 54:1-34. [DOI] [PubMed] [Google Scholar]
- 11.Berkhout, B., and I. Schoneveld. 1993. Secondary structure of the HIV-2 leader RNA comprising the tRNA-primer binding site. Nucleic Acids Res. 21:1171-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown, H. E. V., H. Chen, and A. Engelman. 1999. Structure-based mutagenesis of the human immunodeficiency virus type 1 DNA attachment site: effects on integration and cDNA synthesis. J. Virol. 73:9011-9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cobrinik, D., A. Aiyar, Z. Ge, M. Katzman, H. Huang, and J. Leis. 1991. Overlapping retrovirus U5 sequence elements are required for efficient integration and initiation of reverse transcription. J. Virol. 65:3864-3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cobrinik, D., L. Soskey, and J. Leis. 1988. A retroviral RNA secondary structure required for efficient initiation of reverse transcription. J. Virol. 62:3622-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Damgaard, C. K., H. Dyhr-Mikkelsen, and J. Kjems. 1998. Mapping the RNA binding sites for human immunodeficiency virus type-1 Gag and NC proteins within the complete HIV-1 and -2 untranslated leader regions. Nucleic Acids Res. 26:3667-3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Das, A. T., B. Klaver, B. I. F. Klasens, J. L. B. van Wamel, and B. Berkhout. 1997. A conserved hairpin motif in the R-U5 region of the human immunodeficiency virus type 1 RNA genome is essential for replication. J. Virol. 71:2346-2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Esposito, D., and R. Craigie. 1998. Sequence specificity of viral end DNA binding by HIV-1 integrase reveals critical regions for protein-DNA interaction. EMBO J. 17:5832-5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freund, F., F. Boulme, S. Litvak, and L. Tarrago-Litvak. 2001. Initiation of HIV-2 reverse transcription: a secondary structure model of the RNA-tRNA(Lys3) duplex. Nucleic Acids Res. 29:2757-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang, Y., A. Khorchid, J. Gabor, J. Wang, X. Li, J.-L. Darlix, M. A. Wainberg, and L. Kleiman. 1998. The role of nucleocapsid and U5 stem/A-rich loop sequences in tRNA 3Lys genomic placement and initiation of reverse transcription in human immunodeficiency virus type 1. J. Virol. 72:3907-3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isel, C., C. Ehresmann, G. Keith, B. Ehresmann, and R. Marquet. 1995. Initiation of reverse transcription of HIV-1: secondary structure of the HIV-1 RNA/tRNA(3Lys) (template/primer). J. Mol. Biol. 247:236-250. [DOI] [PubMed] [Google Scholar]
- 21.Isel, C., G. Keith, B. Ehresmann, C. Ehresmann, and R. Marquet. 1998. Mutational analysis of the tRNA3Lys/HIV-1 RNA (primer/template) complex. Nucleic Acids Res. 26:1198-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Isel, C., J.-M. Lanchy, S. F. J. Le Grice, C. Ehresmann, B. Ehresmann, and R. Marquet. 1996. Specific initiation and switch to elongation of human immunodeficiency virus type 1 reverse transcriptase require the post-transcriptional modifications of primer tRNALys3. EMBO J. 15:917-924. [PMC free article] [PubMed] [Google Scholar]
- 23.Isel, C., E. Westhof, C. Massire, S. F. J. Le Grice, B. Ehresmann, C. Ehresmann, and R. Marquet. 1999. Structural basis for the specificity of the initiation of HIV-1 reverse transcription. EMBO J. 18:1038-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacobo-Molina, A., J. Ding, R. G. Nanni, A. D. C. Clark, Jr., X. Lu, C. Tantillo, R. L. Williams, G. Kamer, A. L. Ferris, P. Clark, A. Hizi, S. H. Hughes, and E. Arnold. 1993. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc. Natl. Acad. Sci. USA 90:6320-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klaver, B., and B. Berkhout. 1994. Comparison of 5" and 3" long terminal repeat promoter function in human immunodeficiency virus. J. Virol. 68:3830-3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohlstaedt, L. A., J. Wang, J. M. Friedman, P. A. Rice, and T. A. Steitz. 1992. Crystal structure at 3.5 Å resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science 256:1783-1790. [DOI] [PubMed] [Google Scholar]
- 27.Lanchy, J.-M., C. Isel, G. Keith, S. F. J. Le Grice, C. Ehresmann, and R. Marquet. 2000. Dynamics of the HIV-1 reverse transcription complex during initiation of DNA synthesis. J. Biol. Chem. 275:12306-12312. [DOI] [PubMed] [Google Scholar]
- 28.Le Grice, S. F. J., C. E. Cameron, and S. J. Benkovic. 1995. Purification of human immunodeficiency virus type 1 reverse transcriptase. Methods Enzymol. 262:130-144. [DOI] [PubMed] [Google Scholar]
- 29.Leis, J., A. Aiyar, and D. Cobrinik. 1993. Regulation of initiation of reverse transcription of retroviruses, p. 33-48. In A. M. Skalka and S. P. Goff (ed.), Reverse transcriptase. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Li, Y., Z. Zhang, J. K. Wakefield, S.-M. Kang, and C. D. Morrow. 1997. Nucleotide substitutions within U5 are critical for efficient reverse transcription of human immunodeficiency virus type 1 with a primer binding site complementary to tRNAHis. J. Virol. 71:6315-6322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang, C., X. Li, L. Rong, P. Inouye, Y. Quan, L. Kleiman, and M. A. Wainberg. 1997. The importance of the A-rich loop in human immunodeficiency virus type 1 reverse transcription and infectivity. J. Virol. 71:5750-5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang, C., L. Rong, M. Gotte, X. Li, Y. Quan, L. Kleiman, and M. A. Wainberg. 1998. Mechanistic studies of early pausing events during initiation of HIV-1 reverse transcription. J. Biol. Chem. 273:21309-21315. [DOI] [PubMed] [Google Scholar]
- 33.Marquet, R., C. Isel, C. Ehresmann, and B. Ehresmann. 1995. tRNAs as primer of reverse transcriptases. Biochimie 77:113-124. [DOI] [PubMed] [Google Scholar]
- 34.Metzger, W., T. Hermann, O. Schatz, S. F. Le Grice, and H. Heumann. 1993. Hydroxyl radical footprint analysis of human immunodeficiency virus reverse transcriptase-template primer complexes. Proc. Natl. Acad. Sci. USA 90:5909-5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller, J. T., Z. Ge, S. Morris, K. Das, and J. Leis. 1997. Multiple biological roles associated with the Rous sarcoma virus 5" untranslated RNA U5-IR stem and loop. J. Virol. 71:7648-7656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morris, S., and J. Leis. 1999. Changes in Rous sarcoma virus RNA secondary structure near the primer binding site upon tRNATrp primer annealing. J. Virol. 73:6307-6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rein, A., L. E. Henderson, and J. G. Levin. 1998. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem. Sci. 23:297-301. [DOI] [PubMed] [Google Scholar]
- 38.Rizvi, T. A., and A. T. Panganiban. 1993. Simian immunodeficiency virus RNA is efficiently encapsidated by human immunodeficiency virus type 1 particles. J. Virol. 67:2681-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schatz, O., F. V. Cromme, F. Grüninger-Leitch, and S. F. J. Le Grice. 1989. Point mutations in conserved amino acid residues within the C-terminal domain of HIV-1 reverse transcriptase specifically repress RNase H function. FEBS Lett. 257:311-314. [DOI] [PubMed] [Google Scholar]
- 40.Telesnitsky, A., and S. P. Goff. 1997. Reverse transcriptase and the generation of retroviral DNA, p. 121-160. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 41.Vicenzi, E., D. S. Dimitrov, A. Engelman, T.-S. Migone, D. F. J. Purcell, J. Leonard, G. Englund, and M. A. Martin. 1994. An integration-defective U5 deletion mutant of human immunodeficiency virus type 1 reverts by eliminating additional long terminal repeat sequences. J. Virol. 68:7879-7890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wakefield, J. K., S.-M. Kang, and C. D. Morrow. 1996. Construction of a type 1 human immunodeficiency virus that maintains a primer binding site complementary to tRNAHis. J. Virol. 70:966-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wakefield, J. K., and C. D. Morrow. 1996. Mutations within the primer binding site of the human immunodeficiency virus type 1 define sequence requirements essential for reverse transcription. Virology 220:290-298. [DOI] [PubMed] [Google Scholar]
- 44.Wohrl, B. M., C. Tantillo, E. Arnold, and S. F. Le Grice. 1995. An expanded model of replicating human immunodeficiency virus reverse transcriptase. Biochemistry 34:5343-5356. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, Z., S.-M. Kang, Y. Li, and C. D. Morrow. 1998. Genetic analysis of the U5-PBS of a novel HIV-1 reveals multiple interactions between the tRNA and RNA genome required for initiation of reverse transcription. RNA 4:394-406. [PMC free article] [PubMed] [Google Scholar]