Abstract
The herpes simplex virus type 1 (HSV-1) mutant d109 does not express any of the immediate-early (IE) proteins and persists in cells for a prolonged length of time. As has been shown by Nicholl et al. (J. Gen. Virol. 81:2215-2218, 2000) and Mossman et al. (J. Virol. 75:750-758, 2001) using other mutants defective for IE gene expression, infection with d109 induced the expression of a number of interferon-stimulated genes. Induction of these genes was significantly greater at multiplicities of infection (MOI) of 10 PFU/cell or greater, and the resulting antiviral effect was only seen at MOIs greater than 10 PFU/cell. Using mutants defective for sets of IE genes established that the lack of ICP0 expression was necessary for high levels of interferon-stimulated gene expression in HEL cells. The induction of interferon-stimulated genes by d109 could also be inhibited by infection with an E1−:E3−:E4− adenovirus expressing levels of ICP0 that are comparable to those expressed within the first hour of wild-type virus infection. Lastly, the addition of the proteasome inhibitor MG132 to cells infected with a mutant that expresses ICP0, d106, also resulted in the induction of interferon-stimulated genes. Thus, ICP0 may function through the proteasome very early in HSV infection to inhibit a cellular antiviral response induced by the virion.
Gene expression during lytic infection with herpes simplex virus type 1 (HSV-1) progresses in a regulated cascade, beginning with the induction of immediate-early (IE) genes ICP0, ICP4, ICP22, ICP27, and ICP47 by the virion protein VP16 (4, 8, 29, 30). Expression continues with early gene transcription, followed by DNA replication and subsequently the expression of late genes (29, 30). The five IE proteins, except ICP47, act as the principal regulators for the efficient and coordinated expression of early and late genes. ICP4− and ICP27− mutants have demonstrated the necessity of these proteins for virus replication (14, 43, 56). ICP0− mutants show impaired growth and poor reactivation from latency (7, 11, 24, 57). The mutant d109, deficient in all IE gene expression, does not express any viral proteins and is nontoxic to cells at very high multiplicities of infection (MOI) (58).
HSV-1 mutants deficient in IE gene expression have been shown to induce the expression of interferon-stimulated genes (45, 50). Interferons are proteins or glycoproteins secreted by various cells in response to viral infection or other stimuli. They have antiviral, cell regulatory, and immunomodulatory functions. In many instances, interferon can make cells resistant to infection (44, 46, 61, 64-66). The cellular response to interferon, i.e., the induction of interferon-stimulated genes, often produces inhibitory antiviral effects at different stages of viral replication: entry and uncoating (simian virus 40), transcription (vesicular stomatitis virus), RNA stability (picornaviruses), initiation of translation (adenovirus), maturation, and assembly and release (retrovirus) (reviewed in references 61, 64, and 65).
Some interferon-stimulated genes can be induced by substances other than interferon, including heavy metals, lipopolysaccharide, glucocorticoids, interleukin-1, poly(rI)·poly(rC), double-stranded viral RNA, and adsorption of virus to the cell surface (5, 22, 32, 33, 48, 70, 71). Genes induced by alpha/beta interferon or gamma interferon are often stimulated by viral infection and vice versa (15, 16, 69). Interferon, double-stranded RNA, and viral infection induce different sets of interferon-stimulated genes by using distinct signaling pathways (3, 21, 25, 69, 72). The inducible genes have a similar interferon-stimulated response element, but each interferon-stimulated response element contains unique sequences which could allow differential responses depending on the signaling molecule induced (12, 70, 72).
Interferon-stimulated gene expression has also been detected in cells infected with wild-type HSV-1 when treated with cycloheximide, but little to no interferon-stimulated gene induction was detected during wild-type virus infection (50, 54). It has also been shown that virions must enter cells to induce the expression of interferon-stimulated genes (45). Together, the existing data support the idea that some component of the virion or an event that occurs before viral gene expression results in the induction of interferon-stimulated genes through interferon response factor 3 (IRF-3) (54) and that wild-type virus must therefore express a gene product(s) that inhibits this response.
Many different viruses have evolved mechanisms to escape the antiviral response (reviewed in references 61, 64, and 65). HSV-1 has been shown to be very resistant to the cellular interferon response (40, 49). It produces an inhibitor to the 2",5"-oligoadenylate synthetase-nuclease (9), and the γ134.5 protein of HSV-1 stops the shutoff of protein synthesis by protein kinase R (26, 53). However, treatment of some cell types with interferon before infection with HSV-1 has been shown to decrease the expression of IE genes in a dose-dependent manner (2, 34, 40, 44, 51, 52, 66). It has also been shown that d120, which is defective for ICP4 and is restricted to the expression of IE genes (15), does not induce expression of the interferon response genes, suggesting that one of the other IE proteins may function to inhibit this response (45). Interestingly, in the same study (45), no mutant with a single mutation in any of the five IE genes induced the interferon-stimulated gene ISG54. However, previous studies have shown that pretreatment of cells with interferon severely reduces the plaque-forming ability of ICP0 mutants (46).
In this study, we show that infection by the IE-deficient virus d109, which possesses functional VP16 in the virion, also results in the induction of interferon-stimulated genes and an antiviral state. As IE gene expression has been shown to inhibit the induction of interferon-stimulated genes, we used mutants defective in ICP4 and other sets of IE genes along with an ICP0-expressing adenovirus to identify potential IE gene products individually capable of inhibiting this response. It was established that the expression of ICP0 could inhibit the induction of interferon-stimulated genes and that the activity of the proteasome was involved. The results support a hypothesis that the interferon and antiviral responses induced by the virion are inhibited very early in infection by an activity of ICP0.
MATERIALS AND METHODS
Cells and viruses.
Human embryonic lung (HEL) fibroblasts and Vero cells were from the American Type Culture Collection (CCL-137). Monolayer cultures were maintained by standard cell culture procedures in Dulbecco's modified Eagle's medium with 10% fetal bovine serum as previously described (60). The wild-type HSV-1 strain used for all experiments was KOS.
Earlier studies reported the construction and description of viruses d120 (15), d104, d105, d106, d107, and d109 (27, 28, 58); all are derived from HSV-1 (KOS). The virus RJ1 was constructed by crossing d106 and the virus TOZ. TOZ, like d106, is defective for ICP4, ICP27, ICP22, and, in addition, UL41 by virtue of a β-galactosidase gene (lacZ) inserted into the UL41 locus (35). The ICP4 and ICP27 alleles in TOZ are the same as a previously published ICP4-, ICP27-, and ICP22-defective virus, d95 (74).
Plaque isolates from the progeny of the cross were screened by Southern blot analysis for the incorporation of the UL41 allele from TOZ and the ICP22, ICP47 and thymidine kinase alleles from d107. All viruses were grown and titered on Vero-derived cells stably transfected with trans-complementing HSV-1 IE genes as described previously (59, 60). Adenovirus mutants AdS.10 (E1− E3−), AdS.11D (E1− E3− E4−), and AdS.11E4(ICP0) (E1− E3− E4− ICP0+) were constructed at GenVec Inc., Gaithersburg, Md. (28).
Microarray analysis.
For all of the following, confluent monolayers of HEL cells were infected with the indicated MOI of the HSV mutants. All AdS.10, AdS.11D, and Ads.11E4(ICP0) infections were carried out at 1,000 particles per cell. For comparisons to uninfected cells, HEL cells were simultaneously mock infected and incubated for the same time as the infected cell sample. Total RNA was isolated at the indicated time postinfection using Ultraspec RNA reagent (Biotexc) as per the manufacturer's protocols. Where appropriate, polyadenylated [poly(A)+] RNA was isolated as described previously (27, 28). Poly(A)+ RNA was sent to Incyte Genomics for analysis on human UniGEM V arrays. The data from these experiments were analyzed using GEMtools software (Incyte Genomics).
For experiments using arrays constructed in this study, RNA samples from the two conditions to be compared on each array were differentially labeled with indocarbocyanine (Cy3) and indodicarbocyanine (Cy5). In comparisons involving infected and uninfected cell RNA, mock-derived cDNA was labeled with Cy5, while infected-cell-derived cDNA was labeled with Cy3.
Two methods of labeling were employed. In one of the methods, total RNA from the two samples in a given comparison was reverse transcribed and subsequently differentially labeled following hybridization to the arrays with Cy3 and Cy5 using a Tyramide signal amplification kit (Perkin Elmer) following the manufacturer's protocol. Alternatively, Cy3 and Cy5 (Amersham) were also directly incorporated into cDNA reverse transcribed from poly(A)+ RNA of the two samples in a comparison prior to hybridization, as previously described (62), with minor modifications. Three micrograms of poly(A)+ mRNA was reverse transcribed into Cy3- or Cy5-labeled cDNA using an oligo(dT)12-18 (Amersham) as the primer. After labeling, the probes were passed over a Centrisep column (Princeton Separations) to remove unincorporated nucleotides. For both methods, an Arabidopsis thaliana mRNA mix (Stratagene) was spiked into the labeling reaction in order to later normalize the data. The two methods yielded similar results.
cDNA probes were resuspended in ultrapure H2O (Gibco-BRL). The cDNA probe preparations were denatured prior to hybridization at 65°C for 2 min and then chilled on ice. The probes were suspended in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.2% sodium dodecyl sulfate (SDS) along with 0.5 μg of pdA(12-18) (Amersham) and 0.5 μg of tRNA per μl. They were then dispensed onto a 20-mm by 20-mm array containing target sequences generated as described below and covered with a 22-mm by 22-mm hydrophobic cover slip (Grace Biolabs). Slides were hybridized in a hybridization cassette (Telechem) at 62°C overnight. Twenty-five microliters of 5× SSC and 0.2% SDS was added to both wells in the hybridization cassette to maintain humidity. The posthybridization washes were done at room temperature as follows: 1× SSC and 0.2% SDS for 5 min, two washes of 0.1× SSC and 0.2% SDS for 5 min each, followed by two washes in 0.1× SSC for 30 s each.
Following hybridization and washing, the slides were scanned by an Affymetrix 418 array scanner and quantified using Imagene 4.1 (Biodiscovery). Normalization and analysis were done using Genesight 2.0 (Biodiscovery) and Excel spreadsheets. Prior to inserting gene identifiers into the data, the raw data was processed in the following manner: irregular spots or spots with high background were eliminated, local backgrounds were determined and subtracted from the total signals, a lower limit for signal was established, replicate spots were averaged, the Cy3 and Cy5 signals were normalized using the spike controls, and differential expression ratios were established.
Microarray construction.
All clones were obtained from a Resgen sequence-validated human cDNA library (Research Genetics). Each clone to be incorporated on the array was amplified in a 96-well plate by adding 5 μl of the corresponding transformed Escherichia coli to a 100-μl PCR mixture along with 7.5 U of Yield Ace DNA polymerase (Stratagene), 1 μM each of the universal forward and reverse primers, and 300 μM each nucleotide. The universal forward and reverse primers were: forward, 5"-CTGCAAGGCGATTAAGTTGGGTAAC, and reverse, 5"-GTGAGCGGATAACAATTTCACACAGGAAACAGC. Primers were synthesized with a C6 amino modifier (Glen Research) for covalent attachment to the slides.
PCR was cycled as follows: 2 min at 92°C, followed by 10 cycles of 20 s at 95°C, 58°C for 20 s, and 72°C for 2.5 min. The 10 cycles were followed by 20 cycles of 95°C for 20 s, 55°C for 20 s, and 72°C for 2.5 min (extension time increases by 10 s each cycle). A final extension of 7 min was done at 72°C. PCR products were purified using a 96-well purification kit (Telechem). The DNA concentrations of the purified amplified products were quantified using Picogreen (Molecular Probes) and determined to be in the range of 150 to 250 ng/μl. All PCR products were also electrophoresed on 0.8% agarose gels to examine whether the amplified fragment length corresponds to the insert size as given in the Resgen library for each clone.
Arrays were printed on Superaldehyde slides (Telechem) using an Affymetrix 417 arrayer. Targets were printed in 0.5× microspotting solution (Telechem) in replicates of 5 or 10 with spot spacing of 375 μm and spot diameter of approximately 175 μm. For quality control and normalization purposes, viral DNA, green fluorescent protein (GFP), and Spot Report 10 (Stratagene) were printed in addition to the genes of interest. Slides were processed as described (62). In addition, the slides were UV cross-linked in a Stratalinker (Stratagene) before processing.
Northern blot analysis.
Total RNA was isolated as described above, and 10 μg of each sample was resolved by denaturing formaldehyde-agarose gel electrophoresis, transferred to nitrocellulose membranes, and probed as previously described (31). Probe fragments were generated by PCR amplification of the appropriate human cDNA clone obtained from Incyte Genomics, Inc., or Research Genetics, Inc. 32P-labeled probes were generated from purified PCR fragments by nick translation using [α-32P]dCTP and [α-32P]dGTP.
Western blot analysis.
Total protein was isolated as described previously (58). Proteins were resolved on SDS-acrylamide gels and transferred to polyvinylidene difluoride membranes, and Western blot analysis was performed as previously described (58). Detection reagent used was the ECL Plus kit (Amersham Pharmaceuticals).
RESULTS
The virus d109 has mutations in all five IE genes, and no viral gene expression is detectable following infection of cells in culture (58). In previous studies using small-filter arrays to assess the effects of d109 infection on cellular gene expression, no significant changes were detectable (27). In contrast, the virus d106 does not express ICP4, ICP27, ICP22, and ICP47 but overexpresses ICP0 relative to wild-type virus. This virus has been shown to significantly affect cell survival and the abundance of many cellular transcripts, as determined by expression array analysis (27, 28, 58).
In preliminary experiments with Incyte Genomics microarrays, which have greater than 8,000 human genes, reproducible changes were not evident in d109-infected cells at MOI of 10 PFU/HEL cell or less (data not shown). However, at multiplicities of ≥10 PFU/cell, reproducible changes in cellular mRNA abundance were detected. The mRNAs for 35 of the ∼8,000 genes on the array were reduced in abundance >2-fold, while 33 were increased in abundance >2-fold when the cells were infected at an MOI of 30 PFU/cell, which corresponds to approximately 300 particles/cell (data not shown).
The identities of the induced genes are given in Table 1. These are similar to the genes reported by Mossman et al. (45) and Nicholl et al. (50), and they represent some of the interferon-stimulated genes. Upon increasing the MOI of d109 to 50 PFU/cell, the abundance of 65 mRNAs was reduced >2-fold, while that of 148 was increased >2-fold. In addition to an increased number of differentially expressed genes, the interferon-stimulated genes that were differentially expressed at the lower MOI were differentially expressed to a greater extent at the higher MOI (data not shown).
TABLE 1.
Expression of d109-induced cellular genes in other viral backgrounds
| Genea | Accession no. | Increase or decrease in abundance after infection with indicated virus at h postinfection:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
d109
|
d106
|
d105
|
RJ1
|
||||||||
| 6 | 24 | 6 | 24 | 6 | 24 | 6 | 24 | ||||
| Interferon-induced protein 56 | NM_001548 | 1.2 | 22.2 | 1.1 | 2.2 | 1 | 3 | 1.4 | 1.2 | ||
| Interferon-induced protein 54 | M14660 | 2.6 | 16.3 | ND | 3.2 | 1.6 | 5.4 | 2.1 | 1.7 | ||
| Myxovirus (influenza virus) resistance 1, homolog of murine interferon-inducible protein p78 | AA477235 | 1.1 | 12.7 | 1.1 | 1.5 | 1.2 | 2 | 1.3 | 1.5 | ||
| Pleckstrin | X07743 | −1 | 4.2 | 1.2 | 1.2 | −1.1 | ND | 1.3 | 1.4 | ||
| Myxovirus (influenza virus) resistance 2, homolog of murine | M30818 | 1.1 | 3.6 | 1.1 | 1.1 | 1.1 | 1.5 | 1.2 | 1.4 | ||
| Small inducible cytokine subfamily B (Cys-X-Cys), member 10 | NM_001565 | −1.1 | 3.5 | 1 | 1.2 | 1.1 | ND | 2.5 | 2.6 | ||
| Signal transducer and activator of transcription 1, 91 kDa | NM_007315 | −1.4 | 3.3 | −1.4 | −1.3 | −1.7 | −1.6 | −1.1 | −1.4 | ||
| Hypothetical protein, expressed in osteoblast | F12860 | 1.5 | 2.9 | 1.3 | 1.1 | 1.4 | 1.2 | 1 | 1.1 | ||
| Interferon-induced transmembrane protein 1 (9-27) | J04164 | −1.3 | 2.9 | −1.2 | −1.2 | −1.3 | −1.1 | 1.3 | −1 | ||
| Serine/threonine kinase 4 | NM_006282 | −1 | 2.9 | −1 | 1.2 | −1 | 1.3 | 1.1 | 1.1 | ||
| ESTs | AA928141 | −1.2 | 2.8 | ND | 1.4 | ND | ND | 2.1 | 1.8 | ||
| KIAA0129 gene product | D50919 | 1.1 | 2.7 | 1.2 | 1.1 | −1 | 1.2 | −1 | −1.4 | ||
| Putative transmembrane protein | NM_012342 | 1.4 | 2.7 | 1.6 | 2.4 | 2.7 | 4.1 | 1.3 | 2.5 | ||
| 2"-5" Oligoadenylate synthetase 2 | M87434 | −1 | 2.6 | 1.2 | 1.1 | 1.1 | 1.1 | 1.2 | 1.4 | ||
| Guanosine monophosphate reductase | M24470 | 1 | 2.6 | 1 | 1.2 | 1 | 1.2 | 1.2 | 1.2 | ||
| Human clone 137308 mRNA | U60873 | 1.3 | 2.6 | 1.3 | −1 | 1.8 | 1.2 | 1.1 | −1.1 | ||
| Gamma-interferon-inducible protein 16 | M63838 | −1 | 2.6 | −1 | −1 | −1 | 1.2 | −1.1 | −1.3 | ||
| Interferon-induced protein 35 | U72882 | −1 | 2.6 | −1.1 | −1 | −1 | 1.1 | 1.1 | −1 | ||
| Guanylate binding protein 1, interferon inducible, 67 kDa | M55542 | −1.3 | 2.5 | −1.3 | 1 | −1.6 | 1.2 | 1.2 | −1.2 | ||
| Apolipoprotein L | Z82215 | 1.1 | 2.4 | 1.2 | 1.4 | 1.3 | 1.3 | 1.1 | −1.1 | ||
| Proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional protease 2) | AI923532 | −1 | 2.4 | −1 | 1 | −1.2 | 1.3 | 1.3 | 1.2 | ||
| Caspase 1, apoptosis-related cysteine protease (interleukin-1β convertase) | X65019 | ND | 2.3 | −1.2 | −1.1 | −1.3 | −1.1 | 1.2 | 1.1 | ||
| Nuclear receptor subfamily 1, group I, member 3 | X56199 | −1.1 | 2.3 | −1.1 | 1.1 | −1 | ND | 1.6 | 1.6 | ||
| Stimulated trans-acting factor (50 kDa) | AW407653 | −1 | 2.3 | 1.2 | 1.2 | 1.1 | 1.3 | 1.8 | 1.1 | ||
| Integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) | AI521645 | −1.3 | 2.2 | −1.3 | 1.9 | −2.2 | 2.2 | −1 | 1.9 | ||
| Interferon-induced, hepatitis C-associated microtubular aggregate protein (44 kDa) | NM_006417 | −1 | 2.2 | 1.1 | 1.1 | 1.1 | 1.2 | −1.1 | −1.2 | ||
| Related RAS viral (r-ras) oncogene homolog | NM_006270 | 1.5 | 2.2 | 1.6 | 1.5 | 1.5 | 1.4 | 1.7 | 2.1 | ||
| Sjogren syndrome antigen A1 (52 kDa, ribonucleoprotein autoantigen SS-A/Ro) | M62800 | 1 | 2.2 | 1.2 | 1.4 | 1.1 | 1.6 | 1 | 1.1 | ||
| Uncoupling protein 1 (mitochondrial, proton carrier) | X51952 | ND | 2.2 | 1.3 | 1 | −1.1 | 1.7 | 1.7 | 1.9 | ||
| ESTs | AW468728 | −1 | 2.1 | 1.2 | −1 | 1.1 | ND | 1.8 | 1.8 | ||
| ESTs, weakly similar to delta-like protein 1 precursor (H. sapiens) | AW594704 | 1.3 | 2.1 | 1.2 | 1.7 | −1 | 1.3 | 1.4 | 2.7 | ||
| Interferon-induced protein with tetratricopeptide repeats 4 | AF083470 | 1.5 | 2.1 | 1.3 | 1 | ND | 1.4 | −1.4 | −1 | ||
| Superoxide dismutase 2, mitochondrial | Y00472 | 1.2 | 2.1 | 1.1 | 3.1 | −1.4 | 3 | 1.5 | 1.7 | ||
ESTs, expressed sequence tags; ND, not determined.
Cellular gene expression in d106-infected HEL cells is perturbed to a far greater degree than in d109-infected cells (27, 28). However, the genes that were induced by d109 were not significantly induced by d106 at 6 and 24 h postinfection (Table 1). The set of genes that were induced by d109 were not found to be induced in three other d106 determinations (data not shown). Furthermore, d105 and RJ1 infection also did not result in the induction of interferon-stimulated genes. d105 has the same viral background as d106 except that it does not have the gene for GFP inserted into the deleted ICP27 locus (28). RJ1 has the same background as d106 except that it has a β-galactosidase gene insertion into the UL41 (vhs) locus. Therefore, neither GFP nor the action of UL41 is involved in the lack of interferon-stimulated gene expression in these backgrounds. Rather, it appears that the expression of ICP0, or simply expression from the viral genome, may inhibit the induction of the interferon-stimulated genes.
In order to practically conduct more detailed analysis of the induction of interferon-stimulated genes by HSV, specialized expression arrays were constructed that contained a number of interferon-stimulated genes. A number of interferon-stimulated genes as well as genes that were found not to change with infection on the Incyte Genomics arrays were printed 10 times each on glass slides along with targets for internal spike controls. RNA from d109-infected HEL cells was analyzed using these arrays, and again, d109 was found to induce the expression of interferon-stimulated genes (Table 2). The resulting pattern of induction on the custom arrays was similar to that seen with the Incyte Genomics arrays despite the fact that different clones representing these genes were used in the construction of the two arrays.
TABLE 2.
Differential expression of interferon-induced genes on spotted cDNA microarrays
| Gene | Ratio, d109/mocka |
|---|---|
| Interferon-induced protein 56 | 50.88 |
| Myxovirus (influenza virus) resistance 1, homolog of murine (interferon-inducible protein p78) | 49.24 |
| Interferon-induced protein 54 | 47.52 |
| Hypothetical protein, expressed in osteoblast | 17.08 |
| Interferon regulatory factor 2 | 9.83 |
| Interferon-stimulated protein, 15 kDa | 8.73 |
| Alpha-interferon-inducible protein (clone IFI-6-16) | 8.42 |
| Alpha-interferon-inducible protein 27 | 7.39 |
| Caspase-1, apoptosis-related cysteine protease (interleukin-1 convertase) | 6.68 |
| Interferon stimulated gene (20 kDa) | 5.73 |
| Guanosine monophosphate reductase | 5.01 |
| Stimulated trans-acting factor (50 kDaa) | 4.75 |
| Myxovirus (influenza virus) resistance 2, homolog of murine | 4.67 |
| Interferon regulatory factor 1 | 4.67 |
| Proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional protease 2) | 4.59 |
| Interferon-stimulated transcription factor 3, gamma (48 kDa) | 4.27 |
| ESTs, highly similar to interferon-induced guanylate-binding protein 1 (H. sapiens) | 3.49 |
| Interleukin-6 (beta-interferon) | 3.48 |
| Interferon-induced protein 17 | 3.16 |
The ratio of signals from the d109-infected cell sample to those in the uninfected cell sample that were greater than 3.0.
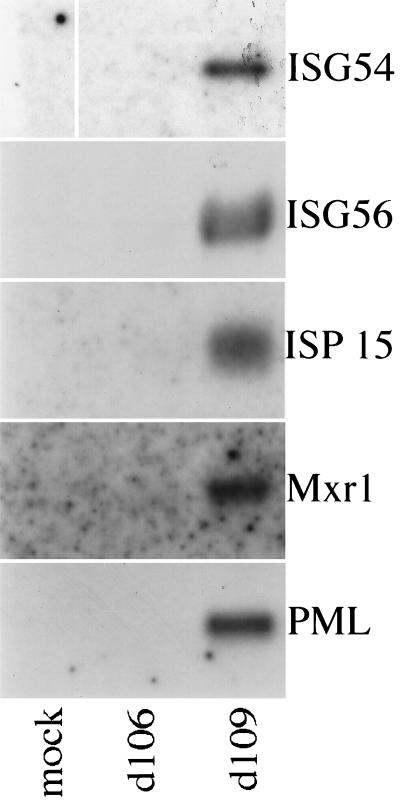
To confirm the induction of the most highly expressed interferon-stimulated genes by d109 infection and examine the induction of another gene (the promyelocytic leukemia [PML] gene) not included on the custom microarray, Northern blot analysis was performed (Fig. 1). As with the custom arrays, d109 infection was found to highly induce expression of ISG54, ISG56, ISG15, and myxovirus resistance gene 1 (Mrx1). Additionally, d109 was also found to induce expression of PML. Infection with d106 did not significantly induce expression of any of these genes by Northern blot analysis.
FIG. 1.
Infection with d109 but not d106 induces expression of interferon response genes. Total mRNA was isolated from mock-infected, d106-infected, and d109-infected cells at 24 h postinfection and analyzed by Northern blot analysis probing for the indicated genes as described in Materials and Methods.
From the previous experiments, it was clear that d109 induced the interferon-stimulated genes at high MOI. Previous studies documented the induction of ISG56, ISG54, myxovirus resistance gene 1, and ISG15 by IE-deficient viruses defective in VP16 activation function (45, 50). However, both used viruses at an MOI of 5, which was insufficient to induce a response measurable by our array experiments. Therefore, we examined the induction of interferon-stimulated genes by d109 as a function of MOI by microarray analysis.
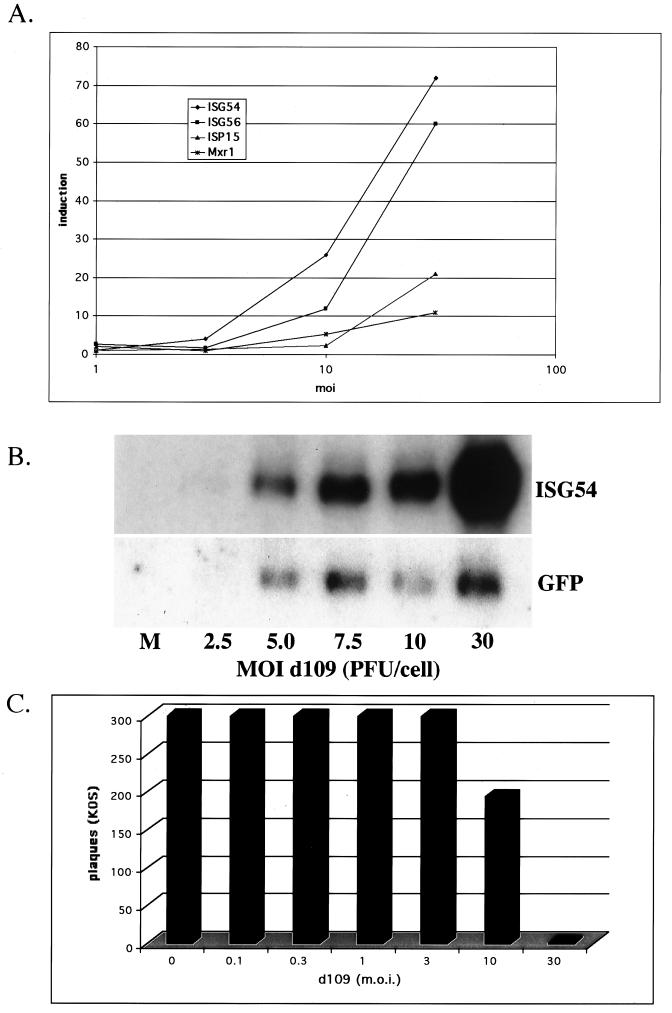
An increase in the abundance of the most highly induced interferon-stimulated genes became detectable at an MOI of 10 and more pronounced at an MOI of 30 (Fig. 2A). This analysis failed to detect reproducible differential expression of most interferon-stimulated genes during d109 infection at a lower MOI. Because small changes are often difficult to observe using microarray analysis, Northern blot analysis of HEL cells infected with d109 at MOI of 2.5, 5, 7.5, 10, and 30 was performed using a probe for the most highly expressed interferon-stimulated gene, ISG54 (Fig. 2B). d109 induced low levels of ISG54 at an MOI of 5 and barely detectable levels at an MOI of 2.5. Similar to the microarray, the induction of ISG54 increased dramatically with multiplicities of infection greater than 10 PFU/cell. In contrast, the amount of GFP RNA increased significantly only between MOI of 2.5 and 5 and did not increase further with increasing MOI.
FIG. 2.
Induction of interferon-stimulated genes and antiviral effect as a function of MOI. (A) Microarray analysis was performed comparing mock-infected cells and cells infected with d109 at the indicated MOI as described in Materials and Methods. Shown are the induction ratios as a function of MOI for the four most highly induced interferon response genes. (B) Abundance of IFI54 and GFP in cells infected with d109 at various MOI was examined by Northern blot analysis. Total mRNA was isolated from mock-infected cells and cells infected with the indicated MOI of d109 at 24 h postinfection. (C) The antiviral effect of d109 is shown as a function of MOI. Monolayers of HEL cells were infected with the indicated MOI of d109 and 24 h later were used in a plaque assay with ≈300 PFU of KOS. Shown are the resulting numbers of plaques that developed 3 days later.
It has been shown previously that the induction of interferon-stimulated genes in cells renders them refractory to wild-type virus plaque-forming ability (45). Induction of interferon-stimulated genes, however, does not always correlate with an effective antiviral response in cells. Thus, we examined the ability of different multiplicities of d109 to inhibit plaque formation of wild-type virus on HEL cells (Fig. 2C). Inhibition of wild-type virus plaque-forming ability, like interferon-stimulated gene induction, increased with MOI. The inhibition of plaque-forming ability was modest at an MOI of 10 but became substantial at an MOI of 30 PFU/cell. There was no antiviral effect at lower MOI. Although it is not clear which interferon-stimulated gene products are responsible for the protective antiviral response, inhibition of plaque-forming ability appears to require a substantial induction of the interferon-stimulated genes by d109.
The data in Table 1 suggest that the expression of ICP0 greatly reduced or eliminated the induction of interferon-stimulated genes. To examine this more closely, a number of IE deletion mutants were used to infect HEL cells, and 24 h later the cultures were harvested, and RNA was extracted and manipulated for array analysis. Arrays were used as targets for cDNA derived from the RNA of d120-, d107-, d106-, d104-, d109-, and mock-infected cells. d120 expresses all of the IE proteins except ICP4 (14). d107 does not express ICP4 and ICP27 (58). d106 does not express ICP4, ICP27, ICP22, and ICP47 (58). d104 does not express ICP4, ICP27, and ICP0 (58). d109 does not express any of the IE proteins (58).
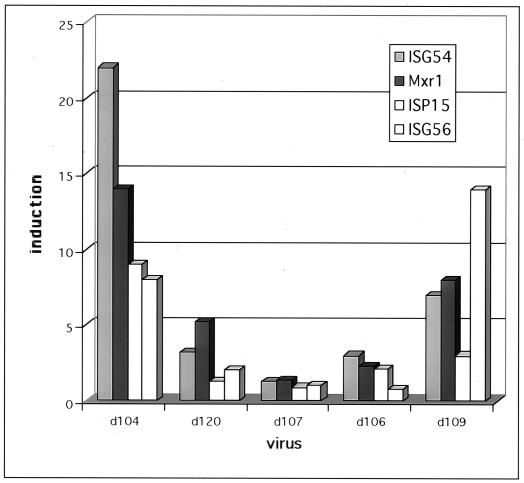
The induction ratios for the four most highly d109-induced interferon-stimulated genes are shown for each virus (Fig. 3). The induction of these genes was evident for the viruses that do not express ICP0 (d104 and d109) and was low from the viruses that do express ICP0 (d120, d107, and d106). Therefore, consistent with the results in Table 1, the results depicted in Fig. 3 also support the notion that expression of ICP0 inhibits the induction of interferon-stimulated genes.
FIG. 3.
Induction of interferon response genes by various HSV mutants. Microarray analysis was performed comparing mock-infected HEL cells and cells infected for 24 h with the indicated mutants (MOI = 30) as described in Materials and Methods. Shown are the induction ratios for each mutant virus infection for the four most highly induced interferon response genes (IFI54, IFI56, ISG15, and Mxr1).
The results of the previous experiment indicate that the expression of ICP0 may preclude the induction of interferon-stimulated genes. However, all of the ICP0-expressing viruses used above overexpress ICP0 relative to wild-type virus. We have previously shown that an E1− E3− E4− adenovirus vector expressing ICP0 from the E4 promoter expresses <0.1% of the amount of ICP0 expressed from d106, is nontoxic to cells, and does not greatly perturb cellular gene expression despite retaining the ability to disrupt ND10 and activate quiescent d109 genomes (28). Therefore, we also examined the ability of this virus to inhibit the d109 induction of the interferon-stimulated genes.
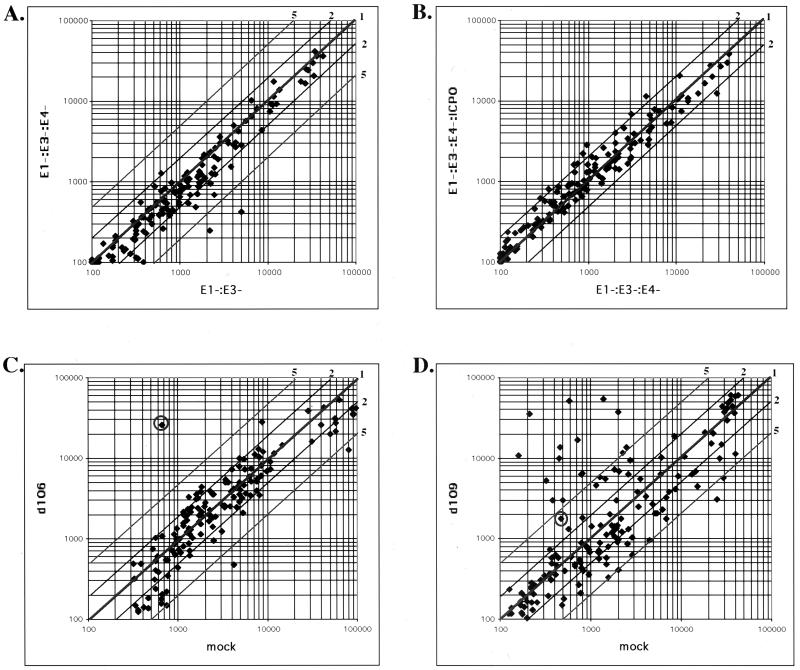
HEL cells were infected with the indicated virus for 24 h and then processed for array analysis. Figure 4 shows four comparisons. The first (Fig. 4A) compares an E1− E3− adenovirus with an E1− E3− E4− adenovirus. There were very few differences seen in this study. Only STAT1 and α-glucoside were induced in the E1− E3− virus relative to the E1− E3− E4− virus. There were no significant differences seen in the comparison of the E1− E3− E4− virus and the E1− E3− E4− ICP0+ virus (Fig. 4B).
FIG. 4.
Comparative expression of interferon response genes by adenovirus and HSV mutants. The effect of infection by AdS.11D (E1− E3− E4−), AdS.11E4(ICP0) (E1− E3− E4− ICP0+), AdS.10 (E1− E3−), d106, and d109 on cellular gene expression was examined by microarray analysis. Total RNA was isolated from infected and mock-infected HEL cells at 24 h postinfection. RNA was labeled and analyzed as described in Materials and Methods. Log-log scale scatter plots of fluorescent intensity are shown, with relative fold increase or decrease represented by diagonal lines. Each spot represents a single gene. The circled spot represents the signal for GFP. (A) Comparative expression in E1− E3− E4− and E1− E3− adenovirus-infected cells. (B) Comparative expression in E1− E3− E4− ICP0+ and E1− E3− E4− adenovirus-infected cells. (C) Comparison of d106- and mock-infected cells. (D) Comparison of d109- and mock-infected cells.
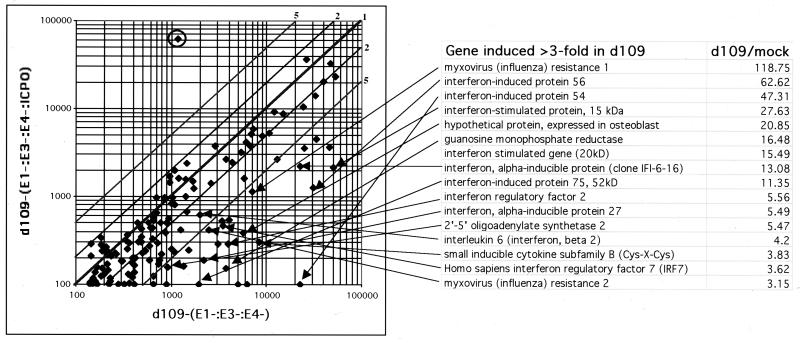
In the comparison of d106-infected to uninfected cells (Fig. 4C), the only gene that was substantially differentially expressed in d106-infected cells was GFP (circled). GFP is expressed from the human cytomegalovirus promoter on the d106 genome. Consistent with the previous data, the set of genes differentially expressed in d109-infected cells corresponded to the interferon-stimulated genes (Fig. 4D). The identities of the induced genes and the magnitudes of their induction are listed in Fig. 5.
FIG. 5.
Inhibition of induced interferon response gene expression by adenovirus-expressed ICP0. HEL cells were infected with AdS.11D (E1− E3− E4−) or AdS.11E4(ICP0) (E1− E3− E4− ICP0+) adenovirus. At 24 h postinfection, the cells were superinfected with d109. Total RNA was isolated 24 h later, and microarray analysis was performed as in Fig. 4. A log-log scale scatter plot shows the relative expression of the interferon response genes in d109-infected cells that were previously infected with the E1− E3− E4− or E1− E3− E4− ICP0+ adenovirus. Each spot represents a single gene. The relative fold increase or decrease is represented by diagonal lines. The circled gene represents GFP expression. The genes induced greater than threefold by d109 relative to mock-infected cells in Fig. 4D are listed along with the fold induction ratios. The arrows extend from the indicated genes on this list to their corresponding signals on the scatter plot.
In a parallel set of infections, HEL cells were first infected with the E1− E3− E4− adenovirus or the E1− E3− E4− ICP0+ adenovirus at an MOI of 1,000 particles per cell and incubated for 24 h. The data in Fig. 4B demonstrate that no gene on the array is differentially expressed between these two infections. Both sets of adenovirus-infected cells were then infected with d109, and 24 h later RNA was isolated from both cultures, which were used in a single comparison on the array depicted in Fig. 5. The abundance of all interferon response genes induced greater than threefold by d109 in Fig. 4D was always greater in the d109-infected sample previously infected with the E1− E3− E4− virus than in that infected with the E1− E3− E4− ICP0+ virus (Fig. 5). Conversely, GFP RNA abundance was 500 times greater in the d109-infected sample that was previously infected with the E1− E3− E4− ICP0+ virus than in the d109-infected sample that was previously infected with the E1− E3− E4− virus. Therefore, the prior expression of ICP0 from the adenovirus is sufficient to both activate gene expression from the d109 genome and inhibit or reduce the induction of the interferon-stimulated genes.
During infection, ICP0 may function by targeting specific cellular proteins for degradation through the proteasome-dependent degradation pathway (18, 20). ICP0 itself has been shown to possess ubiquitin ligase activity (68). Everett and colleagues have also shown that some consequences of ICP0 function can be blocked by addition of the proteasome inhibitor MG132 (18). Thus, if this pathway is also involved in the inhibition of induction of the interferon-stimulated genes, the addition of MG132 to cells undergoing infection in the presence of ICP0 should result in an increase in the abundance of interferon-stimulated gene products.
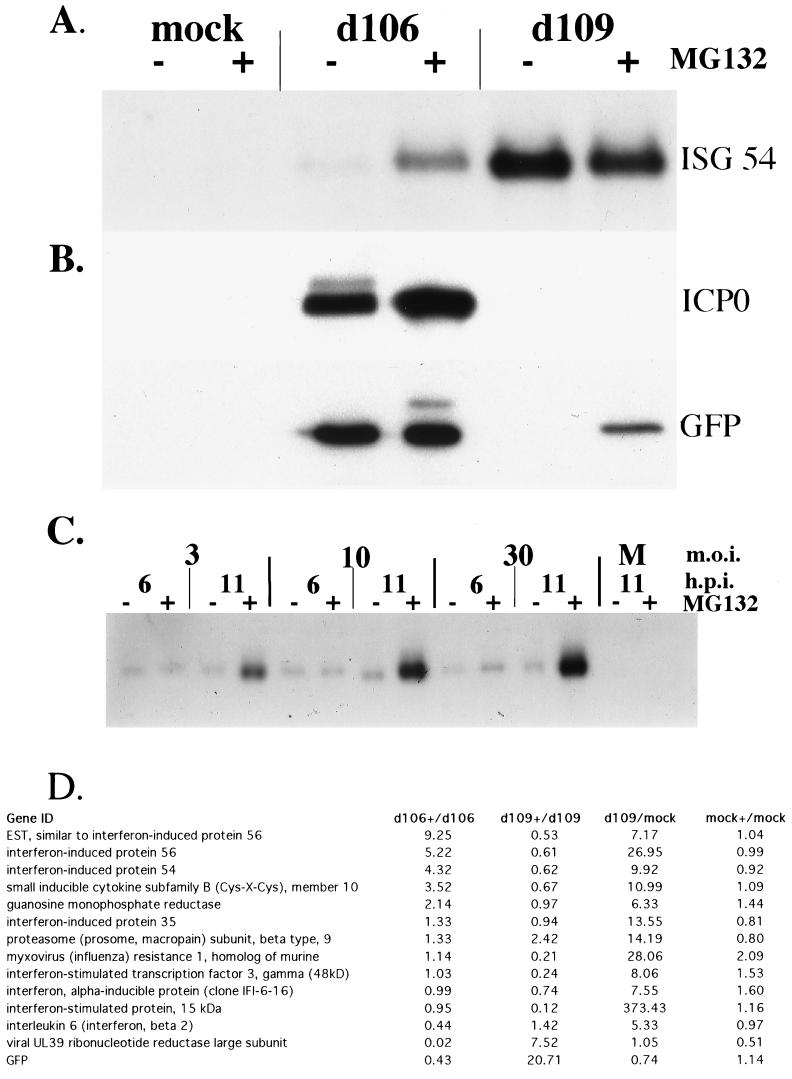
Where indicated, HEL cells were treated with 5 μM MG132 for 30 min prior to being mock infected or infected with d106 or d109. MG132 treatment was continued until RNA or protein was harvested at the indicated time postinfection. The addition of MG132 resulted in elevated levels of ISG54 mRNA in d106-infected cells (Fig. 6A), despite the presence of ICP0 (Fig. 6B). ISG54 was not induced in MG132-treated mock-infected cells. As before, d109-infected cells showed induction of ISG54, which was slightly lower in the MG132-treated cells (Fig. 6A). MG132 treatment also resulted in slightly decreased GFP expression from the human cytomegalovirus promoter in the d106 genome, but stimulated expression in d109-infected cells (Fig. 6B). Interestingly, ICP6 (UL39) expression, which was dramatically lower in MG132-treated d106-infected cells, was also stimulated in d109-infected cells by the addition of MG132 (Fig. 6D).
FIG. 6.
Effect of MG132 on interferon-stimulated gene expression in virus-infected cells. (A) Effect of MG132 treatment on abundance of ISG54 in d106- and d109-infected cells was examined by Northern blot analysis. Cells were treated as indicated with 5 μM MG132 from 30 min prior to infection until RNA isolation. Total mRNA was isolated from mock-infected cells and cells infected with the indicated virus (MOI = 30) at 11 h postinfection. (B) Effect of MG132 treatment on ICP0 and GFP protein expression in d106- and d109-infected cells was examined by Western blot analysis. Cells were treated as in A. Total protein was isolated from mock-infected cells and cells infected with the indicated virus at 11 h postinfection. (C) Effect of MG132 treatment on the abundance of ISG54 in cells infected with d106 at various MOI. Cells were treated as indicated with 5 μM MG132 from 30 min prior to infection until RNA isolation. Total mRNA was isolated from mock-infected cells and cells infected with the indicated MOI of d106 at 6 and 11 h postinfection (h.p.i.). Northern blot analysis was performed, probing for ISG54 message. (D) Microarray analysis was performed on the mRNA from A as described in Materials and Methods. Shown are the induction ratios for comparisons of MG132-treated and untreated cells that have been either mock, d109, or d106 infected. Also shown is a comparison of d109- (MOI = 30) and mock-infected cells.
The effect of virus input on the levels of ISG54 accumulation in MG132-treated d106-infected cells was also examined. ISG54 was induced in MG132-treated d106-infected cells as a function of input MOI (Fig. 6C). Untreated d106-infected cells showed a very low level of ISG54 induction at both 6 and 11 h. At the 11- but not the 6-h time point, the induction was greatly increased in MG132-treated d106-infected cells. This suggests that the amount of ISG54 RNA at early times postinfection may result from an induction event that occurs prior to the accumulation of sufficient ICP0 to inhibit further induction.
Lastly, microarray experiments were conducted to obtain a more general view of the effects of MG132 on the levels of interferon-stimulated genes in d109- and d106-infected cells (Fig. 6D). RNA from MG132-treated d106-infected cells was compared to RNA from untreated d106-infected cells on the same chip. The same type of comparison was conducted for d109- and mock-infected cells. Shown are the data for the interferon-stimulated genes induced fivefold or greater in d109-infected cells relative to uninfected cells. The arrays also contained DNA encoding parts of the genes for GFP and ICP6 (UL39), which are abundantly expressed in d106-infected cells and poorly expressed in d109-infected cells (58). Four genes, including ISG54, were induced by a factor of threefold or greater in MG132-treated d106-infected cells relative to untreated d106-infected cells. MG132 did not affect the expression of these genes in uninfected cells. Therefore, these genes were induced as a function of infection upon inhibition of the proteasome, in the presence of ICP0.
A number of d109-induced genes were not induced in MG132-treated d106-infected cells. Interestingly, the abundance of a number of these RNAs was reduced in MG132-treated d109-infected cells relative to that in untreated d109-infected cells. It is possible that the global effects of inhibiting the proteasome may result in the altered abundance of gene-specific transcription factors that function in the expression of specific interferon-stimulated genes.
DISCUSSION
d109 induced interferon-stimulated genes very similar to other non-IE-expressing mutants, including in1312, used by Nicholl et al., and KM110, used by Mossman et al. (45, 50). Interferon-stimulated gene expression increased with increasing MOI, abruptly becoming more abundant when the MOI exceeded 10 PFU/cell. Interestingly, the plaquing of wild-type virus on d109-infected cells was only significantly inhibited when the MOI of d109 used to establish the antiviral state exceeded 10 PFU/cell. The quantitative differences between this and previous studies (45, 50) may simply be a function of different PFU/particle ratios. Perhaps this cellular response functions to attenuate robust acute infections in vivo and has less influence on low-level and/or latent infections. Alternatively, the cellular response to infection with respect to interferon-stimulated gene induction may differ from cell type to cell type (54).
In previous work, no HSV-1 mutants expressing ICP4 or ICP0 were found to significantly induce expression of interferon-stimulated genes unless inactivated by UV or treated with cycloheximide (45, 50). From Mossman et al. and our microarray data, it is clear that an ICP4-defective virus, d120, cannot induce interferon-stimulated genes (Fig. 3). However, we found that ICP4− mutants that express ICP0, regardless of other IE genes not expressed, also did not significantly induce interferon-stimulated genes. The small amount of interferon-stimulated gene induction seen in d106-infected cells could be that accumulating prior to the accumulation of sufficient ICP0 to inhibit further induction. The only mutant besides d109 found to robustly induce interferon-stimulated genes was d104, which does not express ICP0 but does express ICP22 and ICP47. Consistent with these results, an adenovirus expressing very low levels of ICP0 also inhibited the induction of interferon-stimulated genes by d109. Together, these results suggest that the expression of ICP0 can inhibit the induction of the interferon-stimulated genes by the HSV virion.
ICP0 has been shown to possess ubiquitin ligase activity (68), and some consequences of ICP0 expression have also been shown to be inhibited by inhibition of the proteasome (18). Therefore, as suggested by Everett (17), ICP0 may function by altering the abundance of specific cellular proteins. We have shown that the expression of ICP0 and the function of the proteasome are required for the inhibition of interferon-stimulated gene induction. Given these observations, some possible cellular targets emerge.
Recently, the activation of IRF-3 in HSV-1-infected cells treated with cycloheximide has been reported (54). Activated IRF-3 is a component of both double-stranded RNA-activated factor 1 (DRAF1) and virus-activated factor (VAF) along with CBP/p300 (72, 73). DRAF1 and VAF act to transactivate interferon-stimulated gene expression by binding to interferon-stimulated response element sequences (6, 13, 23). While it is constitutively expressed and mostly cytoplasmic, IRF-3 is not activated until it is phosphorylated and retained in the nucleus by CBP/p300 (36, 39). Further, phosphorylation is a signal for its degradation by the ubiquitin-proteasome pathway (39, 55). IRF-3 activation and nuclear retention is a complicated process that has not been completely elucidated. Thus, it is not clear whether ICP0 could act to increase the degradation rate of activated IRF-3, target another protein involved in the pathway leading to activation of IRF-3, DRAF1, or VAF, or lead to the degradation of a protein wholly unrelated to the IRF-3 pathway.
The interferon response gene PML is involved in regulating gene expression (1, 38, 47, 63, 67) and has also been found to be an interferon-stimulated gene (37). Chelbi-Alix et al. found that with greater PML expression, vesicular stomatitis virus and influenza A virus multiplication could be inhibited up to 100-fold (10). Interestingly, d109 induced the expression of PML (Fig. 3). Deletion of a specific region of PML both altered the punctate localization of PML onto nuclear bodies and eradicated the antiviral properties (10). ICP0 has been shown to alter PML localization in nuclear bodies as a function of active proteasomes (18, 19, 41, 42), and the small amounts of ICP0 from E1− E3− E4− ICP0+ are still able to disrupt ND10 nuclear bodies containing PML protein while promoting viral gene expression (28). Furthermore, while interferon was shown to decrease the expression of IE genes in a dose-dependent manner (2, 34, 40, 44, 51, 52), this decrease was found to correlate with a decrease in the amount of PML disruption by HSV-1 (66). If the destruction of PML by ICP0 is involved in the mechanism preventing interferon-stimulated gene induction, it most likely happens very rapidly in wild-type infection, as the amount of ICP0 necessary to disrupt ND10 is synthesized within the first hour of infection (28).
Cellular interferon responses and antiviral pathways vary greatly between cell types and inducers and use multiple signaling pathways. In addition, the actions of many interferon-stimulated genes remain undercharacterized, and the counteracting viral mechanisms are difficult to characterize. Two of the most studied antiviral pathways, involving the interferon-stimulated genes double-stranded RNA-dependent protein kinase and the 2",5"-oligoadenylate synthetase, are both inhibited by HSV-1 (9, 26, 53). It is clear that HSV can also inhibit the induction of interferon-stimulated genes by the virion, and it has been postulated that multiple HSV gene products may be sufficient for this activity (45, 50). Our studies strongly suggest that one such gene product is ICP0. Therefore, ICP0 expressed very early in infection may simultaneously stimulate viral gene expression and contribute to the inhibition of interferon-stimulated gene induction, thus promoting lytic infection. The ability of ICP0 to inhibit the induction of interferon-stimulated genes requires functional proteasomes, and the specific cellular target(s) of ICP0 action remains to be determined.
Acknowledgments
This work was supported by NIH grants AI44812 and AI30612.
We are grateful to Kimberly Harrison for technical assistance. We also acknowledge Rachel Juhas for the construction and characterization of virus RJ1.
REFERENCES
- 1.Alcalay, M., L. Tomassoni, E. Colombo, S. Stoldt, F. Grignani, M. Fagioli, L. Szekely, K. Helin, and P. G. Pelicci. 1998. The promyelocytic leukemia gene product (PML) forms stable complexes with the retinoblastoma protein. Mol. Cell. Biol. 18:1084-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altinkilic, B., and G. Brandner. 1988. Interferon inhibits herpes simplex virus-specific translation: a reinvestigation. J. Gen. Virol. 69:3107-3112. [DOI] [PubMed] [Google Scholar]
- 3.Bandyopadhyay, S. K., G. T. Leonard, Jr., T. Bandyopadhyay, G. R. Stark, and G. C. Sen. 1995. Transcriptional induction by double-stranded RNA is mediated by interferon-stimulated response elements without activation of interferon-stimulated gene factor 3. J. Biol. Chem. 270:19624-19629. [DOI] [PubMed] [Google Scholar]
- 4.Batterson, W., and B. Roizman. 1983. Characterization of the herpes simplex virion-associated factor responsible for the induction of alpha genes. J. Virol. 46:371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyle, K. A., R. L. Pietropaolo, and T. Compton. 1999. Engagement of the cellular receptor for glycoprotein B of human cytomegalovirus activates the interferon-responsive pathway. Mol. Cell. Biol. 19:3607-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braganca, J., P. Genin, M. T. Bandu, N. Darracq, M. Vignal, C. Casse, J. Doly, and A. Civas. 1997. Synergism between multiple virus-induced factor-binding elements involved in the differential expression of interferon A genes. J. Biol. Chem. 272:22154-22162. [DOI] [PubMed] [Google Scholar]
- 7.Cai, W., T. L. Astor, L. M. Liptak, C. Cho, D. M. Coen, and P. A. Schaffer. 1993. The herpes simplex virus type 1 regulatory protein ICP0 enhances virus replication during acute infection and reactivation from latency. J. Virol. 67:7501-7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell, M. E., J. W. Palfreyman, and C. M. Preston. 1984. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate early transcription. J. Mol. Biol. 180:1-19. [DOI] [PubMed] [Google Scholar]
- 9.Cayley, P. J., J. A. Davies, K. G. McCullagh, and I. M. Kerr. 1984. Activation of the ppp(A2"p)nA system in interferon-treated, herpes simplex virus-infected cells and evidence for novel inhibitors of the ppp(A2"p)nA-dependent RNase. Eur. J. Biochem. 143:165-174. [DOI] [PubMed] [Google Scholar]
- 10.Chelbi-Alix, M. K., F. Quignon, L. Pelicano, M. H. Koken, and H. de The. 1998. Resistance to virus infection conferred by the interferon-induced promyelocytic leukemia protein. J. Virol. 72:1043-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clements, G. B., and N. D. Stow. 1989. A herpes simplex virus type 1 mutant containing a deletion within immediate early gene 1 is latency-competent in mice. J. Gen. Virol. 70:2501-2506. [DOI] [PubMed] [Google Scholar]
- 12.Daly, C., and N. C. Reich. 1995. Characterization of specific DNA-binding factors activated by double-stranded RNA as positive regulators of interferon alpha/beta-stimulated genes. J. Biol. Chem. 270:23739-23746. [DOI] [PubMed] [Google Scholar]
- 13.Daly, C., and N. C. Reich. 1993. Double-stranded RNA activates novel factors that bind to the interferon-stimulated response element. Mol. Cell. Biol. 13:3756-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeLuca, N. A., A. M. McCarthy, and P. A. Schaffer. 1985. Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4. J. Virol. 56:558-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Veer, M. J., M. Holko, M. Frevel, E. Walker, S. Der, J. M. Paranjape, R. H. Silverman, and B. R. Williams. 2001. Functional classification of interferon-stimulated genes identified using microarrays. J. Leukoc. Biol. 69:912-920. [PubMed] [Google Scholar]
- 17.Everett, R. D. 2000. ICP0 induces the accumulation of colocalizing conjugated ubiquitin. J. Virol. 74:9994-10005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Everett, R. D., P. Freemont, H. Saitoh, M. Dasso, A. Orr, M. Kathoria, and J. Parkinson. 1998. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J. Virol. 72:6581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Everett, R. D., and G. G. Maul. 1994. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 13:5062-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Everett, R. D., M. Meredith, A. Orr, A. Cross, M. Kathoria, and J. Parkinson. 1997. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 16:1519-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farrell, P. J., G. C. Sen, M. F. Dubois, L. Ratner, E. Slattery, and P. Lengyel. 1978. Interferon action: two distinct pathways for inhibition of protein synthesis by double-stranded RNA. Proc. Natl. Acad. Sci. USA 75:5893-5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Friedman, R. L., and G. R. Stark. 1985. alpha-Interferon-induced transcription of HLA and metallothionein genes containing homologous upstream sequences. Nature 314:637-639. [DOI] [PubMed] [Google Scholar]
- 23.Genin, P., J. Braganca, N. Darracq, J. Doly, and A. Civas. 1995. A novel PRD I and TG binding activity involved in virus-induced transcription of interferon-A genes. Nucleic Acids Res. 23:5055-5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gordon, Y. J., J. L. McKnight, J. M. Ostrove, E. Romanowski, and T. Araullo-Cruz. 1990. Host species and strain differences affect the ability of an HSV-1 ICP0 deletion mutant to establish latency and spontaneously reactivate in vivo. Virology 178:469-477. [DOI] [PubMed] [Google Scholar]
- 25.Guo, J., K. L. Peters, and G. C. Sen. 2000. Induction of the human protein P56 by interferon, double-stranded RNA, or virus infection. Virology 267:209-219. [DOI] [PubMed] [Google Scholar]
- 26.He, B., M. Gross, and B. Roizman. 1997. The γ134.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1alpha to dephosphorylate the alpha subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc. Natl. Acad. Sci. USA 94:843-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hobbs, W. E., 2nd, and N. A. DeLuca. 1999. Perturbation of cell cycle progression and cellular gene expression as a function of herpes simplex virus ICP0. J. Virol. 73:8245-8255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hobbs, W. E., D. E. Brough, I. Kovesdi, and N. A. DeLuca. 2001. Efficient activation of viral genomes by levels of herpes simplex virus ICP0 insufficient to affect cellular gene expression or cell survival. J. Virol. 75:3391-3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Honess, R. W., and B. Roizman. 1974. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J. Virol. 14:8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Honess, R. W., and B. Roizman. 1975. Regulation of herpesvirus macromolecular synthesis: sequential transition of polypeptide synthesis requires functional viral polypeptides. Proc. Natl. Acad. Sci. USA 72:1276-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Imbalzano, A. N., D. M. Coen, and N. A. DeLuca. 1991. Herpes simplex virus transactivator ICP4 operationally substitutes for the cellular transcription factor Sp1 for efficient expression of the viral thymidine kinase gene. J. Virol. 65:565-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karin, M., A. Haslinger, H. Holtgreve, R. I. Richards, P. Krauter, H. M. Westphal, and M. Beato. 1984. Characterization of DNA sequences through which cadmium and glucocorticoid hormones induce human metallothionein-IIA gene. Nature 308:513-519. [DOI] [PubMed] [Google Scholar]
- 33.Karin, M., R. J. Imbra, A. Heguy, and G. Wong. 1985. Interleukin 1 regulates human metallothionein gene expression. Mol. Cell. Biol. 5:2866-2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klotzbucher, A., S. Mittnacht, H. Kirchner, and H. Jacobsen. 1990. Different effects of interferon gamma and interferon alpha/beta on “immediate early” gene expression of HSV-1. Virology 179:487-491. [DOI] [PubMed] [Google Scholar]
- 35.Krisky, D. M., D. Wolfe, W. F. Goins, P. C. Marconi, R. Ramakrishnan, M. Mata, R. J. Rouse, D. J. Fink, and J. C. Glorioso. 1998. Deletion of multiple immediate-early genes from herpes simplex virus reduces cytotoxicity and permits long-term gene expression in neurons. Gene Ther. 5:1593-1603. [DOI] [PubMed] [Google Scholar]
- 36.Kumar, K. P., K. M. McBride, B. K. Weaver, C. Dingwall, and N. C. Reich. 2000. Regulated nuclear-cytoplasmic localization of interferon regulatory factor 3, a subunit of double-stranded RNA-activated factor 1. Mol. Cell. Biol. 20:4159-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lavau, C., A. Marchio, M. Fagioli, J. Jansen, B. Falini, P. Lebon, F. Grosveld, P. P. Pandolfi, P. G. Pelicci, and A. Dejean. 1995. The acute promyelocytic leukaemia-associated PML gene is induced by interferon. Oncogene 11:871-876. [PubMed] [Google Scholar]
- 38.Lehming, N., A. Le Saux, J. Schuller, and M. Ptashne. 1998. Chromatin components as part of a putative transcriptional repressing complex. Proc. Natl. Acad. Sci. USA 95:7322-7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin, R., C. Heylbroeck, P. M. Pitha, and J. Hiscott. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 18:2986-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lipp, M., and G. Brandner. 1980. Inhibition of herpes simplex virus type 1 specific translation in cells treated with poly(rI)·poly(rC). J. Gen. Virol. 47:97-111. [DOI] [PubMed]
- 41.Maul, G. G., and R. D. Everett. 1994. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J. Gen. Virol. 75:1223-1233. [DOI] [PubMed] [Google Scholar]
- 42.Maul, G. G., H. H. Guldner, and J. G. Spivack. 1993. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0). J. Gen. Virol. 74:2679-2690. [DOI] [PubMed] [Google Scholar]
- 43.McCarthy, A. M., L. McMahan, and P. A. Schaffer. 1989. Herpes simplex virus type 1 ICP27 deletion mutants exhibit altered patterns of transcription and are DNA deficient. J. Virol. 63:18-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mittnacht, S., P. Straub, H. Kirchner, and H. Jacobsen. 1988. Interferon treatment inhibits onset of herpes simplex virus immediate-early transcription. Virology 164:201-210. [DOI] [PubMed] [Google Scholar]
- 45.Mossman, K. L., P. F. Macgregor, J. J. Rozmus, A. B. Goryachev, A. M. Edwards, and J. R. Smiley. 2001. Herpes simplex virus triggers and then disarms a host antiviral response. J. Virol. 75:750-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mossman, K. L., H. A. Saffran, and J. R. Smiley. 2000. Herpes simplex virus ICP0 mutants are hypersensitive to interferon. J. Virol. 74:2052-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mu, Z. M., K. V. Chin, J. H. Liu, G. Lozano, and K. S. Chang. 1994. PML, a growth suppressor disrupted in acute promyelocytic leukemia. Mol. Cell. Biol. 14:6858-6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Navarro, L., and M. David. 1999. p38-dependent activation of interferon regulatory factor 3 by lipopolysaccharide. J. Biol. Chem. 274:35535-35538. [DOI] [PubMed] [Google Scholar]
- 49.Nicholl, M. J., and C. M. Preston. 1996. Inhibition of herpes simplex virus type 1 immediate-early gene expression by alpha interferon is not VP16 specific. J. Virol. 70:6336-6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nicholl, M. J., L. H. Robinson, and C. M. Preston. 2000. Activation of cellular interferon-responsive genes after infection of human cells with herpes simplex virus type 1. J. Gen. Virol. 81:2215-2218. [DOI] [PubMed] [Google Scholar]
- 51.Oberman, F., and A. Panet. 1989. Characterization of the early steps of herpes simplex virus replication in interferon-treated human cells. J. Interferon Res. 9:563-571. [DOI] [PubMed] [Google Scholar]
- 52.Oberman, F., and A. Panet. 1988. Inhibition of transcription of herpes simplex virus immediate early genes in interferon-treated human cells. J. Gen. Virol. 69:1167-1177. [DOI] [PubMed] [Google Scholar]
- 53.Poon, A. P., and B. Roizman. 1997. Differentiation of the shutoff of protein synthesis by virion host shutoff and mutant γ134.5 genes of herpes simplex virus 1. Virology 229:98-105. [DOI] [PubMed] [Google Scholar]
- 54.Preston, C. M., A. N. Harman, and M. J. Nicholl. 2001. Activation of interferon response factor-3 in human cells infected with herpes simplex virus type 1 or human cytomegalovirus. J. Virol. 75:8909-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ronco, L. V., A. Y. Karpova, M. Vidal, and P. M. Howley. 1998. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 12:2061-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sacks, W. R., C. C. Greene, D. P. Aschman, and P. A. Schaffer. 1985. Herpes simplex virus type 1 ICP27 is an essential regulatory protein. J. Virol. 55:796-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sacks, W. R., and P. A. Schaffer. 1987. Deletion mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein ICP0 exhibit impaired growth in cell culture. J. Virol. 61:829-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Samaniego, L. A., L. Neiderhiser, and N. A. DeLuca. 1998. Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J. Virol. 72:3307-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Samaniego, L. A., A. L. Webb, and N. A. DeLuca. 1995. Functional interactions between herpes simplex virus immediate-early proteins during infection: gene expression as a consequence of ICP27 and different domains of ICP4. J. Virol. 69:5705-5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Samaniego, L. A., N. Wu, and N. A. DeLuca. 1997. The herpes simplex virus immediate-early protein ICP0 affects transcription from the viral genome and infected-cell survival in the absence of ICP4 and ICP27. J. Virol. 71:4614-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Samuel, C. E. 1991. Antiviral actions of interferon. Interferon-regulated cellular proteins and their surprisingly selective antiviral activities. Virology 183:1-11. [DOI] [PubMed] [Google Scholar]
- 62.Schena, M. 1999. DNA microarrays: a practical approach. Oxford University Press Inc., New York, N.Y.
- 63.Seeler, J. S., A. Marchio, D. Sitterlin, C. Transy, and A. Dejean. 1998. Interaction of SP100 with HP1 proteins: a link between the promyelocytic leukemia-associated nuclear bodies and the chromatin compartment. Proc. Natl. Acad. Sci. USA 95:7316-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sen, G. C., and P. Lengyel. 1992. The interferon system. A bird's eye view of its biochemistry. J. Biol. Chem. 267:5017-5020. [PubMed] [Google Scholar]
- 65.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 66.Taylor, J. L., D. Unverrich, W. J. O'Brien, and K. W. Wilcox. 2000. Interferon coordinately inhibits the disruption of PML-positive ND10 and immediate-early gene expression by herpes simplex virus. J. Interferon Cytokine Res. 20:805-815. [DOI] [PubMed] [Google Scholar]
- 67.Vallian, S., K. V. Chin, and K. S. Chang. 1998. The promyelocytic leukemia protein interacts with Sp1 and inhibits its transactivation of the epidermal growth factor receptor promoter. Mol. Cell. Biol. 18:7147-7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Van Sant, C., R. Hagglund, P. Lopez, and B. Roizman. 2001. The infected cell protein 0 of herpes simplex virus 1 dynamically interacts with proteasomes, binds and activates the cdc34 E2 ubiquitin-conjugating enzyme, and possesses in vitro E3 ubiquitin ligase activity. Proc. Natl. Acad. Sci. USA 98:8815-8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wathelet, M. G., P. M. Berr, and G. A. Huez. 1992. Regulation of gene expression by cytokines and virus in human cells lacking the type-I interferon locus. Eur. J. Biochem. 206:901-910. [DOI] [PubMed] [Google Scholar]
- 70.Wathelet, M. G., I. M. Clauss, J. Content, and G. A. Huez. 1988. Regulation of two interferon-inducible human genes by interferon, poly(rI)·poly(rC) and viruses. Eur. J. Biochem. 174:323-329. [DOI] [PubMed] [Google Scholar]
- 71.Wathelet, M. G., I. M. Clauss, C. B. Nols, J. Content, and G. A. Huez. 1987. New inducers revealed by the promoter sequence analysis of two interferon-activated human genes. Eur. J. Biochem. 169:313-321. [DOI] [PubMed] [Google Scholar]
- 72.Wathelet, M. G., C. H. Lin, B. S. Parekh, L. V. Ronco, P. M. Howley, and T. Maniatis. 1998. Virus infection induces the assembly of coordinately activated transcription factors on the interferon-beta enhancer in vivo. Mol. Cell 1:507-518. [DOI] [PubMed] [Google Scholar]
- 73.Weaver, B. K., K. P. Kumar, and N. C. Reich. 1998. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol. Cell. Biol. 18:1359-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu, N., S. C. Watkins, P. A. Schaffer, and N. A. DeLuca. 1996. Prolonged gene expression and cell survival after infection by a herpes simplex virus mutant defective in the immediate-early genes encoding ICP4, ICP27, and ICP22. J. Virol. 70:6358-6369. [DOI] [PMC free article] [PubMed] [Google Scholar]