Abstract
Herpes simplex virus type 1 (HSV-1) immediate-early (IE) proteins are required for the expression of viral early and late proteins. It has been hypothesized that host neuronal proteins regulate expression of HSV-1 IE genes that in turn control viral latency and reactivation. We investigated the ability of neuronal proteins in vivo to activate HSV-1 IE gene promoters (ICP0 and ICP27) and a late gene promoter (gC). Transgenic mice containing IE (ICP0 and ICP27) and late (gC) gene promoters of HSV-1 fused to the Escherichia coli β-galactosidase coding sequence were generated. Expression of the ICP0 and ICP27 reporter transgenes was present in anatomically distinct subsets of neurons in the absence of viral proteins. The anatomic locations of β-galactosidase-positive neurons in the brains of ICP0 and ICP27 reporter transgenic mice were similar and included cerebral cortex, lateral septal nucleus, cingulum, hippocampus, thalamus, amygdala, and vestibular nucleus. Trigeminal ganglion neurons were positive for β-galactosidase in adult ICP0 and ICP27 reporter transgenic mice. The ICP0 reporter transgene was differentially regulated in trigeminal ganglion neurons depending upon age. β-Galactosidase-labeled cells in trigeminal ganglia and cerebral cortex of ICP0 and ICP27 reporter transgenic mice were confirmed as neurons by double labeling with antineurofilament antibody. Nearly all nonneuronal cells in ICP0 and ICP27 reporter transgenic mice and all neuronal and nonneuronal cells in gC reporter transgenic mice were negative for β-galactosidase labeling in the absence of HSV-1. We conclude that factors in neurons are able to differentially regulate the HSV-1 IE gene promoters (ICP0 and ICP27) in transgenic mice in the absence of viral proteins. These findings are important for understanding the regulation of the latent and reactivated stages of HSV-1 infection in neurons.
Herpes simplex virus (HSV) causes significant disease in humans, including keratitis, conjunctivitis, encephalitis, and disseminated infections of the newborn (65). Lytic infection occurs initially at peripheral sites and is followed by axonal transport of HSV to sensory ganglion neurons. Neurons undergo either lytic or latent viral infection (59, 60). HSV type 1 (HSV-1) immediate-early (IE) genes are thought to be important in determining the outcome of infection (54). The level of IE gene expression may play a role in cell tropism, establishment of and reactivation from latency (19, 21), and the extent of viral replication and disease. The HSV-1 IE ICP0 protein stimulates expression of early and late viral genes (6, 16, 17, 18, 22, 23, 39, 48, 67). ICP0 has been shown to initiate viral gene expression from a quiescent HSV-1 genome in cultured cells (25, 26, 29, 66, 68), and it appears to play a role in reactivation of the latent HSV-1 genome from sensory ganglion neurons in vivo (7, 35).
Many studies of the mechanism of IE gene regulation have been performed with cultured cells; however, little information exists about activation of the viral IE genes ICP0 and ICP27 in vivo in neurons. In vitro, expression of the viral IE genes has been shown to be activated by protein complexes composed of both viral and host proteins (3, 14, 24, 41, 47, 52, 58). The viral transactivator VP16 forms a complex with the host DNA binding protein Oct-1 and host proteins including HCF to upregulate transcription of IE genes.
It has been hypothesized that regulation of viral IE genes by neuronal proteins may control latent and reactivated infections of HSV-1 (19, 21, 61). Since HSV-1 mutants which have had the VP16 gene deleted can establish a latent infection and reactivate (57) and there is no evidence that VP16 is present in latently infected neurons, it does not appear that VP16 is involved in reactivation from latency. Rather, it appears that nonviral host proteins (neuronal) must fulfill the role of transactivating the IE promoters in the latent genome. It is possible but less likely that viral promoters other than IE promoters are the targets for reactivation of HSV-1. Latency could be maintained either by a decreased level or absence of host transcriptional activators or by an increased level or presence of host repressors of viral IE genes. Neuronal transcription factors (for example, c-Jun, JunD, and Oct-1) are differentially expressed in specific subsets of neurons (27, 28). The level of expression of neuronal transcription factors such as c-Jun varies in neurons during different stages of development and in regenerating neurons following damage by axotomy (28). Infection with HSV-1 induces the expression of genes encoding transcription factors in neurons (62).
A point of much importance to the previously discussed hypothesis of latent HSV-1 regulation is whether in vivo neurons have the capacity to differentially regulate viral IE gene expression (particularly that of ICP0) in the absence of viral proteins. Specifically, it is important to determine experimentally (i) whether host neuronal proteins are capable of activating HSV-1 IE genes in the absence of viral proteins, (ii) whether there are variations in the capacity of host transcriptional proteins to activate HSV-1 IE promoters (ICP0 and ICP27) in different subsets of neurons in animals, and (iii) whether changes in expression of neuronal transcription factors can alter the level of activation of HSV-1 IE promoters in a specific subset of neurons in the absence of viral proteins. It would be expected that the specific array of transcriptional proteins which are available to activate expression of a viral gene would vary in different subsets of neurons and in the same neuronal subset during different stages of differentiation. Development and differentiation of neurons results in changes in expression of transcription factors (28). Examination of different ages of neurons in reporter transgenic mice allows us a convenient test of whether specific neurons can change their capacity to activate HSV-1 IE promoters (particularly ICP0) in the absence of viral proteins.
We previously reported that transcriptional activation of the HSV-1 ICP4 promoter occurs in specific neurons in transgenic mice (43). In order to further study the ability of neurons to activate IE genes of HSV-1, reporter transgenic mice which contain IE gene promoters (ICP0 and ICP27) and a late gene promoter (glycoprotein C [gC]) fused to the β-galactosidase-coding sequence were generated. We found that neurons differentially activated the ICP0 and ICP27 promoters in transgenic mice in the absence of viral proteins. The late gene (gC) promoter was not activated without virus infection in neurons. This suggests that neuronal proteins are capable of specifically regulating the HSV-1 ICP0 and ICP27 promoters. Neuronal regulation of HSV-1 IE genes such as ICP0 may be important in maintenance of latent infection and reactivation of the viral genome from latency.
MATERIALS AND METHODS
Generation and identification of transgenic mice.
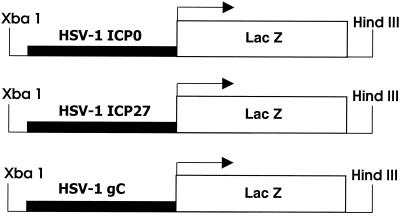
The promoter regulatory regions for IE genes ICP0 and ICP27 and late gene gC of HSV-1 were each fused to the bacterial β-galactosidase-coding sequence by using standard techniques (55) to produce reporter transgenes (Fig. 1). For ICP0, the promoter regulatory region (nucleotides −811 through +148 with respect to the transcription start site of ICP0) (37, 50, 53) was removed by NcoI-SacI digestion from the 1.6-kb BamHI-SacI fragment (46) of the BamHI SP fragment of HSV-1 (51). The NcoI-SacI fragment was made blunt ended and ligated in the proper orientation into the pNlacF plasmid (restriction digested at the SalI site and made blunt ended). The pNlacF plasmid contains the Escherichia coli β-galactosidase-coding sequence and a simian virus 40 nuclear translocation signal (42). The promoter regulatory region of the HSV-1 ICP27 gene (nucleotides −270 through +55 with respect to the transcription start site of ICP27) was removed by HinFI-BamHI digestion from the BamHI B fragment of HSV-1 (37, 50, 51). The HinFI-BamHI fragment containing the ICP27 promoter was blunt ended and ligated in the proper orientation into the pNlacF vector as described above. The promoter regulatory region from the HSV-1 late gene gC (nucleotides −45 through +122 with respect to the transcription start site of gC) (31, 40) was removed by NsiI-EcoRI digestion from a plasmid containing the XbaI E fragment of HSV-1 (8). The NsiI-EcoRI fragment containing the gC promoter was made blunt ended and ligated in the proper orientation into the pNlacF vector as described above. The XbaI-HindIII fragment containing the viral promoter fused to β-galactosidase from each of the three final reporter transgene constructs (Fig. 1) was isolated and purified as previously described (43). For each construct approximately 200 copies were injected (30, 45) into (C57BL/6xC3H) × (C57BL/6xC3H) one-cell embryos. Two transgenic lines were examined in detail for the ICP0 reporter transgene, three transgenic lines were examined for the ICP27 reporter transgene, and three transgenic lines were examined for the gC reporter transgene. Transgenic lines were established from founders through brother-sister matings. For each line, heterozygous transgenic mice and their nontransgenic control littermates were used in experiments. Mice were identified as transgenic or nontransgenic for each of the three reporter transgenes by PCR of tail DNA (45) for the β-galactosidase sequence (43). The primers for the β-galactosidase coding sequence were GCATCGAGCTGGGTAATAAGCGTTGGCAAT and GACACCAGACCAACTGGTAATGGTAGCGAC.
FIG. 1.
Diagrams of the HSV-1 IE gene (ICP0 and ICP27) and late gene (gC) reporter transgenes. The ICP0 promoter DNA fragment included nucleotides −811 through +148 of the ICP0 gene. The ICP27 promoter DNA fragment included nucleotides −270 through +55 of the ICP27 gene. The gC promoter DNA fragment included nucleotides −45 through +122 of the gC gene. These DNA fragments were each fused to the coding sequence for the β-galactosidase gene of E. coli.
Analysis of transgene expression in uninfected mice.
Adult mice, 8 to 10 weeks old, from each of the eight transgenic lines were euthanized, and tissues were removed and stored at −70°C. The following tissues were examined for each of four mice from each transgenic line: brain, trigeminal ganglia, spinal cord, cornea, heart, lung, liver, spleen, kidney, adrenal gland, and small intestine. Whole trigeminal ganglia, cornea, and brain (every other section of the brain) were examined for each animal. Four representative sections of all other tissues for each animal were studied. Trigeminal ganglia and eyes were fixed as whole tissues in 4% paraformaldehyde for 30 min immediately following removal from −70°C. All other tissues were sectioned at 40 μm with a cryotome, adhered to glass slides, and fixed in 4% paraformaldehyde for 30 min. Tissue sections, whole eyes, and whole ganglia were washed in phosphate-buffered saline (PBS) for 5 min and incubated in substrate solution (43) for 14 to 18 h at 37°C. The substrate solution contained 20 mM potassium ferrocyanide, 20 mM potassium ferricyanide, 2 mM MgCl2, 1 mg of 5-bromo-4-chloro-3-indolyl-β-d-galactosidase (X-Gal) per ml, 120 μl of 10% Nonidet P-40, and 100 μl of 1% sodium deoxycholate per 20 ml. Labeled tissue sections and whole tissues (eyes and ganglia) were washed for 5 min in PBS. Cryotome sections, trigeminal ganglia (thinly sliced with a razor blade), and corneas (removed from eyes) were mounted on glass slides, and coverslips were sealed with Permount. In order to reveal specific morphological details, representative 40-μm brain sections were counterstained with 1% neutral red for 30 s and washed three times for 5 min each in PBS before coverslips were applied. Slides were examined for positively labeled cells by light microscopy. Neuroanatomical locations were assigned according to the Mouse Brain Atlas C57BL/J Coronal found at the Mouse Brain Library website (htpp://www.nervenet.org/mbl/).
Analysis of transgene expression in virus-infected mice.
Four mice from each of the eight transgenic lines (two ICP0 lines, three ICP27 lines, and three gC lines) and four nontransgenic control mice were inoculated by the corneal route with 107 PFU of HSV-1 strain F per eye (9, 38, 44). Four mice from each line were mock inoculated by the corneal route of inoculation using medium containing no virus. Mice were euthanized 4 days following inoculation, and corneas and trigeminal ganglia were analyzed as described above for the presence of β-galactosidase-labeled cells.
Colabeling of β-galactosidase-positive cells for a neuron-specific marker in brain and trigeminal ganglia of ICP0 and ICP27 reporter transgenic mice.
Five-millimeter coronal slices of brain and whole trigeminal ganglia were fixed in 4% paraformaldehyde for 12 h, washed in PBS for 5 min, and incubated in substrate solution containing X-Gal (as described above) for 18 to 20 h. Samples containing β-galactosidase-positive cells were embedded in paraffin. Xylene was replaced by Clear-rite 3 in the embedding procedure to reduce the loss of β-galactosidase staining from cells. Six-micrometer sections of brain or trigeminal ganglia were mounted on positively charged glass slides, and tissue sections were deparrafinized using Clear-rite 3. Immunohistochemical localization of the mid-range-molecular-weight neurofilament protein (5, 69) was carried out with a standard biotin-avidin-peroxidase assay as described previously (38, 43, 44, 45). The primary antibody (used at a 1:40 dilution) was a mouse monoclonal antibody directed against neurofilament protein (molecular weight, 160,000) (Sigma). As a control, adjacent sections were incubated with mouse anti-bovine CD3 monoclonal antibody used at a dilution of 1:40. The antibody-biotin-avidin-horseradish peroxidase complexes were visualized by incubation of sections in diaminobenzidine. Endogenous peroxidase activity was blocked by a 30-min incubation in methanol containing 3% hydrogen peroxide. Sections were washed in PBS and incubated in a serum-free protein blocker (Dako Corporation). The M.O.M. immunodetection kit (Vector Laboratories) was used according to the manufacturer's instructions to reduce the background staining associated with using mouse antibody on mouse tissue. The reaction was terminated by washing the slides in distilled water. The slides were then dehydrated, and coverslips were sealed using Permount.
Analysis of transgene expression in trigeminal ganglion neurons of newborn and adult mice.
In order to determine whether neuronal differentiation might influence transcription from the ICP0 or the ICP27 promoter, trigeminal ganglia of newborn and adult mice (8 weeks old) from each of the ICP0, ICP27, and gC lines of mice were analyzed. Four-week-old mice from each of the ICP0-β-galactosidase and ICP27-β-galactosidase transgenic mice lines were also examined. The total number of positive neurons per mouse (two ganglia) was determined for the following number of mice in each age group of each line of reporter transgenic mice: for the 3180 line of ICP0-β-galactosidase transgenic mice, 14 1-day-old mice, 4 4-week-old mice, and 36 8-week-old mice; for the 3054 line of ICP27-β-galactosidase transgenic mice, 12 1-day-old mice, 10 4-week-old mice, and 28 8-week-old mice; and for the 6305 line of ICP4-β-galactosidase transgenic mice, 7 1-day-old mice, 3 4-week-old mice, and 7 8-week-old mice. β-Galactosidase assays were performed as described previously. The previously described ICP4-β-galactosidase transgenic mice (43) were included as a control.
RESULTS
Generation of transgenic mice containing HSV-1 IE (ICP0 and ICP27) and late (gC) gene reporter transgenes.
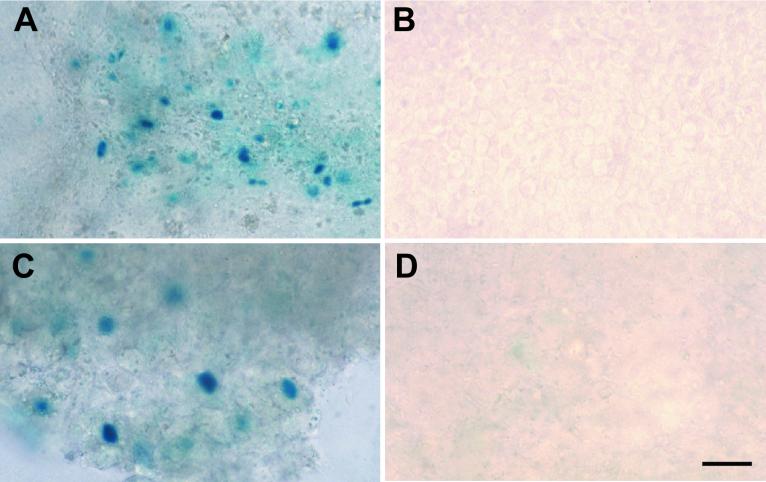
Two founder lines containing the ICP0-β-galactosidase reporter transgene [TgN(HSV0Rp)1wm (Tg3180) and TgN(HSV0Rp)2wm (Tg6825)], three founder lines containing the ICP27-β-galactosidase reporter transgene [TgN(HSV27Rp)1wm (Tg3054), TgN(HSV27Rp)2wm (Tg3058), and TgN(HSV27Rp)3wm(Tg6818)], and three founder lines containing the gC-β-galactosidase reporter transgene [TgN(HSVgCRp)1wm (Tg3401), TgN(HSVgCRp)2wm (Tg3403), and TgN(HSVgCRp)3wm (Tg3405)] were generated and studied in detail. All transgenic lines were screened for expression of the reporter transgene in the presence and absence of HSV-1. Mice from each of the above-described lines were inoculated via the cornea with HSV-1(F) or mock inoculated as described above. Viral infection resulted in moderate numbers of β-galactosidase-positive cells in the cornea and trigeminal ganglia of gC-β-galactosidase transgenic mice (Tg3401, Tg3403, and Tg3405) (Fig. 2A and C and Table 1.) No β-galactosidase-labeled cells were detected in age-matched mock-inoculated gC-β-galactosidase transgenic mice (Tg3401, Tg3403, and Tg3405) or in nontransgenic littermates infected with HSV-1(F) (Fig. 2B and D and Table 1). Virus infection of ICP0-β-galactosidase (Tg3180 and Tg6825) and ICP27-β-galactosidase (Tg3054, Tg3058, and Tg6818) transgenic mice resulted in a moderate number of positive cells in the corneal stroma and an increase in numbers of positive cells in trigeminal ganglia (Table 1 and results not shown). These experiments confirmed that each of the transgenes (ICP0-β-galactosidase, ICP27-β-galactosidase, and gC-β-galactosidase) was appropriately expressed in the presence of viral proteins in both neuronal and nonneuronal cells.
FIG. 2.
The viral reporter transgenes were expressed in the presence of HSV-1 infection in cells of the trigeminal ganglia and cornea. (A) Cornea of a gC reporter transgenic mouse infected with HSV-1 F. (B) Cornea of a mock-infected gC reporter transgenic mouse with no β-galactosidase labeling present. (C) Trigeminal ganglia of a gC reporter transgenic mouse infected with HSV-1(F). (D) Trigeminal ganglia of a mock-infected gC reporter transgenic mouse with no β-galactosidase-labeled cells present. Bar, 40 μm.
TABLE 1.
Expression of β-galactosidase in reporter transgenic mice in the presence and absence of HSV-1
| HSV-1 | Sample | No. of animals positivea
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ICP0
|
ICP27
|
gC
|
Nontransgenic | |||||||||
| Tg3180 | Tg6825 | Tg3054 | Tg3058 | Tg6818 | Tg3401 | Tg3403 | Tg3405 | |||||
| + | Cornea | 4 | 4 | 4 | 4 | 4 | 3 | 3 | 0 | 0 | ||
| Trigeminal ganglia | 4 | 4 | 4 | 4 | 4 | 4 | 4 | 3 | 0 | |||
| − | Cornea | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Trigeminal ganglia | 4 | 4 | 0 | 2 | 3 | 0 | 0 | 0 | 0 | |||
Each number represents the number of mice out of four which contained β-galactosidase-labeled cells in each transgenic line.
Viral proteins are not required for activation of the HSV-1 IE promoters (ICP0 and ICP27) in neurons in transgenic mice.
The brains of uninfected adult ICP0-β-galactosidase transgenic mice (Tg3180 and Tg6825) and ICP27-β-galactosidase transgenic mice (Tg3054, Tg3058, and Tg6818) each contained moderate to large numbers of β-galactosidase-positive cells (Fig. 3A and B and 4A and B and Table 2). The trigeminal ganglia of uninfected adults of both lines of ICP0-β-galactosidase transgenic mice contained low to moderate numbers of β-galactosidase-positive cells (Fig. 5B and E and Table 2). The trigeminal ganglia of uninfected adults of two out of the three lines of ICP27-β-galactosidase transgenic mice contained few to low numbers of β-galactosidase-positive cells (Fig. 5E and Table 2). No β-galactosidase-positive cells were detected in the brains and trigeminal ganglia of uninfected gC-β-galactosidase transgenic mice (Tg3401, Tg3403, and Tg3405) (Fig. 3C and Fig. 4C and Table 2). Nonneural tissues of transgenic mice containing the ICP0-β-galactosidase transgene, the ICP27-β-galactosidase transgene, and the gC-β-galactosidase transgene were negative for β-galactosidase labeling, with two exceptions (Table 3). The Tg3054 line of ICP27-β-galactosidase transgenic mice had scattered positive cells in the renal tubular epithelium in three of the four mice examined (Table 3), and the Tg6818 line of ICP27-β-galactosidase mice had low numbers of positive cells in the myocardium in three of the four mice examined (Table 3). The β-galactosidase-labeled cells in the intestinal walls of ICP0-β-galactosidase and ICP27-β-galactosidase transgenic mice (Table 3) were neurons in the myenteric plexus. Nontransgenic littermates from the above-described transgenic lines contained no positive cells in neural (Fig. 3D and 4D and Table 2) or nonneural (Table 3) tissues.
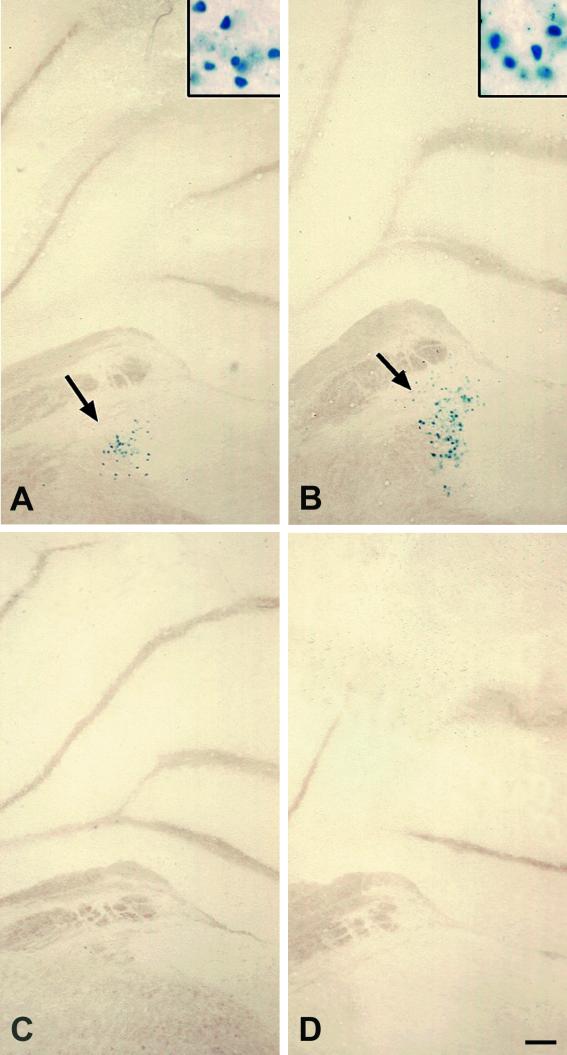
FIG.3.
Expression of β-galactosidase in neurons of the cerebellum in mice containing HSV-1 IE gene (ICP0 and ICP27) and late gene (gC) reporter transgenes. In the absence of HSV-1 proteins, the ICP0 and ICP27 reporter transgenes were expressed in a specific subset of neurons in the cerebellum. (A) Coronal section through the cerebellum of an ICP0 reporter transgenic mouse (Tg3180), showing β-galactosidase-labeled neurons in the vestibular nucleus. The inset shows a higher magnification of the labeled cells indicated by the arrow. (B) Coronal section through the cerebellum of an ICP27 reporter transgenic mouse (Tg3054), showing β-galactosidase-labeled neurons in the vestibular nucleus. The inset shows a higher magnification of the labeled cells indicated by the arrow. (C) Coronal section through the cerebellum of a gC reporter transgenic mouse (Tg3403), showing no β-galactosidase labeling in any cells. (D) Coronal section of cerebellum from a nontransgenic control mouse, showing no β-galactosidase labeling in any cells. Bar, 100 μm (30 μm in insets).
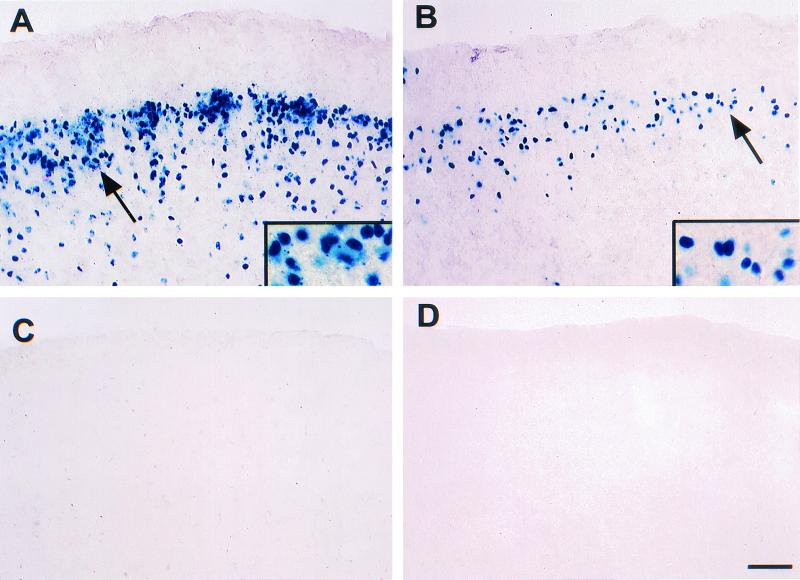
FIG. 4.
Expression of β-galactosidase in neurons of the cerebral cortex in mice containing HSV-1 IE gene (ICP0 and ICP27) and late gene (gC) reporter transgenes in the absence of viral proteins. The cerebral cortex of ICP0 and ICP27 reporter transgenic mice contained large numbers of labeled cells in the external granular layer, few to no labeled cells in the molecular layer, and small numbers of labeled cells in other areas. (A) Coronal section through the cerebrum of an ICP0 reporter transgenic mouse (Tg3180). The inset is a higher magnification of β-galactosidase-labeled neurons indicated by the arrow. (B) Coronal section through the cerebrum of an ICP27 reporter transgenic mouse (Tg3054). The inset picture is a higher magnification of β-galactosidase-labeled neurons indicated by the arrow. (C) Coronal section through the cerebrum of a gC reporter transgenic mouse (Tg3403), showing no labeled cells. (D) Coronal section through the cerebrum of a nontransgenic control mouse, showing no labeled cells. Bar, 40 μm (20 μm in insets).
TABLE 2.
Distribution of β-galactosidase labeling in the nervous systems of mice containing HSV-1 IE gene or late gene reporter transgenes
| Sample | No. of animals positivea
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ICP0
|
ICP27
|
gC
|
Nontransgenic | |||||||
| Tg3180 | Tg6825 | Tg3054 | Tg3058 | Tg6818 | Tg3401 | Tg3403 | Tg3405 | |||
| Cerebellum (purkinje cell layer) | 1 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Vestibular nucleus | 4 | 4 | 4 | 4 | 4 | 0 | 0 | 0 | 0 | |
| Cerebral cortex | 4 | 4 | 4 | 4 | 4 | 0 | 0 | 0 | 0 | |
| Lateral septal nucleus | 4 | 4 | 4 | 4 | 4 | 0 | 0 | 0 | 0 | |
| Cingulum | 4 | 4 | 4 | 4 | 4 | 0 | 0 | 0 | 0 | |
| Hippocampus | 4 | 4 | 2 | 4 | 4 | 0 | 0 | 0 | 0 | |
| Thalamus | 4 | 4 | 4 | 3 | 4 | 0 | 0 | 0 | 0 | |
| Amygdala | 4 | 4 | 4 | 4 | 2 | 0 | 0 | 0 | 0 | |
| Superior colliculus | 1 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Trigeminal ganglia | 4 | 4 | 0 | 2 | 3 | 0 | 0 | 0 | 0 | |
| Spinal cord | 1 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
Each number represents the number of mice out of four which contained β-galactosidase-labeled neurons in the indicated anatomic regions of the nervous system in each transgenic line. Some gray matter regions of the brain contained only a few scattered labeled cells which could not be consistently localized and are not listed. White matter regions of the brain and spinal cord were negative for β-galactosidase labeling.
FIG. 5.
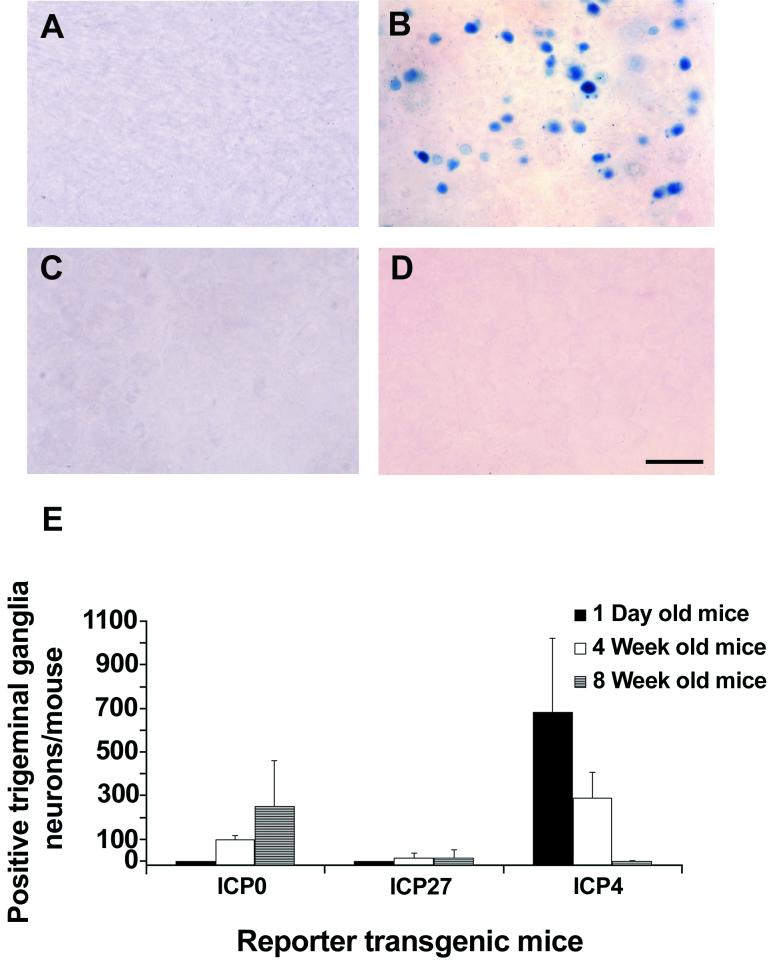
Expression of β-galactosidase in neurons of the trigeminal ganglia in adult, 4-week old, and newborn mice containing HSV-1 reporter transgenes. (A) Trigeminal ganglia of a newborn ICP0 reporter transgenic mouse (Tg3180), showing no β-galactosidase-labeled cells. (B) Trigeminal ganglia of an adult ICP0 reporter transgenic mouse (Tg3180), showing moderate numbers of β-galactosidase-labeled neurons. (C) Trigeminal ganglia of a newborn nontransgenic mouse, showing no β-galactosidase-labeled cells. (D) Trigeminal ganglia of an adult nontransgenic mouse, showing no β-galactosidase-labeled cells. (E) Bar graph showing a comparison of the number of β-galactosidase-labeled neurons in trigeminal ganglia at different ages in ICP0 β-galactosidase (Tg3180), ICP27-β-galactosidase (Tg3054), and ICP4-β-galactosidase (Tg6305) transgenic mice. Results are means ± standard errors of the means. The results for previously described ICP4 reporter transgenic mice were derived from experiments performed concurrently with those of the presently described HSV-1 reporter transgenic mice. Bar, 50 μm.
TABLE 3.
Distribution of β-galactosidase labeling in nonneural tissues of mice containing HSV-1 IE gene or late gene reporter transgenes
| Sample | No. of animals positivea
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ICP0
|
ICP27
|
gC
|
Nontransgenic | |||||||
| Tg3180 | Tg6825 | Tg3054 | Tg3058 | Tg6818 | Tg3401 | Tg3403 | Tg3405 | |||
| Cornea | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Adrenal | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Kidney | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Liver | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Spleen | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Heart | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | |
| Lung | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Intestine | 4 | 4 | 2 | 3 | 0 | 0 | 0 | 0 | 0 | |
Each number represents the number of mice out of four which contained β-galactosidase-labeled cells in the indicated nonneural tissues. The labeled cells in the outer muscle layers of the intestinal wall were myenteric plexus neurons.
ICP27 and ICP0 reporter transgenes are expressed in anatomically distinct subsets of neurons in uninfected adult mice.
Brains from four uninfected mice of each ICP0-β-galactosidase transgenic mouse line (Tg3180 and Tg6825) and ICP27-β-galactosidase transgenic mouse line (Tg3054, Tg3058, and Tg6818) were sectioned coronally and examined as described in Materials and Methods. ICP0-β-galactosidase transgenic mice (Tg3180 and Tg6825) and ICP27-β-galactosidase transgenic mice (Tg3054, Tg3058, and Tg6818) contained small to moderate numbers of positive cells in anatomically defined regions of gray matter in the brain which were determined to be neurons by morphological criteria. Cerebral cortex (external granular layer) (Fig. 3A and B), cingulum, lateral septal nucleus, hippocampus, thalamus, amygdala, and vestibular nucleus (Fig. 4A and B) contained positive cells in ICP0-β-galactosidase transgenic mice (Tg3180 and Tg6825) and ICP27-β-galactosidase transgenic mice (Tg3054, Tg3058, and Tg6818). Neurons in the external granular layer of the cerebral cortex were β-galactosidase positive in both ICP0-β-galactosidase and ICP27-β-galactosidase transgenic mice (Fig. 3A and B), while neurons in the molecular layer and other regions of the cerebral cortex were not labeled or were labeled in much smaller numbers. The superior colliculus, purkinje cell layer of the cerebellum, and spinal cord of ICP0-β-galactosidase transgenic mice (Tg3180 and Tg6825) contained some positive cells, while these regions were negative for β-galactosidase staining in ICP27-β-galactosidase transgenic mice (Tg3054, Tg3058, and Tg6818). White matter regions of the brain, which are composed of nonneuronal cells, contained no positive cells in all transgenic mouse lines (data not shown). No β-galactosidase-positive cells were present in the brains of uninfected gC-β-galactosidase transgenic mice (Tg3054, Tg3058, and Tg6818) (Fig. 3C and 4C).
β-Galactosidase-positive cells in brain and trigeminal ganglia were identified as neurons in colabeling experiments.
β-Galactosidase-positive cells in the cerebral cortex and trigeminal ganglia of the ICP0-β-galactosidase line Tg3180 were colabeled for neurofilament protein, a neuron-specific marker (Fig. 6A and B). β-Galactosidase-positive cells in the cerebral cortex and trigeminal ganglia of the ICP27-β-galactosidase line Tg3054 were also colabeled for neurofilament protein (data not shown). Neurofilament staining (brown) was seen in the neuronal cytoplasm, including the cell bodies and axons, while β-galactosidase label (blue) was located in the nucleus (Fig. 6A and B). Control mouse antibody was used on adjacent sections, and no labeling for neurofilament was detected in either cerebral cortex or trigeminal ganglia (Fig. 6C and D). As expected, β-galactosidase labeling was seen in both cerebral cortex and trigeminal ganglia in a subset of the neurofilament-positive neurons in both ICP0-β-galactosidase transgenic mice (Fig. 6A and C) and ICP27-β-galactosidase transgenic mice (results not shown).
FIG. 6.
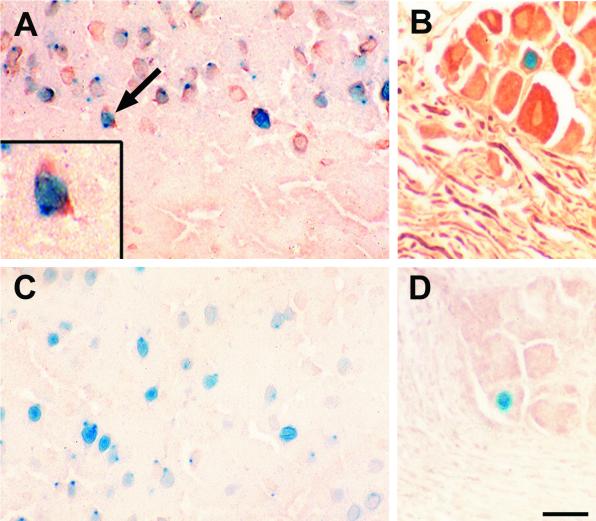
Colabeling of cells for β-galactosidase and a neuronal marker (neurofilament) in the brain and trigeminal ganglia of ICP0 reporter transgenic mice. Sections of cerebrum and trigeminal ganglia from ICP0 reporter transgenic mice (Tg3180) containing β-galactosidase-labeled neurons were double labeled using immunohistochemistry for the neuronal marker neurofilament as described in the text. (A) Section of cerebrum containing neurons double labeled for β-galactosidase and neurofilament. The brown neurofilament staining is present in cytoplasm surrounding the β-galactosidase-positive neuronal nuclei. The inset is a higher magnification of the cell identified by the arrow. Both neurons which express β-galactosidase and neurons which lack β-galactosidase expression are labeled for neurofilament. (B) Section of trigeminal ganglia containing a neuron double labeled for β-galactosidase and neurofilament and several neurofilament-positive and β-galactosidase-negative neurons. The neurofilament-positive linear profiles are axonal processes of neurons. (C) Section of cerebrum containing β-galactosidase-labeled cells which have been reacted with monoclonal anti-bovine CD3 in the immunocytochemical assay as a negative control. (D) Section of trigeminal ganglia containing β-galactosidase-labeled cells that have been reacted with monoclonal anti-bovine CD3 in the immunocytochemical assay as a negative control. Bar, 20 μm (10 μm in the inset).
ICP0 and ICP27 reporter transgenes are differentially expressed in trigeminal ganglion neurons.
The levels of activation of the HSV-1 IE gene (ICP0 and ICP27) and late gene (gC) promoters in trigeminal ganglion neurons in newborn and adult transgenic mice were compared. ICP4-β-galactosidase transgenic mice were included in the experiments as controls (Fig. 5E). In addition, trigeminal ganglia from 4-week-old ICP0-β-galactosidase, ICP27-β-galactosidase, and ICP4-β-galactosidase transgenic mice were analyzed (Fig. 5E). Uninfected trigeminal ganglia of newborn mice containing the ICP0-β-galactosidase reporter transgene (Tg3180), the ICP27-β-galactosidase reporter transgene (Tg3054), and the gC-β-galactosidase transgene (Tg3401) contained no β-galactosidase-labeled cells (Fig. 5A and E and data not shown). The ICP4 promoter was activated in large numbers of trigeminal ganglion neurons of newborn ICP4-β-galactosidase transgenic mice (Tg6305) (Fig. 5 E). Trigeminal ganglia of the adult ICP0-β-galactosidase transgenic mice (Tg3180) contained moderate numbers of β-galactosidase-positive neurons (Fig. 5B and E). Trigeminal ganglia of adult ICP27-β-galactosidase transgenic mice (Tg3054) had low numbers of β-galactosidase-positive neurons (Fig. 5E). Adult gC-β-galactosidase transgenic mice (Tg3401) had no β-galactosidase-positive cells, and adult ICP4-β-galactosidase control mice (Tg6305) had few to no positive cells in trigeminal ganglia (Fig. 5E and data not shown). Results similar to those discussed above were obtained for newborn and adult trigeminal ganglia for the other lines of mice containing each transgene: ICP0-β-galactosidase (Tg6825), ICP27-β-galactosidase (Tg3058 and Tg6818), and gC-β-galactosidase (Tg3403 and Tg3405). The numbers of positive neurons in 4-week-old ICP0-β-galactosidase (Tg3180), ICP27-β-galactosidase (Tg3054), and ICP4-β-galactosidase (Tg6305) transgenic mice were intermediate between adult and newborn mice (Fig. 5E). There also were no β-galactosidase-labeled cells in the trigeminal ganglia of newborn, 4-week-old, or adult nontransgenic control mice (Fig. 5C and D and data not shown). The differences in numbers of β-galactosidase-positive cells between newborn and adult ICP0-β-galactosidase (P = 0.001) and ICP4-β-galactosidase (P = 0.001) transgenic mice were significant (Fig. 5E). The Mann-Whitney rank sum test was used for statistical analysis. These data demonstrate that trigeminal ganglion neurons can differentially express the IE gene (ICP0) promoter, depending upon age.
DISCUSSION
Neuronal transcription factors activate the HSV-1 ICP0 and ICP27 promoters in reporter transgenic mice in the absence of viral proteins. Activation of HSV-1 IE promoters in vivo in the absence of viral proteins is apparently a unique property of neurons. Nonneuronal cells in the nervous system and in nonneural tissues generally did not activate the ICP0 and ICP27 promoters in transgenic mice. Two exceptions were the small numbers of renal tubular epithelial cells which were labeled for β-galactosidase in Tg3054 mice and the cardiac myocytes which were positively labeled in Tg6818 mice. The absence of labeling of these nonneuronal cells in any of the other transgenic mouse lines suggests that these two exceptions were anomalies. Some expression of the ICP0 promoter has been previously observed in nonneuronal cells in vitro (13, 36, 49). However, it is likely that transcriptional regulation of the ICP0 promoter in nonneuronal cells which have undergone multiplication and passage in vitro is different from regulation in the same cells in vivo. These experiments were meant to examine the regulation of the ICP0 promoter in the more relevant in vivo context. A restricted subset of neurons contain the transcription factors which are required to activate the HSV-1 ICP0 and ICP27 promoters in the absence of viral proteins. Specific anatomically defined subsets of neurons as described in Results activated the HSV-1 IE (ICP0 and ICP27) promoters in transgenic mice, while many other types of neurons did not activate these promoters.
Particularly important is the property of a specific subset of neurons to differentially regulate the ICP0 promoter depending upon changes in the neuronal environment. The transcription factors in a specific subset of neurons can be altered by changes in the neuron such as aging or differentiation (28). Expression of the ICP0-β-galactosidase transgene was significantly altered by aging (differentiation) in sensory ganglion neurons. The specific characteristics of sensory neuronal regulation of ICP0 contrasts sharply with those for ICP4 which were described in an earlier study (43). Newborn ganglia did not express the ICP0 promoter, while adult ganglia expressed the promoter in approximately 250 neurons per ganglion pair. In contrast, newborn ganglia expressed the ICP4 promoter in approximately 700 neurons per ganglion pair, while adult ganglia were negative for expression of the ICP4-β-galactosidase transgene. These results suggest that the level of activation of HSV-1 IE gene promoters can be altered by changes in the neuronal environment without any contributions from viral regulatory molecules. Further, these results suggest that the regulatory factors controlling expression of viral IE gene reporter transgenes in trigeminal ganglion neurons are specific for ICP0 and ICP4.
As stated previously, approximately 250 out of 40,000 neurons (11, 15, 56) per ganglion (0.63%) expressed ICP0-β-galactosidase in trigeminal ganglia of adult mice. The subset of ICP0-β-galactosidase-positive neurons probably represents a specific phenotype of neurons which are transcriptionally regulated in a manner different from that of other trigeminal ganglion neurons. Experiments to determine the identity of the ICP0-β-galactosidase-expressing neurons in trigeminal ganglia will be performed. It appears unlikely that the subset of neurons which express the ICP0 reporter transgene would be able to harbor a latent infection with HSV-1. This will also be determined experimentally. The important point of the presently described experiments is to demonstrate that in vivo neurons are able to transcriptionally activate the ICP0 promoter in the absence of viral proteins (including VP16) and that this property can be altered by changes in the physiological environment of the same neurons. These findings support the hypothesis that neuronal transcriptional proteins can regulate the ICP0 promoter, resulting in little or no expression during HSV-1 latency and activation of expression of the ICP0 gene as a result of changes in the properties of the neuron during reactivation.
The major drawback of these experiments is that the viral promoter cannot be examined in its natural context in the viral genome in the correct position of the gene. However, it is important to first determine the potential for neuronal regulation of HSV-1 IE (ICP0 and ICP27) promoters in the absence of viral proteins. Further experiments will be required to verify whether neuronal regulation of the HSV-1 IE promoters in the context of the viral genome varies from what has been observed in transgenic mice. Many experiments which have examined the IE promoters in cultured cells have been performed with the viral promoter inserted either into a plasmid or into the viral genome in a location different from the natural gene (2, 10, 14, 16, 22, 23, 31, 32, 34, 37, 39, 54, 64). Yet much valuable data about the regulation of viral genes in cultured cells has been derived from these studies. Previous studies have used reporter transgenes to study the function of viral promoters in transgenic mice (1, 4, 20, 33, 43, 63). In addition, one study suggests that the latent HSV-1 genome is maintained in a state which is similar to that of chromosomal DNA (12).
In summary, these experiments provide evidence that HSV-1 IE promoters (ICP0 and ICP27) can be regulated in neurons of transgenic mice by neuronal transcription factors in the absence of viral proteins. Further, sensory ganglion neurons can differentially regulate the transgenic ICP0 promoter. It seems particularly significant that the IE promoters of a neurotropic virus (HSV-1) can be differentially regulated in neurons. These results have implications for understanding the regulation of the ICP0 and ICP27 promoters in neurons in the absence of viral proteins. The capacity of sensory ganglion neurons to regulate the ICP0 promoter in the absence of viral proteins suggests the possibility that ICP0 may be important in regulation of the latent HSV genome. Experiments are currently under way to examine whether changes in activation of the ICP0 promoter occur following the types of injury to sensory ganglion neurons which have been shown to reactivate HSV-1 from latency. In addition, these viral promoters may be of use for expression of proteins in specific subsets of neurons in transgenic animals or in gene therapy protocols.
Acknowledgments
We thank Lawrence Butcher, Brandon Reinbold, and Cheri Chapman for technical assistance. Naomi Taus provided advice on and help with experiments. Mark Estes provided anti-bovine CD3 antibody. The transgenic core facilities at the University of Cincinnati and the University of Missouri generated the transgenic founders.
C.M.L. was supported by an NIH postdoctoral fellowship award (EY07007). This work was supported by NIH grants EY11855 and AI01552 to W.J.M.
REFERENCES
- 1.Aiba-Masago, S., S. Baba, R. Y. Li, Y. Shinmura, I. Kosugi, Y. Arai, M. Nishimura, and Y. Tsutsui. 1999. Murine cytomegalovirus immediate-early promoter directs astrocyte-specific expression in transgenic mice. Am. J. Pathol. 154:735-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur, J. L., C. G. Scarpini, V. Conner, R. H. Lachmann, A. M. Tolkovsky, and S. Efstathiou. 2001. Herpes simplex virus type 1 promoter activity during latency establishment, maintenance, and reactivation in primary dorsal root neurons in vitro. J. Virol. 75:3885-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babb, R., C. C. Huang, D. J. Aufiero, and W. Herr. 2001. DNA recognition by the herpes simplex virus transactivator VP16: a novel DNA binding structure. Mol. Cell. Biol. 21:4700-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baskar, J. F., P. P. Smith, G. Nilaver, R. A. Jupp, S. Hoffmann, N. J. Peffer, D. J. Tenney, A. M. Colberg-Poley, P. Ghazal, and J. A. Nelson. 1996. The enhancer domain of the human cytomegalovirus major immediate-early promoter determines cell type-specific expression in transgenic mice. J. Virol. 70:3207-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caccarno, D. V., M. M. Herman, A. Frankfurter, C. D. Katsetos, V. P. Collins, and L. J. Rubinstein. 1989. An immunohistochemical study of neuropeptides and neuronal cytoskeletal proteins in the neuroepithelial component of a spontaneous murine ovarian teratoma. Am. J. Pathol. 135:801-813. [PMC free article] [PubMed] [Google Scholar]
- 6.Cai, W., and P. A. Schaffer. 1992. Herpes simplex virus type 1 ICP0 regulates expression of immediate-early, early, and late genes in productively infected cells. J. Virol. 66:2904-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai, W., T. L. Astor. L. M. Liptak, C. Cho, D. M. Coen, and P. A. Schaffer. 1993. The herpes simplex virus type 1 regulatory protein ICP0 enhances virus replication during acute infection and reactivation from latency. J. Virol. 67:7501-7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Challberg, M. D. 1986. A method for identifying the viral genes required for herpesvirus DNA replication. Proc. Natl. Acad. Sci. USA 83:9094-9098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, E., L. E. Galle, D. J. Maggs, D. M. Estes, and W. J. Mitchell. 2000. Pathogenesis of herpes simplex virus type 1-induced corneal inflammation in perforin-deficient mice. J. Virol. 74:11832-11840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davido, D. J., and D. A. Leib. 1998. Analysis of the basal and inducible activities of the ICP0 promoter of herpes simplex virus type 1. J. Gen. Virol. 79:2093-2098. [DOI] [PubMed] [Google Scholar]
- 11.Davies, A., and A. Lumsden. 1984. Relation of target encounter and neuronal death to nerve growth factor responsiveness in the developing mouse trigeminal ganglia. J. Comp. Neurol. 223:124-137. [DOI] [PubMed] [Google Scholar]
- 12.Deshmane, S. L., and N. W. Fraser. 1989. During latency herpes simplex virus type 1 DNA is associated with nucleosomes in a chromatin structure. J. Virol. 63:943-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Devireddy, L. R., and C. J. Jones. 2000. Olf-1, a neuron-specific transcription factor, can activate the herpes simplex virus type 1-infected cell protein 0 promoter. J. Biol. Chem. 275:77-81. [DOI] [PubMed] [Google Scholar]
- 14.Douville, P., M. Hagmann, O. Georgiev, and W. Schaffner. 1995. Positive and negative regulation at the herpes simplex virus ICP4 and ICP0 TAATGARAT motifs. Virology 207:107-116. [DOI] [PubMed] [Google Scholar]
- 15.ElShamy, W. M., and P. Ernfors. 1996. Requirement of nerotrophin-3 for the survival of proliferating trigeminal ganglion progenitor cells. Development 122:2405-2414. [DOI] [PubMed] [Google Scholar]
- 16.Everett, R. D. 1986. The products of herpes simplex virus type 1 (HSV-1) immediate early genes 1, 2 and 3 can activate HSV-1 gene expression in trans. J. Gen. Virol. 67:2507-2513. [DOI] [PubMed] [Google Scholar]
- 17.Everett, R. D. 1987. A detailed mutational analysis of Vmw110, a transacting transcriptional activator encoded by herpes simplex virus type 1. EMBO J. 6:2069-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Everett, R. D. 2000. ICP0, a regulator of herpes simplex virus during lytic and latent infection. Bioessays 22:761-770. [DOI] [PubMed] [Google Scholar]
- 19.Fraser, N. W., and T. Valyi-Nagy. 1993. Viral, neuronal and immune factors which may influence herpes simplex virus (HSV) latency and reactivation. Microb. Pathog. 15:83-91. [DOI] [PubMed] [Google Scholar]
- 20.Fritschy, J. M., S. Brandner, S. Aguzzui, M. Koedood, B. Luscher, and P. J. Mitchell. 1996. Brain cell type specificity and gliosis-induced activation of the human cytomegalovirus immediate-early promoter in transgenic mice. J. Neurosci. 16:2275-2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia-Blanco, M. A., and B. R. Cullen. 1991. Molecular basis of latency in pathogenic human viruses. Science 254:815-820. [DOI] [PubMed] [Google Scholar]
- 22.Gelman, I. H., and S. Silverstein. 1986. Co-ordinate regulation of herpes simplex virus gene expression is mediated by the functional interaction of two immediate early gene products. J. Mol. Biol. 191:395-409. [DOI] [PubMed] [Google Scholar]
- 23.Gelman, I. H., and S. Silverstein. 1985. Identification of immediate early genes from herpes simplex virus that transactivate the virus thymidine kinase gene. Proc. Natl. Acad. Sci. USA 82:5265-5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grapes, M., and P. O' Hare. 2000. Differences in determinants required for complex formation and transactivation in related VP16 proteins. J. Virol. 74:10112-10121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Halford, W. P., and P. Schaffer. 2001. ICP0 is required for efficient reactivation of herpes simplex virus type 1 from neuronal latency. J. Virol. 75:3240-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harris, R. A., R. D. Everett, X. X. Zhu, S. Silverstein, and C. M. Preston. 1989. Herpes simplex virus type 1 immediate-early protein Vmw110 reactivates latent herpes simplex virus type 2 in an in vitro latency system. J. Virol. 63:3513-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He, X., M. N. Treacy, D. M. Simmons, H. A. Ingraham, I. W. Swanson, and M. G. Rosenfeld. 1989. Expression of a large family of POU-domain regulatory genes in mammalian brain development. Nature 340:35-42. [DOI] [PubMed] [Google Scholar]
- 28.Herdegen, T., and J. D. Leah. 1998. Inducible and constitutive transcription factors in the mammalian nervous system: control of gene expression by jun, fos, and krox, and creb/atf proteins. Brain Res. Rev. 28:370-390. [DOI] [PubMed] [Google Scholar]
- 29.Hobbs, W. E., D. E. Brough, I. Kovesdi, and N. A. DeLuca. 2001. Efficient activation of viral genomes by levels of herpes simplex virus ICP0 insufficient to affect cellular gene expression or cell survival. J. Virol. 75:3391-3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hogan, B., R. Beddington, F. Costantini, and E. Lacy. 1994. Manipulating the mouse embryo: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Homa, F. L., T. M. Otal, J. C. Glorioso, and M. Levine. 1986. Transcriptional control signals of a herpes simplex virus type 1 late (gamma 2) gene lie within bases −34 to +124 relative to the 5" terminus of the mRNA. Mol. Cell. Biol. 6:3652-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones, K. A., and R. Tijan. 1985. Sp1 binds to promoter sequences and activates herpes simplex virus "immediate-early' gene transcription in vitro. Nature 317:179-185. [DOI] [PubMed] [Google Scholar]
- 33.Koedad, M., A. Fichtel, P. Meier, and P. J. Mitchell. 1995. Human cytomegalovirus (HCMV) immediate-early enhancer/promoter specificity during embryogenesis defines target tissues of congenital HCMV infection. J. Virol. 69:2194-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lachman, R. H., M. Sadarangani, H. R. Atkinson, and S. Efstathiou. 1999. An analysis of herpes simplex virus gene expression during latency establishment and reactivation. J. Gen. Virol. 80:1271-1282. [DOI] [PubMed] [Google Scholar]
- 35.Leib, D. A., D. M Coen, C. L. Bogard, K. A. Hicks, D. R. Yager, D. M. Knipe, K. L. Tyler, and P. A. Schaffer. 1989. Immediate-early regulatory gene mutants define different stages in the establishment and reactivation of herpes simplex virus latency. J. Virol. 63:759-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu, R., P. Yang, P. O'Hare, and V. Misra. 1997. Luman, a new member of the CREB/ATF family, binds to herpes simplex virus VP16-associated host cellular factor. Mol. Cell. Biol. 17:5117-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mackem, S., and B. Roizman. 1982. Structural features of the herpes simplex virus α gene 4, 0, and 27 promoter-regulatory sequences which confer a regulation on chimeric thymidine kinase genes. J. Virol. 44:939-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maggs, D., E. Chang, M. Nasisse, and W. J. Mitchell. 1998. Persistence of herpes simplex virus type 1 DNA in chronic conjunctival and eyelid lesions of mice. J. Virol. 72:9166-9172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mavromara-Nazos, P., J. Hubenthal-Voss, J. L. C. McKnight and B. Roizman. 1986. Regulation of herpes simplex virus 1 genes: α gene sequence requirements for transient induction of indicator genes regulated by β or late (γ 2) promoters. Virology 149:152-164. [DOI] [PubMed] [Google Scholar]
- 40.McGeoch, D., M. A. Dalrymple, A. J. Davidson, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 41.McKnight, J. L. C., T. M. Kristie, and B. Roizman. 1987. The binding of the virion protein mediating a gene induction in herpes simplex virus type 1 infected cells to its cis site requires cellular proteins. Proc. Natl. Acad. Sci. USA 84:7061-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mercer, E. H., G. W. Hoyle, R. P. Kapur, R. L. Brinster, and R. D. Palmiter. 1991. The dopamine β-hydroxylase gene promoter directs expression of E. coli lacZ to sympathetic and other neurons in adult transgenic mice. Neuron 7:703-716. [DOI] [PubMed] [Google Scholar]
- 43.Mitchell W. J. 1995. Neurons differentially control expression of a herpes simplex virus type 1 immediate-early promoter in transgenic mice. J. Virol. 69:7942-7950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mitchell, W. J., P. Gressens, J. R. Martin, and R. DeSanto. 1994. Herpes simplex virus type 1 DNA persistence, progressive disease and transgenic immediate early gene promoter activity in chronic corneal infections in mice. J. Gen. Virol. 75:1201-1210. [DOI] [PubMed] [Google Scholar]
- 45.Mitchell, W. J., R. DeSanto, S. Zhang, W. F. Odenwald, and H. Arnheiter. 1993. Herpes simplex virus pathogenesis in transgenic mice is altered by the homeodomain protein Hox 1.3. J. Virol. 67:4484-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mitchell, W. J., R. P. Lirette, and N. W. Fraser. 1990. Mapping of low abundance latency-associated RNA in the trigeminal ganglia of mice latently infected with herpes simplex virus type 1. J. Gen. Virol. 71:125-132. [DOI] [PubMed] [Google Scholar]
- 47.O'Hare, P., and C. R. Goding. 1988. Herpes simplex virus regulatory elements and the immunoglobulin octamer domain bind a common factor and are both targets for virion transactivation. Cell 52:435-445. [DOI] [PubMed] [Google Scholar]
- 48.O'Hare, P., and G. S. Hayward. 1985. Evidence for a direct role for both the 175,000- and 110,000-molecular-weight immediate-early proteins of herpes simplex virus in the transactivation of delayed-early promoters. J. Virol. 53:751-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O'Rourke, D., and P. O'Hare. 1993. Mutually exclusive binding of two cellular factors within a critical promoter region of the gene for the IE110k protein of herpes simplex virus. J. Virol. 67:7201-7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perry, L. J., and D. McGeoch. 1988. The DNA sequences of the long repeat region and adjoining parts of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:2831-2846. [DOI] [PubMed] [Google Scholar]
- 51.Post, L. E., A. J. Conley, E. S. Mocarski, and B. Roizman. 1980. Cloning of reiterated and nonreiterated herpes simplex virus 1 sequences as BamHI fragments. Proc. Natl. Acad. Sci. USA 77:4201-4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Preston, C. M., M. C. Frame, and M. E. M. Campbell. 1988. A complex formed between cell components and an HSV structural polypeptide binds to a viral immediate early gene regulatory DNA sequence. Cell 52:425-434. [DOI] [PubMed] [Google Scholar]
- 53.Resnick, J., B. A. Boyd, and M. L. Haffey. 1989. DNA binding by the herpes simplex virus type 1 ICP4 protein is necessary for efficient down regulation of the ICP0 promoter. J. Virol. 63:2497-2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roizman, B., and A. E. Sears. 1996. Herpes simplex viruses and their replication, p. 2231-2296. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, vol. 2. Lippincott, Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 55.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 56.Sawtell, N. M. 1997. Comprehensive quantification of herpes simplex virus latency at the single-cell level. J. Virol. 71:5423-5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Steiner, I., J. G Spivack, S. L. Deshmane, C. I. Ace, C. M. Preston, and N. W. Fraser. 1990. A herpes simplex virus type 1 mutant containing a nontransinducing Vmw65 protein establishes latent infection in vivo in the absence of viral replication and reactivates efficiently from explanted trigeminal ganglia. J. Virol. 64:1630-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stern, S., M. Tanaka, and W. Herr. 1989. The Oct-1 homeodomain directs formation of a multiprotein-DNA complex with the HSV transactivator VP16. Nature 341:624-630. [DOI] [PubMed] [Google Scholar]
- 59.Stevens, J. G. 1975. Latent herpes simplex virus and the nervous system. Curr. Top. Microbiol. Immunol. 70:31-50. [DOI] [PubMed] [Google Scholar]
- 60.Stevens, J. G. 1989. Human herpes viruses: a consideration of the latent state. Microbiol. Rev. 53:318-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tensor, R. B., W. A. Edris, and K. A. Hay. 1993. Neuronal control of herpes simplex virus latency. Virology 195:337-347. [DOI] [PubMed] [Google Scholar]
- 62.Valyi-Nagy, T., S. L. Deshmane, A. Dillner, and N. W. Fraser. 1991. Induction of cellular transcription factors in trigeminal ganglia of mice by corneal scarification, herpes simplex virus type 1 infection, and explantation of trigeminal ganglia. J. Virol. 65:4142-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van den Pol, A. N., and P. K. Ghosh. 1998. Selective neuronal expression of green fluorescent protein with cytomegalovirus promoter reveals entire neuronal arbor in transgenic mice. J. Neurosci. 18:10640-10651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wagner, E. K., M. D. Petroski, N. T. Pande, P. T. Lieu, and M. Rice. 1998. Analysis of factors influencing kinetics of herpes simplex virus transcription utilizing recombinant virus. Methods 16:105-116. [DOI] [PubMed] [Google Scholar]
- 65.Whitley, R. J. 1996. Herpes simplex viruses, p. 2297-2342. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, vol. 2. Lippincott, Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 66.Wilcox, C. L., R. L. Smith, R. D. Everett, and D. Mysofski. 1997. The herpes simplex virus type 1 immediate-early protein ICP0 is necessary for the efficient establishment of latent infection. J. Virol. 71:6777-6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang, Y.-F., G. B. Devi-Rao, M. Rice, R. M. Sandri-Goldin, and E. K. Wagner. 1987. The effect of elevated levels of herpes simplex virus alpha-gene products on the expression of model early and late genes in vivo. Virology 157:99-106. [DOI] [PubMed] [Google Scholar]
- 68.Zhu, X. X., J. X. Chen, C. S. Young, and S. Silverstein. 1990. Reactivation of latent herpes simplex virus by adenovirus recombinants encoding mutant ICP0 products. J. Virol. 64:4489-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zindy, F., J. J. Cunningham, C. J. Sherr, S. Jogal, R. J. Smeyne, and M. F. Roussel. 1999. Postnatal neuronal proliferation in mice lacking Ink4d and Kip1 inhibitors of cyclin-dependent kinases. Proc. Natl. Acad. Sci. USA 96:13462-13467. [DOI] [PMC free article] [PubMed] [Google Scholar]