Abstract
Peroxisomes are unimembrane, respiratory organelles of the cell. Transport of cellular proteins to the peroxisomal matrix requires a type 1 peroxisomal targeting signal (PTS1) which essentially constitutes a tripeptide from the consensus sequence S/T/A/G/C/N-K/R/H-L/I/V/M/A/F/Y. Although PTS-containing proteins have been identified in eukaryotes, prokaryotes, and parasites, viral proteins with such signals have not been identified so far. We report here the first instance of a virus, the rotavirus, which causes infantile diarrhea worldwide, containing a functional C-terminal PTS1 in one of its proteins (VP4). Analysis of 153 rotavirus VP4-deduced amino acid sequences identified five groups of conserved C-terminal PTS1 tripeptide sequences (SKL, CKL, GKL, CRL, and CRI), of which CRL is represented in approximately 62% of the sequences. Infection of cells by a CRL-containing representative rotavirus (SA11 strain) and confocal immunofluorescence analysis revealed colocalization of VP4 with peroxisomal markers and morphological changes of peroxisomes. Further, transient cellular expression of green fluorescent protein (GFP)-fused VP4CRL resulted in transport of VP4 to peroxisomes, whereas the chimera lacking the PTS1 signal, GFP-VP4ΔCRL, resulted in diffuse cytoplasmic staining, suggesting a CRL-dependent targeting of the protein. The present study therefore demonstrates hitherto unreported organelle involvement, specifically of the peroxisomes, in rotaviral infections as demonstrated by using the SA11 strain of rotavirus and opens a new line of investigation toward understanding viral pathogenesis and disease mechanisms.
Peroxisomes are small vesicle-like subcellular respiratory organelles bound by a single membrane and filled with a dense matrix (6). These organelles form an important constituent of the eukaryotic cell, performing various functions such as hydrogen peroxide degradation, β-oxidation of fatty acids, glyoxylate metabolism, and gluconeogenesis (11, 19, 29). The organelle also contains antioxidant molecules such as ascorbate and glutathione and a battery of antioxidant enzymes such as superoxide dismutase, glutathione reductase, and the principal cellular H2O2-degrading enzyme, catalase (18). Consequently, peroxisomes are sensitive to external stress signals and stress induces peroxisome biogenesis genes (18).
Trafficking of proteins to peroxisomes depends on two types of functional peroxisomal targeting signal (PTS) sequences. The type 1 PTS sequence (PTS1) is a tripeptide (prototype, SKL) that resides either within or at the C terminus (6, 20), and the type 2 sequence (PTS2) resides in the N-terminal half of proteins (13, 28). In nature, conservative functional variants of PTS1 prototype (SKL) tripeptide sequences, such as CKL, CRL, and CRI, were identified in peroxisomal matrix proteins (7, 15). Recently, yeast two-hybrid-based SKL receptor (Pex5p) screening of small peptides with various C-terminal conservative degenerate members of the family of SKL motifs further identified several other functional variants (13, 16, 28). Peroxisomal matrix proteins with a PTS1 sequence were reported to be present in all eukaryotes and in parasites like trypanosomes but not in viruses (6, 8, 25).
In this report we identified for the first time that a virus, the group A rotavirus, encodes a protein (structural protein VP4) which contains the canonical PTS1 or its conserved functional variants at the C terminus and demonstrated that PTS1 is required for targeting rotavirus VP4 to peroxisomes.
Rotaviruses are the causative agents for infantile viral gastroenteritis and account for about 600,000 to 850,000 deaths per year worldwide (2). This virus group belongs to the Reoviridae family and the virion genome contains 11 double-stranded RNA segments and encodes 6 structural and 5 nonstructural proteins (3). One of the structural proteins, VP4, constitutes the spikes projecting from the virion surface (24). Protease cleavage of VP4 renders an N-terminal VP8* subunit (amino acids 1 to 246) and a C-terminal VP5* subunit (amino acids 247 to 776) and enhances infectivity of the virion (9, 10). The PTS1 sequence identified in VP4 as described in this report is present at the C terminus of the VP5* portion of VP4.
Rotavirus infection of MA104 cells and cellular RNA extraction
Simian rotavirus strain SA11 (serotype G3P5) and MA104 cells were obtained from the American Type Culture Collection (ATCC; Manassas, Va.). The strain is designated ATCC SA11. MA104 cells were grown in Dulbecco's modified Eagle's medium supplemented with antibiotics and 10% fetal calf serum in T-25 tissue culture flasks. For immunofluorescence, cells were grown in chamber slides (LabTek). Cells were incubated with SA11 virus at a multiplicity of infection of 1 PFU per cell. Viral infection of cells was carried out essentially as described previously (23). Mock infections were carried out by omitting the virus and replacing it (by volume) with the culture medium to serve as an appropriate control. RNA extraction from mock and infected cells was performed as described previously (22).
VP4-expressing plasmid construction and transfection of cells
The VP4 open reading frame (ORF) coding region and the ORF lacking the C-terminal CRL tripeptide coding sequence were amplified by using appropriate primers and the PCR products were cloned into a mammalian expression plasmid, pEGFP-C2 (enhanced green fluorescent protein [GFP] expression vector; Clontech Labs, Palo Alto, Calif.), as GFP C-terminal fusion proteins. Briefly, 5 μl of total RNA was subjected to reverse transcription-PCR amplification as previously described (22), using a pair of SA11 VP4-specific primers. Primers used to generate the VP4 complete ORF are as follows. The forward primer (no. 7935) 5"-CGC TCA AGC TTC ATG GCT TCA CTC ATT TAT AGA-3" contains a HindIII restriction endonuclease site (underlined sequence) which anneals at nucleotide positions 10 to 30 on the VP4 gene sequence. The reverse primer (no. 4284) 5"-CGC GGA TCC TCA CAA CCT GCA TTG CAT-3" contains a BamHI site (underlined sequence) and the complementary sequence for coding CRL tripeptide (sequence shown in italics) and anneals at nucleotide positions 2340 to 2323 of the VP4 gene. Primers were synthesized based on the published SA11 VP4 gene sequence (21), in which a single nucleotide change was incorporated in the 5" (no. 7935) primer that does not alter the amino acid codon so that it is useful for other strains as well. The VP4 ORF lacking the C-terminal tripeptide (CRL) sequence was produced by using the 5" primer as described above (no. 7935) and a 3" primer (5"-CGC GGA TCC TCA↓ TTG CAT AAT CAG TTG-3") with a deletion of the coding sequence of the CRL tripeptide (position of the deletion indicated by arrowhead) and a BamHI restriction site (underlined) which anneals to nucleotide positions 2328 to 2114. The HindIII and BamHI restriction sites present in the 5" and 3" primers, respectively, were used for cloning into the plasmid. The plasmids were designated pEGFP-VP4CRL and pEGFP-VP4ΔCRL, respectively, to indicate the nature of the VP4 C-terminal coding sequence. Two independent plasmid DNAs for each clone were subjected to nucleotide sequencing as described previously (22). The authenticity of the sequence of cloned ATCC SA11 VP4 genes was confirmed by comparing the sequences with the published SA11 VP4 gene sequence and the sequence was identified to have >99% identity with the published sequence (21). This analysis also confirmed that pEGFP-VP4CRL has the terminal CRL coding sequence in the VP4 gene and pEGFP-VP4ΔCRL lacks the CRL coding sequence as expected of the 3" primers used in the PCR. When COS-7L cells (Gibco BRL) grown in chamber slides reached approximately 80% confluence, they were transfected with purified plasmid DNAs for transient protein expression using Lipofectamine Plus reagent per the manufacturer's instructions (Gibco BRL).
Indirect immunofluorescence (IF) and laser scanning confocal microscopy
Following 24 h of infection or 18 h of transfection, cells were permeabilized using digitonin (20 μg/ml) or 0.5% Triton X-100 as described previously (1, 14). The virus-infected cells were incubated for 90 min with mouse monoclonal antibody (MAb) (1:100 dilution) specific for the VP5* subunit (MAb HS2) of VP4 and either rabbit polyclonal antibody (1:100 dilution) specific for peroxisomal catalase (BioMol, Plymouth Meeting, Pa.) or SKL peptide specific for peroxisomal matrix proteins (Zymed Labs, South San Francisco, Calif.). Following primary antibody incubation, cells were washed with three changes of phosphate-buffered saline at 5-min intervals. Anti-mouse secondary antibody conjugated with Texas red (Vector Labs) and anti-rabbit antibody conjugated with fluorescein isothiocyanate (Vector Labs) at a dilution of 1:200 were used in the secondary incubation for 90 min followed by washes with phosphate-buffered saline as described above. Visualization, analysis, and photography were all performed using a Carl Zeiss laser scanning confocal microscope (model LSM5 PASCAL) equipped with a microprocessor. Images were transferred to the PC version of Adobe Photoshop 5.0 for labeling and printing.
Presence of a putative PTS1 sequence in rotavirus VP4
We identified a tripeptide sequence (CRL) resembling the PTS1 sequence at the C terminus of VP4 of rotavirus strain SA11. The CRL tripeptide belongs to the SKL family, which represents the canonical PTS1. Once it was identified in a rotavirus, we further screened all available 153 rotavirus VP4 deduced amino acid sequences to see whether all these sequences contain a PTS1 representative tripeptide sequence at the C terminus of the protein. To our surprise, all 153 VP4 sequences analyzed have a C-terminal tripeptide sequence representative of the PTS1 sequence (either SKL, CKL, GKL, CRL, or CRI) (Fig. 1). Sixty-two percent of the 153 sequences encode the C-terminal CRL tripeptide (Table 1), while the rest of the sequences fall into the SKL, CKL, GKL, and CRI categories, all of which are well known to represent functional variants of PTS1 sequences (7, 15). Such functionally conserved sequence variability of the PTS1 tripeptide in the VP4 gene sequence suggests that the terminal sequences are not mere sequencing artifacts. On the other hand, there are several VP4 published sequences that we utilized in our analysis in which the PCR primers are known to be limited only to the noncoding region (GenBank accession numbers L26888, V62152, D13401, U35851, and M33516), strongly suggestive of the fact that VP4 ORF sequences at the 3" end are not biased by the 3" PCR primer used but are truly representative of the VP4 RNA sequence. Thus, our analysis constitutes the first report of a PTS1 sequence in a viral protein. Retention of such a signal sequence in the rotavirus protein suggests that rotavirus VP4 is capable of being targeted to peroxisomes and its transport to the organelle may have implications in the cell biology of the virus.
FIG. 1.
Conserved PTS1 in rotavirus VP4. C-terminal 30-amino-acid sequences of rotavirus VP4 from representative strains infecting different hosts. Since most of the animal strains of rotavirus VP4 have 776 amino acids, whereas the human counterpart has only 775 amino acids, the C-terminal amino acid number is represented by 775/776. All the 153 rotavirus VP4 sequences in GenBank had one of the five variants of consensus PTS1 (SKL, CKL, GKL, CRL, and CRI). The accession numbers for the sequences shown are as follows: human (CKL, strain 97"B53), AAG15335; human (SKL, strain 95-87), BAA77555; human (GKL, strain 88-49), BAB32843; bovine (VMRI), AAB18952; porcine (P343), AAB41912; simian (SA11), BAA03850; murine (EC), AAA50484; feline (Cat2), BAA02667; lapine (R2), AAB65815; ovine (sheep rotavirus A), A48480; canine (K9), BAA03545; avian (PO-13), BAA24149.
TABLE 1.
Various types of PTS1 tripeptide sequences identified in VP4 deduced amino acid sequence from different rotavirus strains
| VP4 C terminal PTS1 sequence | No. of sequencesa | Origin of rotavirus strains |
|---|---|---|
| SKL | 1 | Human |
| GKL | 1 | Human |
| CKL | 38 | Human |
| CRLb | 29 | Human |
| 65 | Animal (bovine, simian, porcine, murine, feline, lapine, ovine, canine) | |
| CRI | 19 | Human |
| Bovine | ||
| Equine | ||
| Avian |
The total number of sequences was 153.
The CRL sequences represent 62% of the total.
Demonstration of rotavirus VP4 localization to peroxisomes in the virus-infected cells
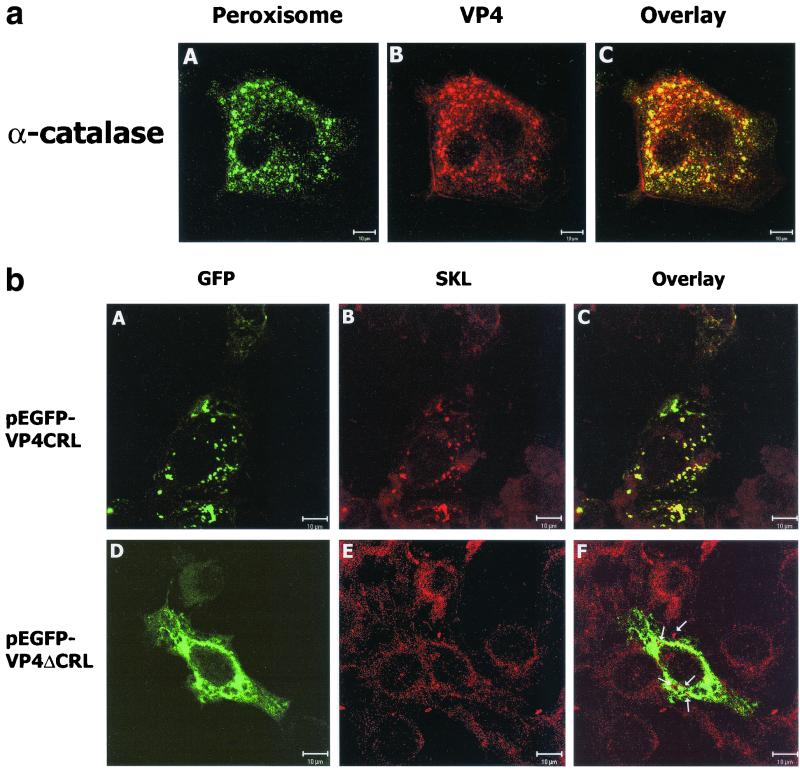
As the C-terminal CRL sequence represents 62% of the rotavirus VP4 sequences available to date, using a tissue culture-adapted CRL-representative rotavirus (strain SA11) and a permissive simian cell line, MA104, cellular localization of VP4 was evaluated. Analysis of virus-infected cells was performed by double IF with anti-VP4 antibodies specific for the VP5* subunit (MAb HS2) and either anticatalase antibodies or anti-SKL antibodies. Our results revealed that HS2 antibodies identified VP4 in small cytoplasmic vesicles and fibrils in addition to the plasma membrane, as is evident in Fig 2a, panel B. The fibrils presumably represent the microtubules, based on a previous study (23). Both anticatalase and anti-SKL (data not shown) antibodies identified the cytoplasmic small vesicles as peroxisomes, as illustrated in Fig. 2a, panel A. Overlay of the double-IF-stained cells identified respective antigens in the majority of the peroxisomes, demonstrating the VP4 colocalization with catalase shown in Fig. 2a, panel C. Besides colocalization of VP4 with the catalase signal in peroxisomes, there are some relatively smaller peroxisomes in which single localization of both VP4 and catalase were observed (Fig. 2a, panel C). This could be due to the fact that at any given time in a given infected cell all the peroxisomes are not actively importing simultaneously both VP4 and catalase from the cytoplasm. Or VP4 and catalase proteins both have different turnover times in the peroxisome, and hence at any given time some of the peroxisomes may contain either VP4 or catalase only. Overall, these results unequivocally demonstrated that VP4 is targeted to the organelle in addition to the other cellular sites.
FIG. 2.
(a) Confocal microscopic images of SA11 rotavirus-infected cells following double-immunofluorescence staining. Peroxisomes were detected by a rabbit polyclonal anticatalase antibody in combination with a secondary fluorescein isothiocyanate-labeled anti-rabbit antibody (A; green). VP4 was detected by primary anti-VP4 mouse MAb (HS2, specific to the VP5* subunit) and a secondary Texas red-conjugated anti-mouse antibody combination (B; red). VP4 antibodies stained small vesicles and fibrils in the cytoplasm and the cell membrane. Peroxisomal localization of rotavirus VP4 is observed by colocalization with a peroxisomal marker, catalase, which was evident by the yellow-orange color in the overlay panel (C). Bar, 10 μm. (b) Cellular distribution of VP4 in pEGFP-VP4CRL and pEGFP-VP4ΔCRL-transfected COS-7L cells. Discrete green fluorescent vesicular structures of GFPVP4CRL are illustrated in panel A, and diffused cytoplasmic distribution of green fluorescence by GFPVP4ΔCRL is shown in panel D. Peroxisomes in the respective cells are identified by anti-SKL antibodies in panels B and E. An overlay illustrating colocalization of GFPVP4CRL with peroxisomal SKL proteins is shown in panel C, whereas in panel F peroxisomes are clearly detected above the green fluorescence and there is no colocalization of GFPVP4ΔCRL with SKL proteins (indicated by arrows). Bar, 10 μm.
In a recent study, it was suggested that in bovine rotavirus strain RF-infected MA104 cells, anti-VP4 antibodies also stained small “annular spots” in addition to the plasma membrane and more so with microtubules, even though the nature of these small vesicles was not further addressed (23). We also initially observed comparable signals of VP4 localization with microtubules similar to the previous study. However, when we optimized the channel intensity to obtain better overall cellular morphology, VP4 signals associated with the fibrils were relatively less intense compared to those of peroxisomes. In addition, different methods used for fixing cells for IF confocal microscopy and the source of VP4 antibodies also might have contributed to minor discrepancies between our observations and previously published data (23). However, the important advancement over the previous study is that previously noted VP4 association with cytoplasmic small vesicles of unknown nature has now been identified to be with peroxisomes in this study by using organelle-specific IF. Thus, these two studies are in fact complementary and provide unequivocal evidence that VP4 is associated with peroxisomes.
Subcellular distribution of GFP-VP4CRL and -VP4ΔCRL protein chimeras
Utilization of a mutant rotavirus lacking a functional VP4 PTS1 or creating a similar virus variant by incorporating mutations at the PTS1 site of the VP4 gene is ideal to test the functional role of protein PTS1-dependent targeting of VP4 directly to the organelle. However, neither natural variants of rotavirus with VP4 PTS1 mutations nor a reverse genetics approach for this virus are available. Therefore, such studies are not possible in the rotavirus field. In addition, rotaviruses failed to infect a human fibroblast cell line (control) and its peroxisomal-deficient counterparts (kindly provided by Stephen Gould, Johns Hopkins University), as they all turned out to be nonpermissive for rotavirus infection (unpublished data from our laboratory). Since cellular integrins α2β1, α4β1, and α1β2 are involved in rotavirus cell entry (17), we speculate that deficiency or low expression levels of these specific integrins on the surface of these cell lines might contribute to their nonpermissiveness for rotavirus infection. This left us with no alternative but to study VP4 transport by transient expression methods as described below. Using GFP-fused VP4 expression plasmids with or without the CRL motif (pEGFP-VP4CRL and pEGFP-VP4ΔCRL), COS-7L cells were transfected. Cells expressing pEGFP-VP4CRL showed a punctate, cytoplasmic distribution similar to that of rotavirus-infected cells (Fig. 2b, panel A) which colocalized with the peroxisomal marker identified by anti-SKL antibody (Fig. 2b, panel C), whereas pEGFP-VP4ΔCRL-expressing cells exhibited a diffused cytoplasmic fluorescence pattern of VP4 (Fig. 2b, panel D) which did not colocalize with the peroxisomal marker (Fig. 2b, panel F). This clearly demonstrated that rotavirus VP4 alone (in the absence of the viral infection) with its C-terminal PTS1 can translocate to peroxisomes, and if the PTS1 sequence is removed, such property of VP4 is abrogated. In a previous study by another group (unrelated to peroxisomal targeting of VP4), when VP4 was fused at its C terminus to GFP and the protein chimera was expressed in the cell, it was seen diffused in the cytoplasm, although it contained PTS1 (23). This could be explained by the differences between the plasmid construction of VP4 as GFP fusion proteins in the present study and those reported previously (23). In the present study, GFP is fused to the N terminus of VP4, thus leaving a free C terminus tripeptide PTS1 sequence. This free C-terminal PTS1 is required for its function. In contrast, in the previous study, the GFP was fused to the C terminus of VP4, and even though the VP4 contained PTS1, it was not functional as it was not available as a free C terminus (23).
Certain human metabolic disorders, such as Zellweger syndrome, neonatal adrenoleukodystrophy, infantile Refsum disease, and rhizomelic chondrodysplasia punctata, are associated with the peroxisomal defect in protein import and/or biogenesis (12). Since rotaviruses as a group have evolutionarily conserved a PTS1 sequence in VP4 as identified in this report, it remains to be evaluated whether the viral infection and targeting of VP4 to peroxisomes of the enterocytes of infants have any disease consequences, especially in children with inherited peroxisomal disorders. Recently, the human immunodeficiency virus (HIV) Nef protein devoid of a PTS motif was shown to translocate to peroxisomes by binding to a peroxisomal matrix protein containing a PTS1 motif, the human thioesterase II in the cytoplasm (5). However, the functional significance of the viral protein transport itself to the organelle in HIV disease remains unknown, though the Nef binding of thioesterase was shown to down regulate CD4 cells (5).
Conservation of a functional PTS1 in rotavirus VP4 and its targeting to the peroxisomes suggest that the virus has evolved a strategy to utilize the organelle in controlling or modulating the functions of this specialized organelle to its own advantage. Among the several known functions of the peroxisome which may be implicated in the rotavirus life cycle, of particular interest are its roles in lipid metabolism and posttranslational modifications of proteins. This could be substantiated by the facts that (i) lipids have been shown to play an important role in rotavirus infectivity (14, 26, 27) and (ii) some of the rotaviral proteins, such as VP2 and VP6, reportedly undergo fatty acid modifications which are essentially controlled by peroxisomes (4, 30). The fact that diverse viruses such as rotavirus and HIV evolved to transport viral proteins to the peroxisomes by unique mechanisms signifies the emerging importance of peroxisomes in viral pathogenesis and possibly in disease manifestation.
Acknowledgments
We acknowledge Harry Greenberg, Stanford University, for kindly providing a gift of VP4 MAbs (HS1 and HS2) and Kathryn Carbone, FDA, for use of the confocal microscopy facility. The generous gift of PBD cell lines, from which we learned that the cells are nonpermissive for rotavirus infection, from S. J. Gould is highly appreciated.
This work was supported by a Center Director's Targeted Research Award, CBER, FDA, to C.D.A. and by an Oak Ridge Institute for Science and Engineering fellowship to K.V.K.M. and I.S.
REFERENCES
- 1.Biardi, L., and S. K. Krisans. 1996. Compartmentalization of cholesterol biosynthesis. Conversion of mevalonate to farnesyl diphosphate occurs in the peroxisomes. J. Biol. Chem. 271:1784-1788. [DOI] [PubMed] [Google Scholar]
- 2.Bishop, R. F. 1998. Natural history of human rotavirus infection. Arch. Virol. 12(Suppl.):119-128. [DOI] [PubMed] [Google Scholar]
- 3.Both, G. W., A. R. Bellamy, and D. B. Mitchell. 1994. Rotavirus protein structure and function. Curr. Top. Microbiol. Immunol. 185:67-105. [DOI] [PubMed] [Google Scholar]
- 4.Clark, B., and U. Desselberger. 1988. Myristylation of rotavirus proteins. J. Gen. Virol. 69:2681-2686. [DOI] [PubMed] [Google Scholar]
- 5.Cohen, G. B., V. S. Rangan, B. K. Chen, S. Smith, and D. Baltimore. 2000. The human thioesterase II protein binds to a site on HIV-1 Nef critical for CD4 down-regulation. J. Biol. Chem. 275:23097-23105. [DOI] [PubMed] [Google Scholar]
- 6.de Hoop, M. J., and G. Ab. 1992. Import of proteins into peroxisomes and other microbodies. Biochem. J. 286:657-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Zoysa, P. A., and I. F. Connerton. 1994. The function and specificity of the C-terminal tripeptide glyoxysomal targeting signal in Neurospora crassa. Curr. Genet. 26:430-437. [DOI] [PubMed] [Google Scholar]
- 8.Ding, M., C. Clayton, and D. Soldati. 2000. Toxoplasma gondii catalase: are there peroxisomes in toxoplasma? J. Cell Sci. 113:2409-2419. [DOI] [PubMed] [Google Scholar]
- 9.Espejo, R. T., S. Lopez, and C. Arias. 1981. Structural polypeptides of simian rotavirus SA11 and the effect of trypsin. J. Virol. 37:156-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Estes, M. K., D. Y. Graham, and B. B. Mason. 1981. Proteolytic enhancement of rotavirus infectivity: molecular mechanisms. J. Virol. 39:879-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flatmark, T., A. Nilsson, J. Kvannes, T. S. Eikhom, M. H. Fukami, H. Kryvi, and E. N. Christiansen. 1988. On the mechanism of induction of the enzyme systems for peroxisomal beta-oxidation of fatty acids in rat liver by diets rich in partially hydrogenated fish oil. Biochim. Biophys. Acta 962:122-130. [DOI] [PubMed] [Google Scholar]
- 12.Gould, S. J., and D. Valle. 2000. Peroxisome biogenesis disorders: genetics and cell biology. Trends Genet. 16:340-345. [DOI] [PubMed] [Google Scholar]
- 13.Gould, S. J., G. A. Keller, N. Hosken, J. Wilkinson, and S. Subramani. 1989. A conserved tripeptide sorts proteins to peroxisomes. J. Cell Biol. 108:1657-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guerrero, C. A., S. Zarate, G. Corkidi, S. Lopez, and C. F. Arias. 2000. Biochemical characterization of rotavirus receptors in MA104 cells. J. Virol. 74:9362-9371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunt, M. C., S. E. Nousiainen, M. K. Huttunen, K. E. Orii, L. T. Svensson, and S. E. Alexson. 1999. Peroxisome proliferator-induced long chain acyl-CoA thioesterases comprise a highly conserved novel multigene family involved in lipid metabolism. J. Biol. Chem. 274:34317-34326. [DOI] [PubMed] [Google Scholar]
- 16.Lametschwandtner, G., C. Brocard, M. Fransen, P. van Veldhoven, J. Berger, and A. Hartig. 1998. The difference in recognition of terminal tripeptides as peroxisomal targeting signal 1 between yeast and human is due to different affinities of their receptor Pex5p to the cognate signal and to residues adjacent to it. J. Biol. Chem. 273:33635-33643. [DOI] [PubMed] [Google Scholar]
- 17.Londrigan, S. L., M. J. Hewish, M. J. Thomson, G. M. Sanders, H. Mustafa, and B. S. Coulson. 2000. Growth of rotaviruses in continuous human and monkey cell lines that vary in their expression of integrins. J. Gen. Virol. 81:2203-2213. [DOI] [PubMed] [Google Scholar]
- 18.Lopez-Huertas, E., W. L. Charlton, B. Johnson, I. A. Graham, and A. Baker. 2000. Stress induces peroxisome biogenesis genes. EMBO J. 19:6770-6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masters, C. 1997. Gluconeogenesis and the peroxisome. Mol. Cell. Biochem. 166:159-168. [DOI] [PubMed] [Google Scholar]
- 20.McCammon, M. T., J. A. McNew, P. J. Willy, and J. M. Goodman. 1994. An internal region of the peroxisomal membrane protein PMP47 is essential for sorting to peroxisomes. J. Cell Biol. 124:915-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitchell, D. B., and G. W. Both. 1989. Complete nucleotide sequence of the simian rotavirus SA11 VP4 gene. Nucleic Acids Res. 17:2122. [DOI] [PMC free article] [PubMed]
- 22.Mohan, K. V., and C. D. Atreya. 2000. Comparative sequence analysis identified mutations outside the NSP4 cytotoxic domain of tissue culture-adapted ATCC-Wa strain of human rotavirus and a novel interspecies variable domain in its C terminus. Arch. Virol. 145:1789-1799. [DOI] [PubMed] [Google Scholar]
- 23.Nejmeddine, M., G. Trugnan, C. Sapin, E. Kohli, L. Svensson, S. Lopez, and J. Cohen. 2000. Rotavirus spike protein VP4 is present at the plasma membrane and is associated with microtubules in infected cells. J. Virol. 74:3313-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaw, A. L., R. Rothnagel, D. Chen, R. F. Ramig, W. Chiu, and B. V. Prasad. 1993. Three-dimensional visualization of the rotavirus hemagglutinin structure. Cell 74:693-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sommer, J. M., Q. L. Cheng, G. A. Keller, and C. C. Wang. 1992. In vivo import of firefly luciferase into the glycosomes of Trypanosoma brucei and mutational analysis of the C-terminal targeting signal. Mol. Biol. Cell 3:749-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Superti, F., L. Seganti, M. Marchetti, M. L. Marziano, and N. Orsi. 1992. SA-11 rotavirus binding to human serum lipoproteins. Med. Microbiol. Immunol. 181:77-86. [DOI] [PubMed] [Google Scholar]
- 27.Superti, F., M. L. Marziano, G. Donelli, M. Marchetti, and L. Seganti. 1995. Enhancement of rotavirus infectivity by saturated fatty acids. Comp. Immunol. Microbiol. Infect. Dis. 18:129-135. [DOI] [PubMed] [Google Scholar]
- 28.Swinkels, B. W., S. J. Gould, A. G. Bodnar, R. A. Rachubinski, and S. Subramani. 1991. A novel, cleavable peroxisomal targeting signal at the amino-terminus of the rat 3-ketoacyl-CoA thiolase. EMBO J. 11:3255-3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van den Bosch, H., R. B. Schutgens, R. J. Wanders, and J. M. Tager. 1992. Biochemistry of peroxisomes. Annu. Rev. Biochem. 61:157-197. [DOI] [PubMed] [Google Scholar]
- 30.Yalovsky, S., M. Rodriguez-Concepcion, and W. Gruissem. 1999. Lipid modifications of proteins--slipping in and out of membranes. Trends Plant Sci. 4:439-445. [DOI] [PubMed] [Google Scholar]