Abstract
The solitary long terminal repeats (LTRs) of ERV-9 endogenous retrovirus contain the U3, R, and U5 regions but no internal viral genes. They are middle repetitive DNAs present at 2,000 to 4,000 copies in primate genomes. Sequence analyses of the 5" boundary area of the erythroid β-globin locus control region (β-LCR) and the intron of the embryonic axin gene show that a solitary ERV-9 LTR has been stably integrated in the respective loci for at least 15 million years in the higher primates from orangutan to human. Functional studies utilizing the green fluorescent protein (GFP) gene as the reporter in transfection experiments show that the U3 region of the LTRs possesses strong enhancer activity in embryonic cells of widely different tissue origins and in adult cells of blood lineages. In both the genomic LTRs of embryonic placental cells and erythroid K562 cells and transfected LTRs of recombinant GFP plasmids in K562 cells, the U3 enhancer activates synthesis of RNAs that are initiated from a specific site 25 bases downstream of the AATAAA (TATA) motif in the U3 promoter. A second AATAAA motif in the R region does not serve as the TATA box or as the polyadenylation signal. The LTR-initiated RNAs extend through the R and U5 regions into the downstream genomic DNA. The results suggest that the ERV-9 LTR-initiated transcription process may modulate transcription of the associated gene loci in embryonic and hematopoietic cells.
The solitary long terminal repeats (LTRs) of human endogenous retroviruses comprise approximately 5% of the human genome and belong to the category of middle repetitive DNAs characterized as retrotransposons (14, 19, 24, 35). These solitary LTRs contain the U3 enhancer and promoter region, the transcribed R region whose 5" end marks the initiation site of retroviral RNA synthesis, and the U5 region (27) but no internal gag, pol, and env genes. During primate evolution, the LTRs were apparently self-replicated and inserted into various host chromosomal sites. The functional roles in the host genomes of these transposed, repetitive DNAs are not clear. The LTR retrotransposons have been suggested to be selfish DNAs that do not serve relevant host functions (8). However, recent findings indicate that the solo LTRs can provide enhancers and promoters for cis-linked genes and regulate host gene transcription (9, 10, 22, 25, 26, 29).
The human genome contains approximately 50 copies of the ERV-9 endogenous retrovirus and an additional 3,000 to 4,000 copies of solitary ERV-9 LTRs (15, 17, 19, 35, 36). Compared with the LTRs of other families of endogenous retroviruses, the ERV-9 LTRs exhibit an unusual sequence feature: the U3 regions contain from 5 to 17 tandem repeats of 37 to 41 bases (17, 18) with recurrent GATA (23), CCAAT (28), and CCACC (21) motifs potentially capable of binding to cognate transcription factors expressed in embryonic and hematopoietic cells. This suggests that the ERV-9 enhancer and promoter could be active in those cells.
To gain further insight into the stability and functional significance of the ERV-9 LTRs, here we have mapped the erythroid β-globin and embryonic axin gene loci in primates by using human primers in PCR. We found a solitary ERV-9 LTR that is conserved in identical locations in the 5" boundary area of the β-globin gene locus and in the axin gene in the higher primates orangutan, gorilla, chimpanzee, and human, whose ancestors diverged over an evolutionary period of 15 million years (12). In the lower primates gibbon and monkey, whose ancestors diverged from the human ancestor 18 and 25 million years ago, respectively (12), the globin and axin LTRs are absent in the respective gene loci. However, other ERV-9 LTRs are detectable in the monkey genome. These results indicate that copies of the ERV-9 LTRs present in the lower primates were inserted into the globin and axin gene loci in the common ancestor of the higher primates 15 to 18 millions years ago and have remained stably integrated in the host sites during the ensuing years of primate evolution.
To assess the functional significance of the ERV-9 LTR retrotransposons, we have developed a simple and quantitative transfection assay using the green fluorescent protein (GFP) gene as the reporter and fluorescence-activated cell sorter (FACS) analyses to determine the ERV-9 LTR enhancer activity in a wide spectrum of human cells and cell lines. The results show that the U3 region of the globin LTR in both human and chimpanzee and of the axin LTR in human possesses strong enhancer activity in cells of hematopoietic lineages and even stronger enhancer activity in many embryonic cell lines of widely different tissue origins. RNA analyses using rapid amplification of cDNA 5" ends (5"RACE) (11) demonstrate that the U3 enhancer initiates transcription from a specific site downstream of the AATAAA (TATA) motif in the U3 promoter. A second AATAAA motif in the R region of the LTR does not serve as the TATA box or as the polyadenylation signal for the LTR-initiated RNAs. The LTR RNAs extend through the R and U5 regions into the GFP reporter gene in integrated recombinant constructs and into the downstream genomic DNA in the endogenous genomes of embryonic cells and adult erythroid cells. The possible functional significance of the ERV-9 LTR enhancer in regulating transcription of the cis-linked gene loci during early ontogeny and hematopoietic differentiation is discussed.
MATERIALS AND METHODS
Mapping the ERV-9 LTRs in the 5" boundary area of the β-LCR and in the axin gene in primates by PCR using human primers.
Genomic DNAs used as the template in PCRs were isolated from primate blood samples obtained from the Yerkes Primate Center of Emory University. Three sets of overlapping primer pairs located upstream of the LTR, spanning the LTR, and downstream of the LTR were synthesized according to the known human DNA sequences of the 5" boundary area of the β-globin locus control region (β-LCR) (accession no. AF064190). The respective locations of the primer pairs in AF064190 are as follows: I, positions 2239 to 2260 and 3260 to 3281; II, 3247 to 3271 and 4419 to 4443 (spanning the ERV-9 LTR); and III, 4432 to 4452 and 4900 to 4924. In the human axin locus (accession no. AC005202), the forward and reverse primer pairs spanning the ERV-9 LTR are as follows: positions 12646 to 12669 and 14456 to 14478. The primate amplicons were checked for authenticity first by the digestion patterns of restriction enzymes that cleaved the corresponding human amplicons and second by DNA sequencing, using the cycle sequencing technique with fluorescent dideoxy terminators as described previously (18). For the sequencing reaction, the PCR amplicons were used either directly or after having been inserted into plasmid vectors.
Slot blots.
Membranes containing genomic DNAs from various primate and nonprimate sources were hybridized to the 5"HS5 LTR probe at 60°C overnight in buffer solution without carrier DNA (7% sodium dodecyl sulfate, 1% bovine serum albumin, 1 mM EDTA, 250 mM Na2HPO4 [pH 8]). After hybridization, the membranes were washed four times in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate at room temperature and twice for 30 min in 0.5× SSC at 60°C. Signal intensity was quantified with a PhosphorImager (Molecular Dynamics).
Construction of recombinant GFP plasmids.
The GFP plasmids (see Fig. 5a) were made from pEGFP-C1 vector (Clontech) which was digested with AseI and NheI to generate the vector backbone containing the GFP reporter gene and the simian virus 40 poly(A) signal downstream of the GFP gene. The inserts were generated by PCR with forward and reverse primers containing the corresponding AseI and NheI ends either from a phage template spanning the 5" boundary area of the human β-LCR (18) or from human K562 or chimpanzee genomic DNAs. To generate the following PCR DNAs spanning the human and chimpanzee β-globin (E-P-r), the positions of forward and reverse PCR primers in AF064190 were 2650 to 2672 and 3965 to 3987, and those for the axin (E-P-r) in AC005202 were 12908 to 12931 and 13931 to 13955. The (E-P-r)-fragments contained the first 55 bases of the R region upstream of the AATAAA motif in the R region (see Fig. 5a). The authenticity of the PCR fragment was confirmed by DNA sequencing. The reference GFP plasmid was made by recircularizing the vector with an AseI-NheI adapter.
FIG. 5.
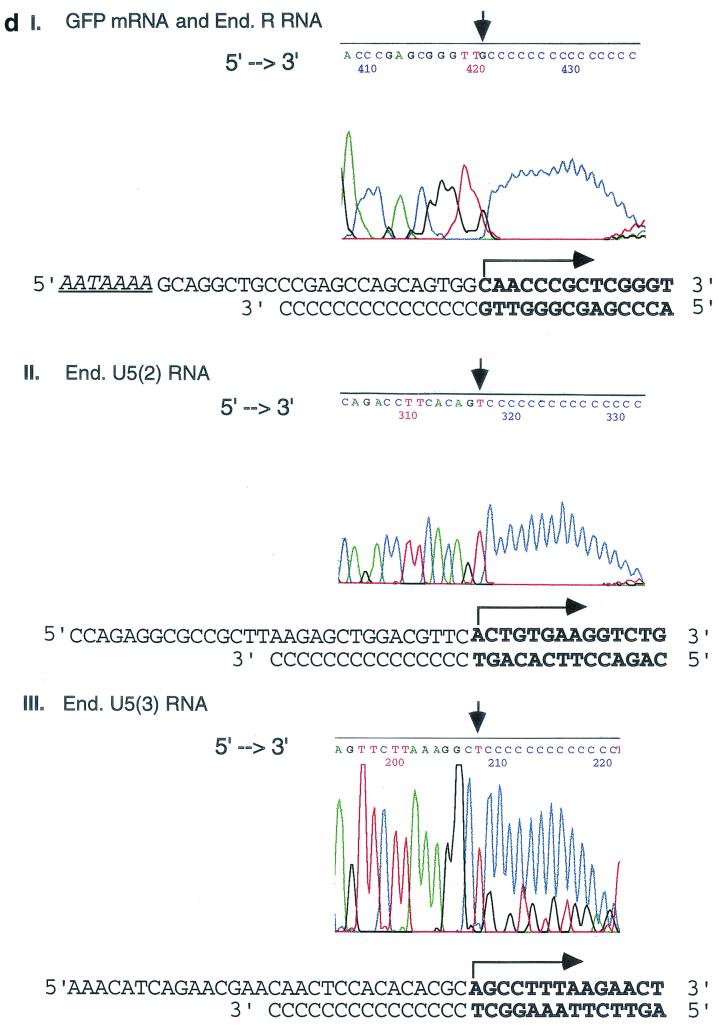
Transcriptional initiation sites of 5"HS5 LTR in transfected plasmids and in the endogenous human genomes mapped by 5"RACE. (a) Mapping the 5" ends of RNAs transcribed from 5"HS5 (E-P-R)-GFP plasmid integrated into K562 cells. Top, plasmid map. E, P, R, and GFP, same as in Fig. 1b. Angled arrow, transcriptional initiation site in the LTR marking the 5" border of the R region. Left-to-right arrows, GFP mRNA. Two right-to-left arrows, cDNA reverse transcribed from the GFP mRNA and DNA amplified from the cDNA template after 35 cycles of PCR, respectively. The PCR fragment depicted was the subsequently sequenced DNA strand; the arrows are aligned with the plasmid map on top. (C)n, poly(dC) tails added to the 3" ends of the cDNAs by the terminal deoxynucleotide transferase enzyme. Arrowheads at the 5" ends of the cDNA or the PCR fragment, reverse primers used for cDNA synthesis, PCR amplification, or DNA sequencing (seq.). Numbers, sizes in nucleotides of the cDNAs estimated from the locations of the initiation sites of the GFP mRNAs and of the PCR fragments determined from gel electrophoresis and DNA sequencing (see panels c and d). (b) Mapping of the 5" ends and initiation sites of the 5"HS5 LTR RNAs transcribed from the endogenous genome of K562 cells. Angled arrows, locations of the three transcriptional initiation sites mapped by 5"RACE; R with ∗, RNA initiation site found also in placental cells (see panel c, lane P); U5 with ∗∗, RNA initiation site found also in HeLa cells (see panel c, lane H). Left-to-right arrows, LTR R, U5(2), and U5(3) RNAs. The 5" ends of these three RNAs are located at the 5" borders of the R region and the second and third repeats of the U5 region, respectively. The 3" ends of the RNAs were drawn to the locations of the reverse primers used in the cDNA synthesis, although the actual 3" ends of the RNAs may be located further downstream. Right-to-left arrows and other designations, same as in panel a. (c) Gel electrophoresis of PCR fragments used for sequencing. The PCR fragments were generated from the following RNAs: Lane K1, GFP mRNA transcribed from the integrated 5"HS5 (E-P-r)-GFP plasmid; lanes K2, P, H, and K3, endogenous (End.) RNAs of nontransfected K562 cells, placental cells, and HeLa cells and a duplicate sample of the K562 RNAs, respectively. The band in lane K1 was generated by 35 cycles of PCR; the bands in lanes K2, P, H, and K3 were generated by 2 × 35 cycles of PCR with nested primers; and the 580-bp band in lane K3 was skewed upward due to a tear in the gel. Numbers on the right margins, sizes of the PCR DNAs in base pairs; lanes M, size markers; numbers on the left margins, sizes of the size marker bands in base pairs (the top three bands are 1,500, 1,250, and 1,000 bp, respectively; shorter bands are spaced 100 bp apart). (d) Electropherograms showing the locations of the 5" ends of GFP mRNA and the endogenous R RNA (panel I), the endogenous U5(2) RNA (panel II), and the U5(3) RNA (panel III). The electropherogram presented in panel I is generated by endogenous K562 5"HS5 LTR R RNA (the identical electropherograms of GFP mRNA and placental 5"HS5 LTR R RNA are not shown). 5"→3", the 5"→3" direction of the DNA sequences in the electropherograms. The vertical arrows mark the 3" ends of the cDNAs abutting the poly(dC) tails; the corresponding RNA initiation sites are marked with angled arrows in the DNA templates shown below the electropherograms. Boldface letters, transcribed bases in the sense DNA strand (top strands) and in the complementary cDNAs (bottom strands). For complete DNA sequences of the R and U5 regions, see reference 18.
Transfection assays and fluorescent flow cytometry analyses.
Circular GFP plasmids (10 μg each) or linearized plasmids (20 μg each, cleaved at a unique ApaLI site upstream of the AseI cloning site in the vector) were transfected by electroporation in duplicate or triplicate into 4 × 106 host cells in 400 μl of medium without fetal calf serum at 240 V and 960 μF in a Gene Pulser II (Bio-Rad). Enhancer-promoter activities of the transfected plasmids were analyzed 48 h later in a FACSCalibur with Cellquest software (Becton Dickinson). A total of 2 × 104 live cells were analyzed for each sample; necrotic cells were excluded from the FACS analyses by propidium iodide staining. The expression levels of the GFP gene in the transfected plasmids were calculated as shown in Fig. 4b. The calculated GFP levels were then corrected with respect to the copy numbers of the transfected GFP gene, which were determined by PCR as described below. Transfected cells were harvested at the time of FACS analyses and treated with DNase I to remove extracellular plasmid DNA. Cellular DNAs were isolated as described previously (30). The DNAs (0.2 μg) were amplified for 19 cycles with a GFP primer pair (positions 616 to 631 and 956 to 981 in the pEGFP-C1 sequence map) (Clontech). The PCR conditions were as described in “RT-PCRs” below. The intensities of the PCR bands were quantified with an IS1000 Image Analyzer (Alpha Innotech). The copy numbers of the transfected GFP gene based on PCR band intensities were obtained from a standard curve of PCR band intensities, generated under PCR conditions identical to those for the test samples, from aliquots of K562 DNA containing a range of 20 to 120 copies of pEGFP plasmid per cell. Under the PCR conditions used, the PCR band intensities were proportional to the copy numbers of the pEGFP plasmid and were thus within the linear range of PCR amplifications. The calculated GFP copy numbers were next normalized with respect to an internal control PCR band generated from the same test samples with a β-actin primer pair (Stratagene) after 24 cycles to correct for possible sampling errors in the PCR. Human cell lines used as transfection hosts were obtained from the American Type Culture Collection and grown in specified media (31). Human peripheral blood erythroid progenitor cells (CFU-E) were purified and grown as described previously (34).
FIG. 4.
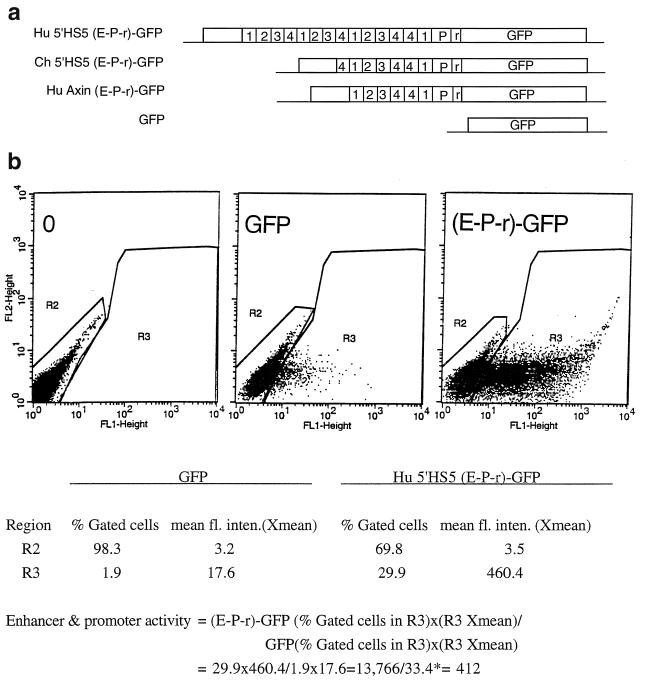
Enhancer activities of human and chimpanzee 5"HS5 LTRs and human axin LTR determined by transfection assays. (a) Maps of the transfected GFP plasmids. Hu 5"HS5 (E-P-r)-GFP and Ch 5"HS5 (E-P-r)-GFP, human and chimpanzee 5"HS5 LTR enhancer and promoter coupled to the GFP gene. The reference GFP plasmid contains no enhancer and promoter sequences 5" of the GFP gene. (b) Calculation of the enhancer-promoter activity of the transiently transfected human 5"HS5 (E-P-r)-GFP plasmid. Left, middle, and right panels, dot plots by FACS analyses of K562 cells transfected with Tris buffer, reference GFP plasmid, and (E-P-r)-GFP plasmids, respectively. x axis, GFP fluorescence intensities of the transfected cells; y axis, FL2 channel. The dot plots are the same whether FL2 or side scatter was used as the y axis in the Cellquest program. However, using FL2 as the y axis produced more compact and thus more easily gated fluorescent and nonfluorescent cell populations. R2 region, nonfluorescent cells; R3 region, fluorescent cells. The table below the dot plots shows quantitative analysis by the Cellquest program of the dot plot data. Xmean, mean fluorescence intensities (fl. inten.) of the gated cells. Below the table is a calculation of the enhancer-promoter activity of human 5"HS5 (E-P-r)-GFP in K562 cells. The enhancer-promoter activity after normalization with respect to the ratio of the copy number of transfected (E-P-r)-GFP plasmid to that of transfected GFP plasmid is 412/1.6 = 258. ∗, in transfections where this number was less than 1 for the reference GFP plasmid, a value of 1 was used in the calculation to obtain minimum enhancer-promoter activities.
K562 cells containing integrated plasmids.
The 5"HS5 and axin (E-P-R)-GFP plasmids were linearized at the unique ApaLI site in the vector before transfection into K562 cells by electroporation. K562 cell populations containing integrated plasmids were cultured and selected with G418 as previously described (3). The average per-cell copy numbers of the integrated plasmids were determined by Southern blotting (32) and/or by the PCR method as described above.
RNA isolation.
Total cellular RNAs were isolated with the Totally RNA kit (Ambion) from nontransfected Bewo, K562, and HeLa cells and transfected K562 cells containing integrated plasmids and also from the chorionic trophoblasts of fresh placentas of newborn infants (6). The isolated RNAs were treated with RNase-free DNase I to eliminate possible DNA contamination before being used as templates in reverse transcription-PCR (RT-PCR) and 5"RACE.
5" RACE.
The 5" RACE kit (Gibco BRL) was used according to the vendor's protocol. In brief, cDNAs were first synthesized from the total cellular RNAs by using reverse primers specific to the GFP gene or the HS5 site (see Fig. 5a). Polydeoxycytosines were then added to the 3" ends of the cDNAs by using terminal deoxynucleotide transferase. The cDNAs with the poly(dC) tails were then amplified with 35 cycles of PCR using a nested gene-specific reverse primer and a universal anchored forward primer, poly(dG). Following this, another 35 cycles of PCR were carried out, using a second set of nested, gene-specific reverse primers and the universal anchored forward primer. After the second round of PCR, the amplicons were purified by agarose gel electrophoresis and sequenced by the Molecular Biology Core Laboratory using the cycle sequencing technique. The positions of the gene-specific, nested reverse primers used for cDNA synthesis, two rounds of PCR amplifications, and DNA sequencing, respectively, were as follows. In the GFP gene (see Fig. 5), the primers used were at positions 826 to 850, 736 to 757, 617 to 640, and 617 to 640 (see Clontech manual on pEGFP-C1 for corresponding primer sequences). In the endogenous DNA region between the 5"HS5 LTR (located at positions 3250 to 4349 [accession number AF064190]) and the HS5 site (located at positions 5472 to 6710), the primers used were at positions 6267 to 6289, 5522 to 5545, 4950 to 4974, and 4470 to 4493 or 4355 to 4379 (accession number AF064190).
RT-PCRs.
Two to five micrograms of the endogenous total cellular RNAs isolated from various cells was used as the template in each RT reaction; aliquots of cDNAs transcribed from 400 ng of RNAs were used in the subsequent PCRs with appropriate primer pairs. The RT step was carried out with Moloney murine leukemia virus reverse transcriptase (Gibco-BRL) at 42°C for 60 min. The PCR conditions were as follows: denaturation at 94°C for 1 min, annealing at 58°C for 1 min, and extension at 72°C for 1 min, repeated for 32 cycles, if not otherwise specified. PCR products (5 μl of 50 μl) were analyzed by electrophoresis in 2% agarose gels. For Fig. 8, to detect polyadenylated RNAs, the reverse primer used for cDNA synthesis was (T)33 (5" 33[T]-C/G/A-C/G/A/T 3"). The coordinates in AF064190 of the PCR primers were as follows: F1, 3247 to 3271; F2, 4003 to 4028; G1, 4469 to 4493; F3, 5522 to 5545; and G2, 6267 to 6289. In nested PCRs for Fig. 5c, 3 μl of 50 μl of first-round PCR products after 25 cycles was used as templates for the nested, second-round PCR for an additional 25 cycles. For Fig. 7 to determine the transcriptional direction of the 5"HS5 LTR and downstream genomic DNA up to the HS5 site, the coordinates in AF064190 of the forward and reverse primers in primer pairs 1 to 5 were as follows: 1, 4003 to 4028 and 4469 to 4493 (same as F2-G1 in Fig. 6); 2, 4482 to 4502 and 4950 to 4974; 3, 4950 to 4974 and 5522 to 5542; 4, 5522 to 5545 and 6267 to 6289 (same as F3-G2 in Fig. 6); and 5, 6267 to 6289 and 6695 to 6717. To detect sense RNA colinear with the transcriptional direction of the β-LCR and the β-like globin genes, the reverse primers in each primer pair were used in the RT step to synthesize cDNAs; to detect antisense RNAs, the forward primers in each primer pair were used in the RT step. For Fig. 8 to determine the transcriptional direction of the axin LTR and the axin gene, the coordinates in AC005202 of the forward and reverse primers in primer pairs 1 to 4 were as follows: 1, 18040 to 18064 and 18477 to 18501; 2, 13969 to 13994 and 14605 to 14629; 3, 13969 to 13994 and 14221 to 14244; and 4, 11549 to 11573 and 11941 to 11965.
FIG. 8.
Transcriptional direction of the human axin gene locus. (a) Map of the second axon and second intron of the human axin locus. Angled arrows, transcriptional directions of the axin gene and the intronic ERV-9 LTR. Horizontal arrows, RNAs detected by RT-PCR primer pair 1, located in exon 2; by primer pairs 2 and 3, which spanned the ERV-9 LTR and part of intron 2 between the LTR and the second intron; and by primer pair 4, which spanned a further-downstream region of the second intron. Right-to-left arrows, sense RNAs colinear with the transcription direction of the axin gene. Left-to-right arrow, antisense RNAs transcribed from within the ERV-9 LTR and extended into intron 2 DNA. Numbers below the arrows, sizes of the RT-PCR bands in base pairs. (b) RT-PCR products. Lanes 1 to 4, RT-PCR bands generated by primer pairs 1 to 4, respectively. + lanes, sense RNAs colinear with the axin gene. − lanes, LTR-initiated RNAs in the antisense direction to the axin gene. White dots, anticipated sizes of the RT-PCR bands. Lanes M, 100-base DNA size markers from 100 to 1,000 bp. The top two bands are 1,250 and 1,500 bp, respectively.
FIG. 7.
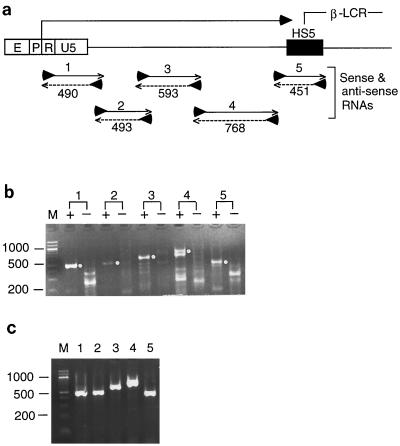
Transcriptional direction of the RNAs between the 5"HS5 LTR and HS5 site. (a) The RNAs transcribed from DNA between the 5"HS5 LTR and HS5 site of the β-LCR were exclusively in the sense direction in nontransfected K562 cells as determined by RT-PCRs. Horizontal left-to-right arrow, sense RNAs transcribed from the LTR into the HS5 site as amplified with primer pairs 1 to 5. Numbers below the PCR products, sizes of the RT-PCR bands in base pairs. (b) Gel electrophoresis of RT-PCR products synthesized with locus-specific primers. Lanes 1 to 5, PCR bands amplified from cDNAs by primer pairs 1 to 5, respectively. + lanes, PCR bands generated from the sense R RNAs using the reverse primer of primer pairs 1 to 5 in the RT step. − lanes, PCR bands generated from the antisense RNAs using the forward primers of primer pairs 1 to 5 in the RT step. White dots on the right margins, PCR bands anticipated to be generated by primer pairs 1 to 5. (c) PCR products amplified from cDNAs synthesized with random hexamer primers in the RT step. Lanes 1 to 5, PCR bands amplified from the cDNAs by primer pairs 1 to 5, respectively.
FIG. 6.
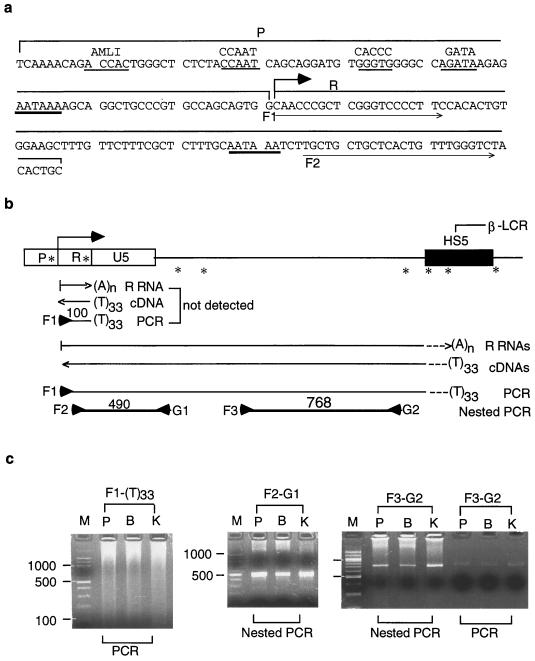
Polyadenylated 5"HS5 LTR RNAs detected by RT-PCR. (a) DNA sequences of the U3 promoter (P) and R regions of 5"HS5 LTR. The two AATAAA motifs with heavy underlines are the TATA box in the promoter and the potential polyadenylation signal in the R region, respectively. Angled arrow, LTR transcriptional initiation site. Underlined bases in the promoter region, binding sites for transcription factors AML1 (GTGGT), CCAAT, CACCC, and GATA (21, 23, 28). Thin horizontal arrows, F1 and F2 forward primers used in the RT-PCRs to amplify LTR cDNAs. (b) The AATAAA motif in the R region did not serve as the polyadenylation signal to terminate the LTR R RNAs. Top, genomic map of the region between the 5"HS5 LTR and the HS5 site. Angled arrow, transcriptional initiation site of the LTR R RNA. ∗, locations of the AATAAA motifs in the LTR and six additional AATAAA motifs or potential polyadenylation signals between the 5"HS5 LTR and the 3" end of the HS5 site. Left-to-right arrows, polyadenylated LTR R RNAs; dotted lines, different DNA sequences present in the 3" ends of the R RNAs generated by different potential polyadenylation signals in the region. (A)n, poly(A) tails of unknown lengths. (T)33, oligo(dT) primer with 33 Ts used as the reverse primer in cDNA synthesis and PCR. Horizontal line bracketed by forward F1 and reverse (T)33 primers, PCR products generated from the cDNAs by primer pair F1-(T)33. Thick lines bracketed by arrowheads, second-round nested PCRs amplified from the F1-(T)33 PCR products by nested primer pairs F2-G1 and F3-G2. The positions of all horizontal arrows, lines, and arrowheads are aligned with the genomic map of the region at the top. (c) Gel electrophoresis of PCR and nested PCR products. Left panel, PCR bands generated by the F1-(T)33 primer pair from LTR R RNAs of placenta (lane P), Bewo (lane B), and K562 (lane K) cells. Lane M, DNA size markers, the same as in Fig. 2c. The PCR bands were generated from cDNA templates after 25 PCR cycles. Center and right panels, the nested PCR bands were amplified from the F1-(T)33 PCR products after 25 additional PCR cycles by nested primer pair F2-G1 or F3-G2.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the 5"HS5 ERV-9 LTR in the β-globin gene locus are as follows: human, AF064190; chimpanzee, AF139840; gorilla, AF094515; orangutan, AF141972; gibbon, AF141973; and monkey, AF141975. Those for the ERV-9 LTR in the axin gene locus are as follows: human, AC005202; chimpanzee, AF227995; gorilla, AF227994; orangutan, AF227993; gibbon, AF227992; and monkey, AF227991.
RESULTS
Conservation of the ERV-9 LTRs during primate evolution.
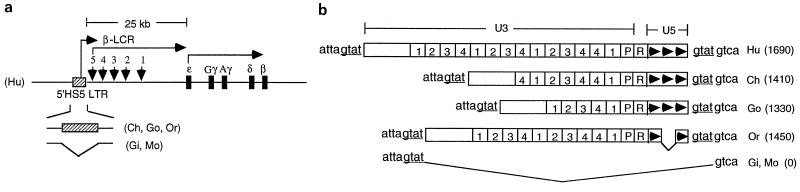
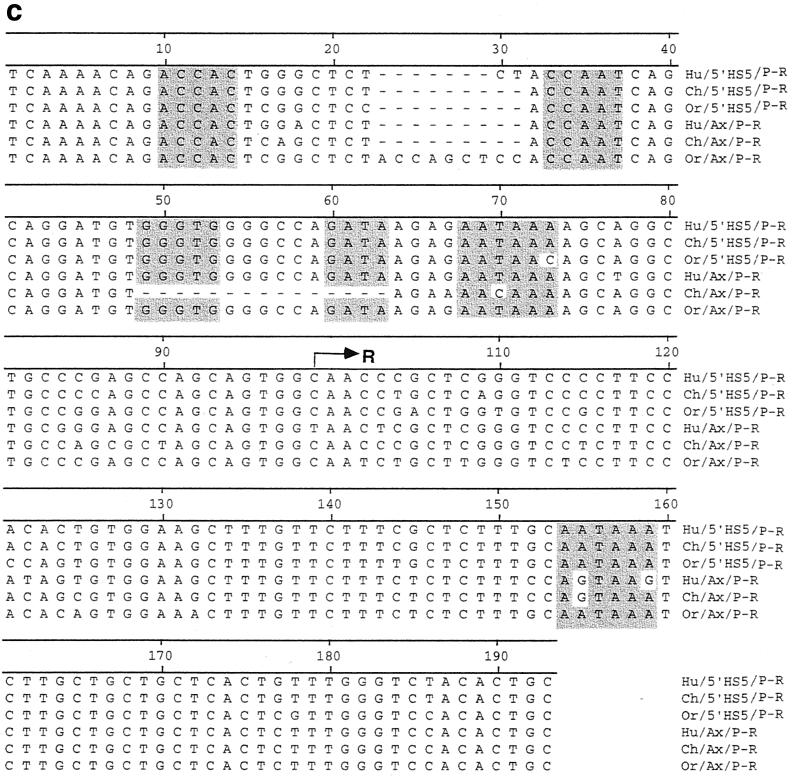
We reported earlier that an ERV-9 LTR is located in the 5" boundary area of the β-globin gene locus, proximal to the HS5 site in the β-LCR of human and gorilla genomes (18). Here, we have mapped the 5" boundary area of the β-LCR in higher and lower primates from chimpanzee to monkey by PCRs using human primers. Sequence analyses of the PCR amplicons show that the 5"HS5 ERV-9 LTR is conserved, with over 90% sequence identity between human and the higher primates chimpanzee, gorilla, and orangutan; the 5"HS5 LTR is absent in the lower primates gibbon and monkey (Fig. 1). The U3 regions of the 5"HS5 LTRs in chimpanzee, gorilla, and orangutan contain fewer U3 enhancer repeats than the human LTR (Fig. 1b). However, the U3 enhancer repeats were always in phase with those in the human LTR; i.e., no partial repeats were ever found in the LTRs, and the order of the subtype repeats 1-2-3-4 was always conserved. The U3 enhancer and promoter regions and the R region, which contain potentially functional sequence motifs such as the identifiable transcription factor binding motifs GTGGT, CCAAT, CACCC, and GATA (21, 23, 28), the AATAAA motif in the U3 promoter (the potential TATA box for initiating synthesis of LTR RNAs), and the AATAAA motif in the R region (the potential polyadenylation signal for LTR RNAs) (see Fig. 2c, Fig. 6a, and reference 18), are 95 to 100% conserved (Fig. 2c). The U5 regions contained three 72-bp repeats, except for the orangutan U5 region, which contained only two U5 repeats (Fig. 1b). The 5"HS5 LTRs in the higher primates are integrated into an identical host site, as indicated by the identical four-base repeat GTAT in the host genome flanking the LTR (Fig. 1b). In the lower primates, the 5"HS5 LTR was not detectable; however, the host integration site GTAT as well as further-upstream and -downstream genomic DNAs are present (Fig. 1b). These findings indicate that the 5"HS5 LTR was inserted into the primate genome during evolution between gibbon and orangutan approximately 15 to 18 million years ago and has been stably integrated in the genomes of the higher primates, including humans (18).
FIG. 1.
Conservation of a solitary ERV-9 LTR in the β-globin gene locus during primate evolution. (a) Map of the β-globin gene locus in primates. Hu, Ch, Go, Or, Gi, and Mo, β-globin gene loci in human, chimpanzee, gorilla, orangutan, gibbon, and monkey, respectively. Hatched box, ERV-9 LTR. Vertical arrows, the five DNase I-hypersensitive sites defining the β-LCR. Black bars, embryonic ɛ-, fetal γ-, and adult δ- and β- globin genes. Angled arrows, direction of transcription of the ERV-9 LTR, β-LCR, and globin genes. Bent line, absence of the ERV-9 LTR in gibbon and monkey. (b) Structure of the 5"HS5 ERV-9 LTR in primates. Boxes marked 1, 2, 3, and 4, the four subtypes of the 40-bp enhancer repeats in U3 (18). Arrowheads, the 72-bp U5 repeats in U5. attagtat and gtatgtca flanking the LTR, DNA bases in the primate genomes flanking the integration site of the 5"HS5 LTR. Numbers in parentheses, lengths in DNA bases of the respective primate LTRs.
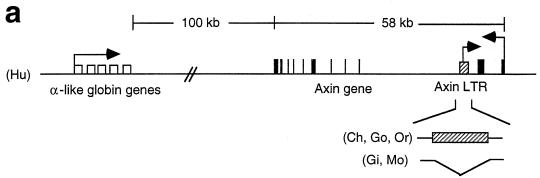
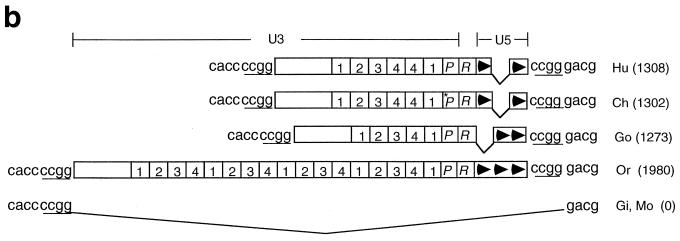
FIG. 2.
Conservation of a solitary ERV-9 LTR in the axin gene locus during primate evolution. (a) Map of the axin gene locus in primates. Unfilled boxes, α-like globin genes. Black bars, the 11 exons of the axin gene. Other designations are the same as for Fig. 1a. The 300-kb locus in 16p 13.3 from the axin gene to the α-globin gene was assembled from GenBank files under accession numbers AC005202, AC004652, Z99754, Z69667, Z69075, Z69890, Z69706, Z84721, Z69666, Z84813, and Z84722. (b) Structure of the axin ERV-9 LTR. Designations are the same as for Fig. 1b. (c) Alignments of the U3 promoter and R regions of the 5"HS5 LTR and axin LTR in human, chimpanzee, and orangutan. Highlighted bases, ACCAC (GTGGT), CCAAT, GGGTG (CACCC) GATA, and AATAAA sequence motifs. Arrow, transcription initiation site of LTR RNAs marking the 5" boundary of the R region.
Using the BLAST program, we found a solitary ERV-9 LTR in the human axin gene locus located downstream of the α-globin gene cluster on chromosome 16. Sequence alignments of the human axin cDNA (accession number AF009674) with the GenBank sequence files spanning the axin gene locus (see Fig. 2a legend) showed that the human axin gene contains 11 exons and spans 58 kb of DNA. An ERV-9 LTR is located, in the antisense orientation, in the second intron at a location 4 kb from exon 2 of the axin gene (accession number AC005202) (Fig. 2a). The human axin LTR bears extensive sequence identity of over 90% with the human 5"HS5 LTR, although the axin LTR is shorter, containing six U3 enhancer repeats and two U5 repeats. As in the 5"HS5 LTR, the identifiable transcription factor binding motifs GTGGT, CCAAT, CACCC, and GATA and the AATAAA box in the U3 region of the axin LTR are 95 to 100% conserved during primate evolution from orangutan to human (Fig. 2c). However, a number of deletions and base mutations are observed: in the chimpanzee U3 promoter, the 15 bases spanning the CACCC and the GATA motifs are deleted and the AATAAA motif (TATA box) is mutated to AACAAA, indicating that the U3 promoter in the chimpanzee axin LTR was considerably weakened. In addition, the second AATAAA motif in the R region of orangutan is mutated to AGTAAA in gorilla and chimpanzee and to AGTAAG in human (Fig. 2c). Like the 5"HS5 LTR, the axin LTR is conserved in an identical location in the higher primates from orangutan to human (Fig. 2b) and thus has been stably integrated in the primate genome for at least 15 million years.
The 5"HS5 and axin LTRs are not found in the lower primates gibbon and monkey; however, ERV-9 LTRs are detectable in the monkey genome. Slot blots of primate DNAs show that the monkey genome contains approximately 2,000 copies of the ERV-9 LTRs (Fig. 3). The conservation of the ERV-9 LTRs in the primate genomes for at least 25 million years suggests that the ERV-9 LTRs are not detrimental to the hosts and may be conserved during primate evolution to serve useful cellular functions.
FIG. 3.
ERV-9 LTRs are present in both the higher and the lower primates. (a) Slot blots of primate and nonprimate genomic DNAs. The membranes containing the DNA samples were hybridized to the human 5"HS5 LTR probe. (b) Copy numbers of ERV-9 LTRs in primates and nonprimates relative to the haploid copy numbers in human.
Development of a new transfection assay utilizing the GFP gene as the reporter and FACS analyses for quantitative analysis of enhancer and promoter activities.
To investigate the function of the 5"HS5 LTR, we developed a simple and sensitive transfection assay utilizing the GFP gene as the reporter followed by FACS analyses to assess the enhancer and promoter activities of the 5"HS5 LTR in a wide spectrum of human cells. Recombinant constructs containing the U3 enhancer and promoter linked to an enhancerless and promoterless GFP gene (Fig. 4a) were electroporated into the appropriate host cells. The enhancer-promoter activities of the inserts as reflected by the GFP fluorescence of the transfected cells were determined from a combination of the following parameters: (i) the percentage of fluorescent cells in the transfected-cell population and (ii) the mean fluorescence intensity of the fluorescent cells and the relative copy numbers of the transfected plasmids (Fig. 4b).
The mean fluorescence intensity of the fluorescent cells is a measure of the combined strengths of the enhancer and promoter coupled to the GFP gene, since the GFP gene in the enhancerless and promoterless reference GFP plasmid exhibited very weak mean fluorescence intensity (Fig. 4b). In the dot plots of FACS analysis, the percentage of fluorescent cells in the R3 region reflects the transfectability of the cells, which theoretically should be a constant for a specific cell type even when transfected with different plasmids. However, we observed that the percentages of fluorescent cells correlated positively with the enhancer and promoter strengths (the mean fluorescence intensities) of the transfected plasmids (see percentages of fluorescent cells of each cell type transfected with different plasmids in Table 1). Hence, we took the product of the mean fluorescence intensity of the fluorescent cells and the percentage of the fluorescent cells as a quantitative measure of the combined enhancer and promoter strengths of the transfected plasmid. In calculating the relative enhancer-promoter activities of the test plasmids, the activity of the enhancerless and promoterless GFP plasmid was used as the reference standard (Fig. 4b).
TABLE 1.
Enhancer and promoter activities of the human 5"HS5 LTR, chimpanzee 5"HS5 LTR, and human axin LTR in the (E-P-r)-GFP plasmids and the reference GFP plasmida determined by transient-transfection assays
| Cell lineb | Activityc
|
|||
|---|---|---|---|---|
| Human 5"HS5 | Chimpanzee 5"HS5 | Human axin | GFP | |
| Bewo | 1,014 ± 15 (6.4, 388, 2.4) | 514 ± 28 (5.3, 223, 2.3) | 2,300 ± 450 (11, 795, 3.8) | 1.0 (0.06, 16.4, 1) |
| 293 | 1,920 ± 520 (44, 522, 1.9) | NDd | ND | 1.0 (0.13, 48.3, 1) |
| WRL68 | 623 ± 130 (33.8, 239, 1.6) | 456 ± 60 (21.2, 175, 1) | ND | 1.0 (0.14, 57.5, 1) |
| CFU-E | 374 ± 64 (5.4, 76.4, 1.2) | ND | ND | 1.0 (0, 0, 1) |
| K562 | 248 ± 70∗ (21, 130, 2) | 170 ± 45∗ (23, 80, 1.9) | 176 ± 50∗ (25, 85, 2.2) | 1.0 (0.5, 11, 1) |
| Jurkat | 132 ± 30∗ (25, 184, 1.9) | 60 ± 16∗ (19, 87, 1.5) | ND | 1.0 (0.5, 34, 1) |
| HepG2 | 82 ± 25 (16.5, 429, 1.0) | ND | ND | 1.0 (1.7, 51, 1) |
| H1299 | 24 ± 3 (11.2, 105, 2) | ND | ND | 1.0 (0.9, 27, 1) |
| HeLa | 14 ± 6 (60, 70, 1.6) | ND | 10 ± 2 (50, 70, 1.8) | 1.0 (15, 13, 1) |
See Fig. 5a.
Human primary cells and transformed cell lines used as transfection hosts are as follows: Bewo, placental trophoblasts from choriocarcinoma; 293, transformed embryonic kidney; WRL68, transformed embryonic liver; CFU-E, erythroid progenitor cells from adult peripheral blood; K562, erythroleukemia; Jurkat, acute T-cell leukemia; HepG2, hepatoma; H1299, lung carcinoma; HeLa, cervical carcinoma.
Values marked with an asterisk are means ± standard deviations (n = 4); all other values are arithmetic averages (n = 2). The three numbers in parentheses are, respectively, the percentage of fluorescent cells 48 h after electroporation, the mean fluorescent intensity of the fluorescent cells, and the copy number ratio of the test plasmids in the electroporated cell population relative to the reference GFP plasmid in control transfections.
ND, not determined.
This quantitative functional assay is simple to perform. Transfected cells in numbers from 20,000 to 50,000 without further biochemical manipulation can be directly analyzed by FACS in minutes to obtain statistically meaningful transfection results.
The U3 enhancer of ERV-9 LTRs is active in embryonic cells and erythroid cells.
Using the transfection assays described above, we determined the U3 enhancer and promoter activities of the human and chimpanzee 5"HS5 LTR and the human axin LTR in a wide variety of human cells. The (E-P-r)-GFP plasmids contained the U3 enhancer and promoter and the 5" half of the R region before the second AATAAA motif (see Fig. 6a). The transfection results show that the enhancer and promoter activities of the ERV-9 LTRs were 2- to 10-fold higher in embryonic cells derived from placenta, embryonic kidney, and liver than in hematopoietic cells of erythroid and lymphoid lineages and were 10- to 100-fold higher in embryonic cells than in some adult nonhematopoietic cells (Table 1). The (P-r)-GFP plasmids containing only the U3 promoter activated the GFP reporter gene to levels approximately one-fifth to one-quarter of those of the (E-P-r)-GFP plasmids (data not shown).
The U3 enhancer initiates mRNA synthesis of the cis-linked GFP gene from a site 25 bases downstream of the AATAAA motif (TATA box) in the U3 promoter.
To further investigate the LTR enhancer and promoter activities in the (E-P-r)-GFP construct, we used the 5"RACE technique (11) to map the 5" ends of the GFP RNAs transcribed from the 5"HS5 (E-P-r)-GFP plasmid integrated into K562 cells. In particular, we wished to determine whether the LTR enhancer-promoter activated synthesis of GFP mRNA from the presumptive AATAAA (TATA) box in the U3 promoter as identified by sequence analysis.
5"RACE showed that a single PCR band of 140 bases anticipated to be generated by the nested primer pairs from the GFP mRNA was indeed observed (Fig. 5a and c, lane K1). DNA sequencing of this PCR band showed that the GFP mRNA was initiated from a specific site located 25 bases downstream of the AATAAA (TATA) box in the U3 promoter (Fig. 5d, panel I). This LTR RNA initiation site thus defines the 5" border of the R region in the 5"HS5 LTR (27).
In the endogenous genomes of erythroid and embryonic cells, the 5"HS5 LTR initiates RNA synthesis from the same site 25 bases downstream of the AATAAA box in the U3 promoter, and the LTR-initiated RNAs extend through the intervening DNA into the HS5 site
Correlating with the RNA initiation site in integrated plasmids, the 5"HS5 LTR in the endogenous genome of nontransfected K562 cells also initiated RNA synthesis from the same site at the 5" border of the R region (Fig. 5b). The cDNA reverse transcribed from this endogenous R RNA produced a nested PCR band of the anticipated size of 580 bp in duplicate 5"RACE reactions (Fig. 5c, lanes K2 and K3). DNA sequence analyses of this band confirmed that the endogenous R RNA, like the GFP mRNA transcribed from transfected plasmids, was initiated from the same base located 25 bases downstream of the AATAAA box in the U3 promoter (Fig. 5d, panel I).
In K562 cells, two additional endogenous RNA initiation sites were detected in the U5 region, generating the U5(2) and U5(3) RNAs (Fig. 5b). These two U5 RNAs produced, respectively, the PCR bands of 370 and 260 bp (Fig. 5c, lanes K2 and K3). DNA sequencing of these two bands showed that the 5" ends of the U5(2) and U5(3) RNAs were located at the respective 5" ends of the second and third U5 repeats (Fig. 5d, panels II and III). However, unlike the R RNA, whose 5" end was reproducibly mapped to the C base 25 nucleotides (nt) downstream of the AATAAA box in the U3 promoter, the U5 RNAs were transcribed from regions that did not contain identifiable AATAAA (TATA) boxes located 25 to 30 bases upstream of their respective 5" ends (Fig. 5d, panels II and III). These U5 RNAs were not reproducibly detectable: the U5(2) RNA producing the PCR band of 370 bp was not detected in duplicate K562 RNA samples (compare lanes K2 and K3 in Fig. 5c). Moreover, their 5" ends were not fixed, varying between K562 and other cell types within a range of 20 bases in the 5" borders of the respective U5 repeats (5" end analyses of U5 RNAs in these latter cell types are not shown).
The R and U5 RNAs all extended into the HS5 site in K562 cells, since the PCR bands of 580, 370, and 260 bp were generated from cDNA templates that were synthesized from a reverse primer located within the HS5 site (Fig. 5b and c, lanes K2 and K3).
In nontransfected placental trophoblasts, the endogenous R RNA initiated from the 5" border of the R region produced a single detectable PCR band of 580 bp (Fig. 5c, lane P). DNA sequence analysis showed that, as in K562 cells, this R RNA was initiated from the identical C base located 25 bases downstream of the AATAAA box in the U3 promoter (electropherogram not shown). This indicates that the U3 enhancer and promoter of the endogenous 5"HS5 LTR were active in placental trophoblasts, which confirms the transfection results (shown above) that the 5"HS5 LTR enhancer and promoter in transfected GFP plasmids were active in the Bewo placental trophoblast cell line (Table 1).
In HeLa cells, the R RNA was not detectable and did not produce the PCR band of 580 bp (Fig. 5c, lane H), indicating that the U3 enhancer and promoter of the 5"HS5 LTR in the endogenous genome of HeLa cells were not active. This finding is again consistent with the transfection result that the 5"HS5 (E-P-r)-GFP plasmid exhibited weak enhancer-promoter activities in HeLa cells (Table 1). However, the U5 region in the endogenous 5"HS5 LTR apparently initiated the transcription of U5(2) RNA from the second U5 repeat, which generated the PCR band of 370 bp (Fig. 5c, lane H). This indicates that the U5 region in HeLa cells, as in K562 cells, may be transcriptionally active and capable of initiating RNA synthesis independent of identifiable AATAAA boxes located 25 to 30 bases upstream of the apparent transcriptional initiation sites.
In summary, RNA analyses by 5"RACE indicate that the 5"HS5 LTR is transcriptionally active in placental trophoblasts and erythroid K562 cells and initiates sense RNA synthesis in these cells from a specific site 25 bases downstream of the AATAAA motif in the U3 promoter.
The 5"HS5 LTR RNAs are polyadenylated, but the AATAAA motif in the R region located downstream of the U3 promoter does not serve as a polyadenylation signal for the LTR RNAs.
In the 5"HS5 LTR, a second AATAAA motif is present in the R region, at a location 80 bases downstream of the AATAAA (TATA) motif in the LTR promoter (Fig. 6a). This second AATAAA motif did not serve as the TATA box in initiating transcription of the 5"HS5 LTR RNAs, since RNAs initiated from this AATAAA motif would have generated in 5"RACE a nested PCR band of 480 bp, which was not detected (Fig. 5b and c). Such duplicated AATAAA motifs in the promoter and R regions are found not only in the solitary ERV-9 LTRs (7, 17, 18) but also generally in both the 5" and the 3" LTRs of retroviruses (5). In retroviruses, the second AATAAA box in the 3" LTR, but not the one in the 5" LTR, has been reported to serve as the polyadenylation signal for retroviral RNAs (2). In the 5"HS5 LTR of the endogenous K562 genome, if the AATAAA motif in the R region served as a poly(A) signal, it would have produced very short, polyadenylated LTR RNAs of approximately 100 nt which consisted of 55 nt of RNA between the transcriptional initiation site and the AATAAA poly(A) signal in the R region (Fig. 6a) plus approximately 20 additional bases between the poly(A) signal and the polyadenylation site (2, 5) and a poly(A) tail of 33 nt complementary to the (T)33 primer used in the reverse transcription step (Fig. 6b). To detect such short, polyadenylated RNAs of 100 nt, we carried out the following RT-PCR experiments.
The templates for the RT-PCRs were total cellular RNAs isolated from nontransfected placental cells and the Bewo and K562 cell lines, in which the 5"HS5 LTR enhancer and promoter were active as shown by transfection and 5"RACE results. The polyadenylated RNAs were first transcribed into cDNAs with the reverse primer (T)33. The cDNAs were then amplified in PCR with the (T)33 reverse primer and the F1 forward primer located at the transcriptional initiation site in the R region (Fig. 6b). In gel electrophoresis, PCR bands of 100 bp were not produced by RNAs isolated from any one of the cell types, although much longer PCR bands were detectable (Fig. 6c, left panel). To confirm that the long PCR fragments generated with the F1-(T)33 primer pair indeed were amplified from the 5"HS5 LTR locus, we performed a second-round nested PCR using the nested primer pair F2-G1 to amplify the F1-(T)33 PCR products (Fig. 6c). The nested PCR band of 490 bp anticipated to be generated by the F2-G1 primer pair was indeed produced (Fig. 6c, middle panel). DNA sequencing showed that this 490-bp band contained the 5"HS5 LTR as well as the unique, downstream genomic DNA and was specific to the 5"HS5 LTR locus (18). In addition, the nested PCR using primer pair F3-G2, which spanned the unique genomic DNA further downstream of the F2-G1 region, also produced the anticipated nested PCR band of 768 bp from the F1-(T)33 PCR products (Fig. 6c, right panel). This F3-G2 band was not amplified directly from shorter cDNA templates synthesized from the (T)33 reverse primer, since PCRs using the cDNAs as the direct templates produced barely detectable PCR bands of 768 bp (Fig. 6c, right panel, last three lanes). These results indicate that the polyadenylated LTR RNAs spanned both the F2-G1 and F3-G2 regions and thus the entire region between the 5"HS5 LTR and the HS5 site.
Together, these results indicate that in placental, Bewo, and K562 cells, the RNAs transcribed from the 5"HS5 LTR were polyadenylated but the AATAAA motif in the R region of the 5"HS5 LTR did not serve as the poly(A) signal, so that the polyadenylated LTR RNAs extended into the downstream genomic DNA. A corollary observation of the absence of the short 100-bp polyadenylated LTR RNA is that the AATAAA motifs in the R regions of many other solitary ERV-9 LTRs in the human genome also did not serve as the polyadenylation signal to terminate the ERV-9 LTR RNAs.
The 5"HS5 LTR- and axin-initiated RNAs are transcribed predominantly in a direction toward the downstream genomic DNA.
Both the 5"RACE and the RT-PCR studies indicate that the 5"HS5 LTR RNAs were transcribed in the sense direction toward the HS5 site, colinear with the direction of transcription of the further downstream β-like globin genes (Fig. 5b and 6b). This suggests that the LTR enhancer, like the further-downstream HS2 enhancer in the β-LCR (16, 32), initiated transcription predominantly in the sense direction. To confirm this, we carried out the following RT-PCRs to determine whether LTR RNAs were also transcribed in the antisense direction from the HS5 site into the 5"HS5 LTR. We used five overlapping primer pairs that spanned the 2.7 kb of DNA from the R region to the 3" end of the HS5 site (Fig. 7a). To synthesize cDNAs from the sense transcripts, reverse primers 1 to 5 were used in the RT step; to synthesize cDNAs from the antisense transcripts, forward primers 1 to 5 were used in the RT step (Fig. 7a). The cDNAs were then amplified separately in PCRs with the respective primer pairs 1 to 5. As expected, the sense transcripts produced RT-PCR bands of the anticipated lengths (+ lanes in Fig. 7b), but the antisense transcripts did not produce RT-PCR bands of the anticipated lengths (− lanes in Fig. 7b).
The shorter bands in the + lanes in Fig. 7b were not amplified from shorter sense transcripts, nor were shorter bands in the − lanes amplified from shorter antisense transcripts of the region. They were spurious bands produced in RT-PCRs when the same primer was used in both cDNA synthesis and PCR amplification. In RT-PCRs using random hexamers as primers for cDNA synthesis, which should be able to anneal to and transcribe both the sense and the antisense RNAs, followed by PCR with primer pairs 1 to 5, the spurious shorter bands were not detected, and only the bands of the anticipated lengths produced from the sense RNAs were detected (Fig. 7c). These results indicate that the 5"HS5 LTR and the genomic DNA downstream of it were transcribed exclusively in the sense direction toward the downstream HS5 site.
In the human axin gene locus, the ERV-9 LTR is located in the second intron in reverse orientation to the transcription direction of the axin gene (Fig. 2a and 8a). RT-PCR studies indicate that the axin LTR enhancer still initiated RNA synthesis predominantly in the direction toward the downstream genomic DNA, i.e., in an antisense direction to the transcription of the axin gene (Fig. 8b, lanes 3). This antisense transcription did not extend beyond 400 bases downstream of the axin LTR, since the direction of transcription of the further-downstream axin intron and exon DNA was exclusively in the sense direction of axin gene transcription (Fig. 8b, lanes 1 and 2). These results indicate that both the 5"HS5 LTR and the axin LTR enhancer initiated transcription into the downstream genomic DNAs regardless of the orientation of the LTR with respect to the associated gene loci.
DISCUSSION
In this study we show that both the 5"HS5 LTR and the axin ERV-9 retrotransposons were inserted into the ancestral genome of orangutan and have been stably integrated into the respective gene loci in the higher primates from orangutan to human for over 15 million years. It has been reported that the ERV-9 LTR in the ZNF80 gene locus is not found in the genome of orangutan but is present in the genomes of gorilla, chimpanzee, and human (7). This indicates that the ERV-9 LTR retrotransposons were inserted into separate loci of the primate genomes at different times during evolution. The absence of these ERV-9 LTRs in the lower primates gibbon and monkey, which presumably have properly regulated globin and axin genes, suggests that these retrotransposons in the higher primates may be selfish DNAs serving no relevant host function. However, ERV-9 LTRs are detectable in the monkey genome at approximately 2,000 copies and are apparently associated with many other monkey gene loci. The conservation of the ERV-9 LTRs in the primates for at least 25 million years from monkey to human suggests that the ERV-9 LTRs are not detrimental to the hosts and may be conserved to provide additional levels of transcriptional control for the associated genes during primate evolution.
Transfection studies showed that the 5"HS5 and axin LTRs possessed strong enhancer and promoter activities that exhibited tissue preference. The ERV-9 LTR enhancer activities in embryonic cells were 2- to 10-fold higher than those in hematopoietic cells of erythroid and lymphoid lineages and were over 100-fold higher than those in some adult nonhematopoietic cells. In the endogenous genomes of embryonic placental cells and erythroid K562 cells and in plasmids integrated into K562 cells, the U3 enhancer activated RNA synthesis from a specific site located 25 bases downstream of the AATAAA motif (TATA box) in the U3 promoter. The specific location of the transcriptional initiation site downstream of a TATA box suggests that the LTR RNAs were transcribed by RNA polymerase II (pol II). The LTR RNAs extended through a second AATAAA motif in the R region into the downstream genomic DNA and the HS5 site. This second AATAAA motif thus did not serve as a TATA box or as a polyadenylation signal for LTR transcription.
In the endogenous genome of erythroid K562 cells, the 5"HS5 LTR RNAs extended through the R and U5 region into the HS5 site exclusively in the sense direction colinear with the direction of transcription of the β-LCR (1, 16, 32) and the further downstream β-like globin genes. The sense LTR RNAs were polyadenylated, indicating again that the LTR RNAs were transcribed by pol II, since pol II through its unique C-terminal domain has been reported to be instrumental in polyadenylation of the RNAs it transcribes (20). These rare, endogenous LTR RNAs, which are detectable only after PCR amplifications, do not appear to be mRNAs encoding translatable gene products, as the 1.1-kb DNA between the ERV-9 LTR and the HS5 site and the DNA in the second intron of the axin gene carry no long open reading frames and do not appear to contain a gene (GenBank accession numbers AF064190 and AC005202).
The β-LCR defined by DNase I-hypersensitive sites HS1 to HS5 is conserved during mammalian evolution from mouse to human and serves an indispensable role in transcriptional activation of the β-like globin genes in erythroid cells (13). The HS2 enhancer in the β-LCR located further downstream of the 5"HS5 LTR has also been reported to initiate HS2 enhancer transcription preferentially in the sense direction toward the far-downstream globin promoters and genes (1, 16, 32). These and other observations suggest that the enhancer-initiated transcription process plays a role in mediating enhancer function over distance. We are currently studying the effects of deletion of the 5"HS5 LTR and thus abolition of the 5"HS5 LTR-initiated transcription process on transcription of the downstream LCR and the β-like globin genes during ontogeny and hematopoietic differentiation.
In the mouse axin gene locus, a murine endogenous retrovirus, an intracisternal A particle (IAP) whose LTRs possess enhancer-promoter activities (4), has been reported to regulate transcription of the cis-linked axin gene and cause the Fused and Knobbly mutations in mice (33). In the Fused mutation, the insertion in intron 6 of an IAP in the antisense orientation to the axin gene creates a gene that produces wild-type transcripts as well as mutant transcripts that initiate from the LTRs of the IAP. In the Knobbly mutation, the insertion of an IAP also in the antisense orientation in exon 7 interrupts transcription of axin mRNA and precludes the production of wild-type axin protein. These mutations are manifested by a dominant gain-of-function phenotype of a kinked tail in heterozygotes. Homozygous Knobbly mutants are embryonic lethal, showing duplication of the embryonic axis and neuroectodermal and cardiac defects (33). These findings strongly suggest that the ERV-9 LTR enhancer inserted in the second intron of the human axin gene in reverse orientation to the gene may modulate the transcription of the human axin gene in embryonic and hematopoietic cells through synthesis of the antisense LTR RNAs.
In the human genome, the ERV-9 LTRs are middle repetitive DNAs present at 3,000 to 4,000 copies. Many of these LTRs share extensive sequence identities of over 90% with the 5"HS5 and axin LTRs, as revealed by BLAST searches of the GenBank database. It remains to be determined whether these ERV-9 LTRs possess similar enhancer and promoter activities and regulate the transcription of the cis-linked genes during ontogeny and hematopoietic differentiation.
Acknowledgments
We thank G. LaMantia for the kind gift of the phage Fix 1.2 clone of the ZNF80 gene locus, M. Goodman and S. Page for the gibbon DNA, C. Leithner for DNA sequencing, J. Luna for purification of CFU-E, and S. Y. Li for assistance with electronic graphics.
This work was supported in part by NIH grants DK-1-5555 (to S.K.) and HL 39948 and 62308 (to D.T.).
REFERENCES
- 1.Ashe, H. L., J. Monks, M. Wijgerde, P. Fraser, and N. J. Proudfoot. 1997. Intergenic transcription and transinduction of the human β-globin locus. Genes Dev. 11:2494-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashe, M. P., P. Griffin, W. James, and N. J. Proudfoot. 1995. Poly(A) site selection in the HIV-1 provirus: inhibition of promoter-proximal polyadenylation by the downstream major splice donor site. Genes Dev. 9:3008-3025. [DOI] [PubMed] [Google Scholar]
- 3.Cavallesco, R., and D. Tuan. 1997. Modulatory subdomains of the HS2 enhancer differentially regulate enhancer activity in erythroid cells at different developmental stages. Blood Cells Mol. Dis. 23:8-26. [DOI] [PubMed] [Google Scholar]
- 4.Christy, R., and R. C. Huang. 1988. Functional analysis of the long terminal repeats of intracisternal A particle genes: sequences within the U3 region determine both the efficiency and direction of a promoter activity. Mol. Cell. Biol. 8:1093-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coffin, J., S. Hughes, and H. Varmus. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 6.Cross, J. C., Z. Werb, and S. J. Fisher. 1994. Implantation and the placenta: key pieces of the development puzzle. Science 266:1508-1518. [DOI] [PubMed] [Google Scholar]
- 7.Di Cristofano, A., M. Strazzullo, T. Parisi, and G. La Mantia. 1995. Mobilization of an ERV9 human endogenous retroviral element during primate evolution. Virology 213:271-275. [DOI] [PubMed] [Google Scholar]
- 8.Doolittle, W. F., and C. Sapienza. 1980. Selfish genes, the phenotype paradigm and genome evolution. Nature 284:601-603. [DOI] [PubMed] [Google Scholar]
- 9.Fan, H. 1994. Retroviruses and their role in cancer, p. 313-362. In J. Levy (ed.), The Retroviridae, vol. 3. Plenum Press, New York, N.Y. [Google Scholar]
- 10.Feuchter, A. E., J. D. Freeman, and D. L. Mager. 1992. Strategy for detecting cellular transcripts promoted by human endogenous long terminal repeats: identification of a novel gene (CDC4L) with homology to yeast CDC4. Genomics 13:1237-1246. [DOI] [PubMed] [Google Scholar]
- 11.Frohman, A. 1993. Rapid amplification of complementary DNA ends for generation of full-length complementary DNAs: thermal RACE. Methods Enzymol. 218:340-356. [DOI] [PubMed] [Google Scholar]
- 12.Goodman, M., C. Porter, J. Czelusniak, S. Page, H. Schneider, J. Shoshani, G. Gunnell, and C. Groves. 1998. Toward a phylogenetic classification of primates based on DNA evidence complemented by fossil evidence. Mol. Phylogenet. Evol. 9:585-598. [DOI] [PubMed] [Google Scholar]
- 13.Hardison, R., J. L. Slightom, D. L. Gumucio, M. Goodman, N. Stojanovic, and W. Miller. 1997. Locus control regions of mammalian beta-globin gene clusters: combining phylogenetic analyses and experimental results to gain functional insights. Gene 205:73-94. [DOI] [PubMed] [Google Scholar]
- 14.Henikoff, S., E. Greene, S. Pietrokovski, P. Bork, T. Attwood, and L. Hood. 1997. Gene families: the taxonomy of protein paralogs and chimeras. Science 278:609-614. [DOI] [PubMed] [Google Scholar]
- 15.Henthorn, P. S., D. L. Mager, D. L. Huisman, and O. Smithies. 1986. A gene deletion ending within a complex array of repeated sequences 3" to the human beta-globin gene cluster. Proc. Natl. Acad. Sci. USA 83:5194-5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kong, S., D. Bohl, C. Li, and D. Tuan. 1997. Transcription of the HS2 enhancer toward a cis-linked gene is independent of the orientation, position, and distance of the enhancer relative to the gene. Mol. Cell. Biol. 17:3955-3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.La Mantia, G., D. Maglione, G. Pengue, A. Di Cristofano, A. Simeone, L. Lanfrancone, and L. Lania. 1991. Identification and characterization of novel human endogenous retroviral sequences preferentially expressed in undifferentiated embryonal carcinoma cells. Nucleic Acids Res. 19:1513-1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Long, Q., C. Bengra, C. Li, F. Kutlar, and D. Tuan. 1998. A long terminal repeat of the human endogenous retrovirus ERV-9 is located in the 5" boundary area of the human β-globin locus control region. Genomics 54:542-555. [DOI] [PubMed] [Google Scholar]
- 19.Lower, R., J. Lower, and R. Kurth. 1996. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc. Natl. Acad. Sci. USA 93:5177-5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCracken, S., N. Fong, K. Yankulov, S. Ballantyne, G. Pan, J. Greenblatt, S. D. Patterson, M. Wickens, and D. L. Bently. 1997. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385:357-361. [DOI] [PubMed] [Google Scholar]
- 21.Miller, I. J., and J. J. Bieker. 1993. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Kruppel family of nuclear proteins. Mol. Cell. Biol. 13:2776-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schulte, A. M., S. Lai, A. Kurtz, F. Czubayko, A. T. Riegel, and A. Wellstein. 1996. Human trophoblast and choriocarcinoma expression of the growth factor pleiotrophin attributable to germ-line insertion of an endogenous retrovirus. Proc. Natl. Acad. Sci. USA 93:14759-14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shivdasani, R., and S. H. Orkin. 1996. The transcriptional control of hematopoiesis. Blood 87:4025-4039. [PubMed] [Google Scholar]
- 24.Smit, A. F. 1996. The origin of interspersed repeats in the human genome. Curr. Opin. Genet. Dev. 6:743-748. [DOI] [PubMed] [Google Scholar]
- 25.Stavenhagen, J. B., and D. M. Robins. 1988. An ancient provirus has imposed androgen regulation on the adjacent mouse sex-limited protein gene. Cell 55:247-254. [DOI] [PubMed] [Google Scholar]
- 26.Strazzullo, M., T. Parisi, A. Di Cristofano, M. Rocchi, and G. La Mantia. 1998. Characterization and genomic mapping of chimeric ERV9 endogenous retroviruses--host gene transcripts. Gene 5:77-83. [DOI] [PubMed] [Google Scholar]
- 27.Temin, H. M. 1981. Structure, variation and synthesis of retrovirus long terminal repeat. Cell 27:1-3. [DOI] [PubMed] [Google Scholar]
- 28.Tenen, D. G., R. Hromas, J. D. Licht, and D. E. Zhang. 1997. Transcription factors, normal myeloid development, and leukemia. Blood 90:489-519. [PubMed] [Google Scholar]
- 29.Ting, C. N., M. P. Rosenberg, C. M. Snow, L. C. Samuelson, and M. H. Meisler. 1992. Endogenous retroviral sequences are required for tissue-specific expression of a human salivary amylase gene. Genes Dev. 6:1457-1465. [DOI] [PubMed] [Google Scholar]
- 30.Tuan, D., W. Solomon, Q. Li, and I. M. London. 1985. The “β-like-globin” gene domain in human erythroid cells. Proc. Natl. Acad. Sci. USA 82:6384-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tuan, D., W. Solomon, I. M. London, and D. P. Lee. 1989. An erythroid-specific, developmental-stage-independent enhancer far upstream of the human “β-like globin” genes. Proc. Natl. Acad. Sci. USA 86:2554-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tuan, D., S. Kong, and K. Hu. 1992. Transcription of the hypersensitive site HS2 enhancer in erythroid cells. Proc. Natl. Acad. Sci. USA 89:11219-11223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vasicek, T. J., L. Zeng, X. J. Guan, T. Zhang, F. Constantini, and S. M. Tilghmann, S. M. 1997. Two dominant mutations in the mouse fused gene are the result of transposon insertions. Genetics 147:777-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wickrema, A., S. B. Krantz, J. C. Winkelmann, and M. C. Bondurant. 1992. Differentiation and erythropoietin receptor gene expression in human erythroid progenitor cells. Blood 80:1940-1949. [PubMed] [Google Scholar]
- 35.Wilkison, D., D. Mager, and J. Leong. 1994. Endogenous human retroviruses, p. 465-535. In J. Levy (ed.), The Retroviridae, vol. 3. Plenum Press, New York, N.Y. [Google Scholar]
- 36.Zucchi, I., and D. Schlessinger. 1992. Distribution of moderately repetitive sequences pTR5 and LF1 in Xq24-q28 human DNA and their use in assembling YAC contigs. Genomics 12:264-275. [DOI] [PubMed] [Google Scholar]