Abstract
Here, we describe a new HLA-B*3501-restricted cytotoxic T lymphocyte (CTL) epitope in the influenza A virus (H3N2) nucleoprotein, which was found to exhibit a high degree of variation at nonanchor residues. The influenza virus variants emerged in chronological order, and CTLs directed against old variants failed to recognize more recent strains of influenza A virus, indicating an escape from CTL immunity.
CD8+ cytotoxic T lymphocytes (CTLs) contribute to the control of viral infections by recognizing antigenic peptides of viral proteins presented by major histocompatibility complex (MHC) class I molecules on infected cells. The specific recognition of these MHC-peptide complexes by CTLs may lead to the elimination of virus-infected cells. One of the mechanisms exploited by viruses to evade recognition by CTLs (19, 23, 28) involves antigenic variation in CTL epitopes or mutations in sequences flanking these epitopes. Antigenic variation in CTL epitopes, resulting in the evasion of immune surveillance by specific CTLs, has been described for several viruses that cause chronic infections, including Epstein-Barr virus (5, 8, 9, 12), human immunodeficiency virus (4, 7, 13, 14, 22, 26), hepatitis B virus (1, 2), and hepatitis C virus (6, 35).
Recently, mutations were also found at the anchor residue of an HLA-B*2705-restricted epitope of the influenza A virus nucleoprotein (NP), which consisted of amino acids 383 to 391 (NP383-391). Both the R384G and the R384K mutations abolished class I-restricted presentation and allowed for escape from CTL recognition (34). Thus, these viruses, which cause acute infections in a significant portion of the human population annually, can escape from immune surveillance by CTLs in addition to escaping from neutralizing antibodies (antigenic drift and antigenic shift).
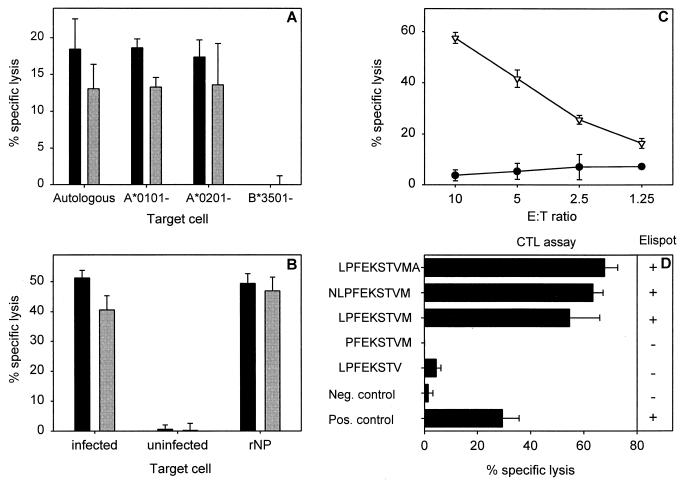
For the identification of new CTL epitopes, CD3+- and CD8+-T-cell clones were generated by limiting dilution (34) from the peripheral blood mononuclear cells (PBMC) of an HLA-A*0101-, HLA-A*0201-, HLA-B*0801-, and HLA-B*3501-positive donor after in vitro stimulation with influenza virus Resvir-9, a reassortant vaccine strain of A/Nanchang/933/95 (3). For the initial specificity testing of the T-cell clones, an enzyme-linked immunospot assay was used, as described previously (3), with cells from the infected autologous B-lymphoblastoid cell line (BLCL) as stimulator cells. Clone 1980-1 was found to recognize a yet unidentified epitope in an HLA-B*3501- and HLA-B*3503-restricted fashion, as demonstrated with cells from the HLA-matching and -nonmatching infected BLCL as target cells (Fig. 1A; also data not shown). This clone was specific for the NP of influenza A virus (H3N2), since it lysed target cells incubated with recombinant NP (rNP) (Fig. 1B), as described previously (3, 33). By using a CTL epitope prediction program (25) (http://www.umds.ac.uk/tissue), putative HLA-B*3501-restricted epitopes in the NP of influenza A virus (H3N2) were predicted. The 9-mer with the highest ranking, LPFEKSTVM (NP418-426/1980), was synthesized and was found to be recognized by the T-cell clone 1980-1 in a CTL assay (Fig. 1C). Removal of the C-terminal methionine abolished recognition by clone 1980-1 (Fig. 1D). A similar result was observed when the lysine at the N terminus of the 9-mer epitope LPFEKSTVM was removed. Addition of one amino acid at the N or C terminus of the epitope did not significantly improve recognition of target cells, indicating that NP418-426 was the minimal epitope.
FIG. 1.
Characterization of an HLA-B*3501-restricted epitope and CTL clone (1980-1). (A) HLA class I restriction of CTL clone 1980-1 was determined by partially mismatched target cells (BLCL) infected with influenza A (H3N2) virus (Resvir-9) in a 51Cr release assay at effector-to-target cell (E:T) ratios of 10:1 (black bars) and 5:1 (grey bars) as described previously (3). Autologous BLCL cells (HLA-A*0101, HLA-A*0201, HLA-B*0801, and HLA-B*3501) and BLCL cells mismatched for a single HLA class I molecule (HLA-A*0101−, HLA-A*0201−, and HLA-B*3501−) were used. All BLCL cells expressed HLA-B*0801. The percent lysis of uninfected target cells was subtracted from the percent lysis of target cells infected with influenza virus. (B) Protein specificity of an HLA-B*3501-restricted CTL clone. HLA-B*3501+ target cells were infected with influenza A (H3N2) virus (Resvir-9) or were left uninfected and used as positive and negative controls, respectively. In addition, HLA-B*3501+ target cells were incubated overnight with 100 μg of bacterially expressed rNP of A/Netherlands/18/94 (A/H3N2)/ml as previously described (33). The target cells were used in a 51Cr release assay with CTL clone 1980-1 at E:T ratios of 10:1 (black bars) and 5:1 (grey bars). (C) A 9-mer peptide (NP418-426; LPFEKSTVM) (▿), predicted with an HLA binding prediction program (25), was loaded onto HLA-B*3501+ BLCL cells and used as target cells in a 51Cr release assay with the T-cell clone 1980-1 at the indicated E:T ratios. Untreated BLCL cells (•) were included as a negative control. (D) Minimal epitope mapping of HLA-B*3501-restricted epitope. Peptides were synthesized based on the initial 9-amino-acid sequence LPFEKSTVM that was extended or truncated at the C- or N-terminal end of the NP418-426 epitope in order to determine the minimal epitope. BLCL cells loaded with 5 μM concentrations of the different peptides were used as target cells in a CTL assay at an E:T ratio of 5:1 or applied to stimulate the CTL clone (1980-1) in an enzyme-linked immunospot assay. A plus indicates gamma interferon (IFN-γ) production by the CTL clone, while a minus indicates no production of IFN-γ. Influenza virus (Resvir-9)-infected BLCL cells and uninfected BLCL cells were included in both assays as positive and negative controls, respectively. Percent lysis is given as the mean ± standard deviation, and the results are representative of multiple experiments.
Experiments with rNPs from different virus strains indicated that clone 1980-1 did not react with the rNP obtained from A/Hong Kong/2/68 (H3N2) (data not shown). Therefore, known NP418-426 sequences of influenza A viruses obtained from the influenza sequence database (http://www.flu.lanl.gov) were compared. It was found that the amino acid sequence in the epitope varied at four positions and that these variants emerged in chronological order. All variants and their designations are shown in Table 1. Although the strains used for NP sequence comparison were not controlled for passage history of the strains (eggs grown or passaged in mammalian cell lines), it is unlikely that this has biased the analysis, since no immediate selective pressure on the NP was observed by adaptation in eggs, as was the case for the hemagglutinin (29).
TABLE 1.
Variation in HLA-B35-restricted influenza A virus epitope NP418-426
| Sequencea | Yr(s) of isolation | Virus subtype | No. of virusesb | Epitope name |
|---|---|---|---|---|
| LPFDRPTIM | Before 1933 | H1N1 | 1 | |
| -----T--- | 1934 | H1N1 | 1 | |
| ----KT--- | 1940-1957 | H1N1/H2N2 | 6 | |
| ----K---- | 1957-1972 | H2N2/H3N2 | 7 | NP418-426/1957 |
| ----KS--- | 1972-1978 | H3N2 | 12 | NP418-426/1972 |
| ----KS-V- | 1977 | H3N2 | 1 | |
| ---EKS--- | 1983 | H3N2 | 1 | |
| ---EKS-V- | 1980-present | H3N2 | 35 | NP418-426/1980 |
A dash indicates identity with amino acid at same position in first sequence.
Number of viruses with the reported sequence in the influenza sequence database (http://www.flu.lanl.gov). Influenza A viruses (H1N1) were excluded from the analysis from 1977, the year that viruses of this subtype were reintroduced.
A T2 cell line expressing HLA-B*3501 (30) was used to assess the affinity of the NP418-426 epitopes for the HLA-B*3501 molecule as previously described (32). The concentration of peptide necessary to inhibit the signal (mean fluorescence intensity [MFI]) of the fluorescein isothiocyanate-labeled reference peptide LPSCFLADVEF (20) by 50% (IC50) was determined. The mean IC50 of the NP418-426/1980 epitope was 1.1 μM, while the mean IC50s of the NP418-426/1972 and NP418-426/1957 epitopes were 1.5 and 1.6 μM, respectively; this indicates that all three epitopes bound strongly and in the same order of magnitude to HLA-B*3501 (Table 2), which suggests that the three peptides represent CTL epitopes. For comparison, the IC50 of a previously described HLA-B35-restricted influenza A virus epitope, M1128-135, was determined and found to be higher than the highest concentration used (20 μM).
TABLE 2.
Binding affinity of NP418-426 peptide to HLA-B*3501
| Peptide | Sequence | IC50 (μM)a |
|---|---|---|
| NP418-426/1957 | LPFDKPTIM | 1.3-2.0 |
| NP418-426/1972 | LPFDKSTIM | 1.1-2.0 |
| NP418-426/1980 | LPFEKSTVM | 0.9-1.3 |
| M1128-135b | ASCMGLIY | >20 |
Capacity of binding to HLA-B*3501 is expressed as the concentration (micromolar) of peptide able to inhibit binding of the fluorescein isothiocyanate-labeled reference peptide by 50% (32) based on two assays.
HLA-B35-restricted influenza virus epitope from the matrix protein with a reported low binding affinity (10).
The frequency of peptide-specific CTL precursors was determined for the NP418-426/1980, NP418-426/1972, and NP418-426/1957 variants of the epitope and found to differ considerably among four HLA-B35+ donors (Table 3). The average frequency of NP418-426/1980-specific cells was 1 in 7,426, ranging from 1 in 4,073 to 1 in 15,485, which is relatively high compared to the frequencies of other CTL epitopes of influenza A virus (3), indicating that NP418-426/1980 is an immunodominant epitope. The mean frequency of NP418-426/1972-specific cells was much lower (1 in 16,226); however, one donor exhibited a high number of NP418-426/1972-specific cells. For the NP418-426/1957 epitope, low to undetectable numbers of specific cells were detected.
TABLE 3.
NP418-426 peptide-specific CTL frequencies in PBMC
| Donor no. | Frequency of peptide-specific CTL
|
||
|---|---|---|---|
| NP418-426/1980 | NP418-426/1972 | NP418-426/1957 | |
| 5017 | 1/1,885 | 1/17,921 | —a |
| 5972 | 1/4,073 | 1/5,054 | 1/86,550 |
| 2384 | 1/15,485 | 1/17,329 | 1/10,108 |
| 5991 | 1/7,542 | 1/24,600 | 1/24,600 |
| Mean | 1/7,246 | 1/16,226 | 1/40,419 |
No specific spots detected.
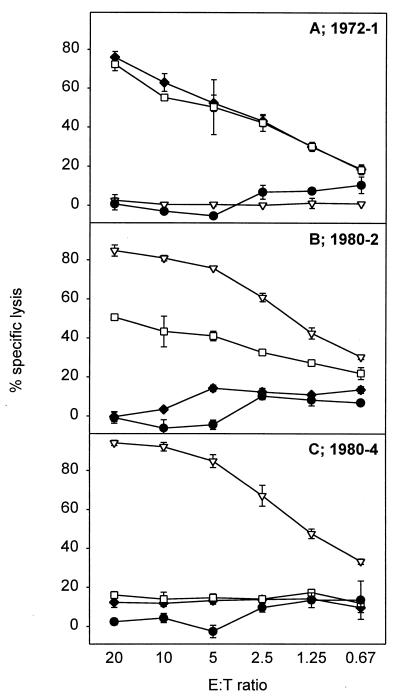
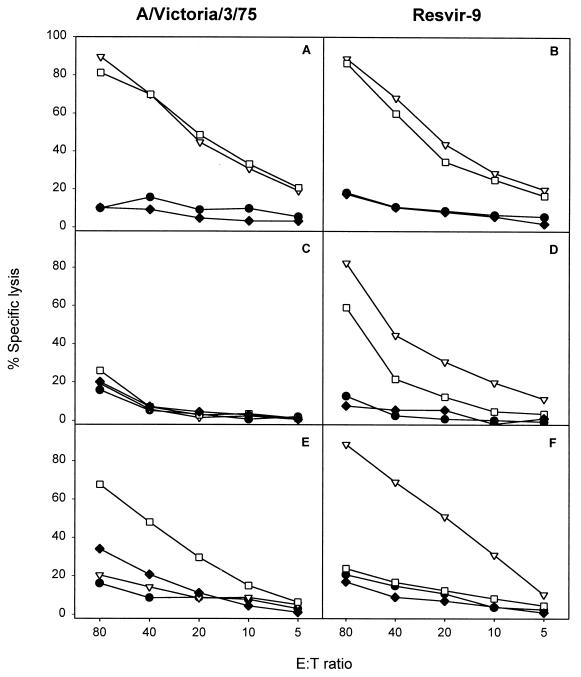
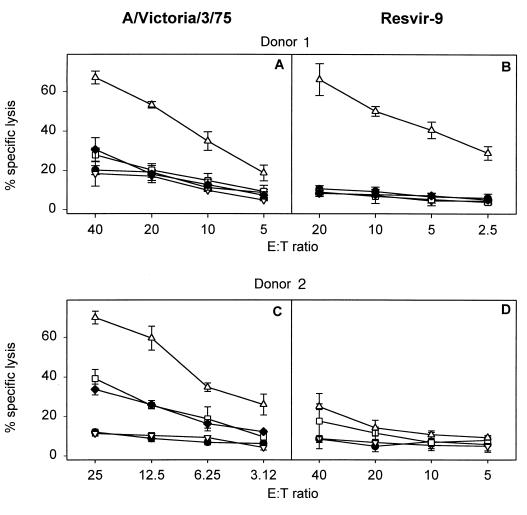
Based on the prevalence of the sequences (Table 1), we decided to focus on the NP418-426/1980, NP418-426/1972, and NP418-426/1957 epitopes. Since mutations in the NP418-426 epitope emerged in an evolutionary fashion, we speculated that CTL immunity directed against these epitopes was the basis for the selection and/or emergence of mutant viruses. To test this hypothesis, more T-cell clones were raised against historic (A/Victoria/3/75) and recent (Resvir-9) viruses by using PBMC from HLA-B35+ donors obtained in the year 2001. CTL clone 1972-1 directed against the NP418-426/1972 epitope failed to recognize the NP418-426/1980 epitope (Fig. 2A). Similarly, three out of five CTL clones (1980-1, 1980-4, and 1980-5) directed against the NP418-426/1980 epitope failed to react with the NP418-426/1957 and NP418-426/1972 epitopes (Fig. 2C; also data not shown). With the other two CTL clones (1980-2 and 1980-3), some cross-reactivity was observed with the epitopes in older influenza A virus strains (Fig. 2B; also data not shown). The absence of recognition of the NP418-426/1980 epitope by CTLs directed against older variants was also demonstrated with PBMC obtained from two donors stimulated in vitro with influenza virus A/Victoria/3/75 (Fig. 3E; also data not shown). These PBMC did, however, recognize the homologous NP418-426/1972 epitope. PBMC of some donors displayed cross-reactivity with both variants after stimulation with A/Victoria/3/75 or Resvir-9 (Fig. 3). This cross-reactivity can be explained by the expansion of cross-reactive CTLs in PBMC, a phenomenon previously described in C57BL/10 mice infected with different strains of influenza A virus (15). Finally, one donor (donor 5017) did not respond at all after stimulation with A/Victoria/3/75 (Fig. 3C).
FIG. 2.
NP418-426 variant epitope specificity of CTL clones. Target cells (BLCL) were incubated overnight with 5 μM concentrations of the HLA-B*3501-restricted NP418-426 epitope variants, namely, NP418-426/1980 (▿), NP418-426/1972 (□), and NP418-426/1957 (⧫), or were left untreated (•) and used as a negative control. Clones 1972-1 (A), 1980-2 (B), and 1980-4 (C), obtained from different donors, were added at different effector-to-target cell ratios, and specific lysis was calculated. The results, given as the percent specific lysis (mean ± standard deviation), are representative of multiple assays.
FIG. 3.
Epitope specificity of the polyclonal response following in vitro stimulation of PBMC cryopreserved in the year 2001 with influenza A viruses (H3N2) containing different variants of the epitope. PBMC of HLA-B35+ donors, aged between 30 and 50 years, were stimulated in vitro with A/Victoria/3/75 (A, C, and E), containing the NP418-426/1972 variant, or Resvir-9 (B, D, and F), containing the NP418-426/1980 epitope, as described previously (3). After 8 days, the effector cells were tested for lytic activity against target cells incubated with the peptides corresponding to NP418-426/1980 (▿), NP418-426/1972 (□), and NP418-426/1957 (⧫). Untreated target cells (•) were included in each assay as a negative control. Mean percentages of specific lyses of two independently repeated experiments are shown for donors 5972 (A and B), 5017 (C and D), and 5991 (E and F).
To exclude the influence of consecutive natural infections with various influenza A viruses on the NP418-426-specific CTL response, we obtained HLA-A2+ HLA-B35+ PBMC cryopreserved between the years 1982 and 1984. Since this occurred shortly after the introduction of the NP418-426/1980 variant epitope, the chance of an infection with an NP418-426/1980 variant virus in these donors was relatively small. The NP418-426/1980 epitope was not recognized by the PBMC of these donors (donors 1 and 2) (Fig. 4) after stimulation with both a recent strain of influenza virus (Resvir-9) and A/Victoria/3/75, which contained the NP418-426/1972 epitope. The lack of NP418-426/1980-specific activity was not caused by the absence of virus-specific CTL activity, since the NP418-426/1972 variant epitope and the conserved HLA-A*0201-restricted M158-66 epitope were recognized.
FIG. 4.
CTL responsiveness after stimulation of PBMC cryopreserved between 1982 and 1984 with influenza A viruses (H3N2) containing the NP418-426/1980 or NP418-426/1972 variant epitope. PBMC from two HLA-A2+ HLA-B35+ donors (donors 1 and 2) were stimulated in vitro with A/Victoria/3/75 (A and C), containing the NP418-426/1972 variant epitope, or Resvir-9 (B and D), containing the NP418-426/1980 variant epitope. The lytic activities of the effector cells were determined after 8 days of culture against target cells incubated with 5 μM concentrations of peptides corresponding to NP418-426/1980 (▿), NP418-426/1972 (□), NP418-426/1957 (⧫), or M158-66 epitope (▵), and a negative control (•) at different effector-to-target cell ratios. NP418-426/1957 was not determined for PBMC of donor 2 after stimulation with Resvir-9. The mean percentage of specific lysis ± standard deviation is given for one experiment.
Based on these findings, we argue that in addition to the variation in the HLA-B8- and HLA-B*2705-restricted epitopes, NP380-388 and NP383-391, respectively, the variation in this newly identified HLA-B*3501-restricted NP418-426 epitope is driven by CTL immunity, leading to escape from recognition by these CTLs. The variations in the NP380-388 and NP383-391 epitopes were found at the anchor residues (R384G mutation) of the respective 9-mers (34). In the NP418-426 epitope, the anchor residues for binding to HLA-B35 were perfectly conserved in all virus strains isolated and sequenced since 1933. It can be speculated that variation at these residues is restricted by functional constraints. For example, an R267A mutation within an HLA-A3-restricted epitope has been shown to affect RNA binding by NP (11). Since the NP of A/Hong Kong/2/68, containing the NP418-426/1957 epitope, and the NP of A/Netherlands/18/94, containing the NP418-426/1980 epitope, function equally well (34) and both viruses grow to comparable titers, it is unlikely that the mutations in the NP418-426 epitope were selected based on the improved fitness of these viruses.
HLA-B35-positive individuals constitute a significant portion of the human population, ranging from 5% in Orientals to 10% in Caucasians (21). The immune pressure mediated by CTLs in these individuals recognizing the NP418-426 epitope may have contributed to the emergence of escape mutant viruses from the quasi species of influenza viruses and their continued circulation. As a result of the immunodominant nature of the epitope, the CTL response might have been oligoclonal in HLA-B35+ individuals, allowing for the selection of mutant viruses. Alternatively, other unknown epitopes may overlap with NP418-426, further contributing to the selection pressure. The variants are maintained in HLA-B35-negative individuals because the mutations in the NP418-426 epitope did not reduce the fitness of these viruses. It is of interest that the infection of mice transgenic for a single T-cell receptor (TCR) specific for the H-2Db-restricted NP366-374 epitope of influenza A virus resulted in the emergence of viruses containing amino acid mutations in this epitope, which impaired the presentation of viral peptides by MHC class I molecules or interfered with T-cell receptor recognition (27). Other mutations at nonanchor residues have been described only for sporadic virus isolates, and their effect on T-cell recognition was not studied (24). A mutation in a CTL epitope (NS1122-130) in the (not naturally occurring) high-yield vaccine strain of influenza virus A/Texas/36/91 (31) inhibited recognition by a CTL clone. Other mechanisms based on mutations in CTL epitopes contributing to reduction or elimination of CTL recognition have been described, like “original antigenic sin” (18), antagonism (16, 17), and anergy. Although original antigenic sin and anergy were not investigated in great detail, in one of the PBMC samples, obtained in 1982, a response was found that was specific for the NP418-426/1972 epitope but not for the homologous NP418-426/1980 epitope used for stimulation.
In conclusion, a new variable CTL epitope was identified in the NPs of influenza A viruses. Epidemiological and immunological evidence indicates that the variation found in this epitope was the result of antigenic drift resulting from immune pressure mediated by specific CTLs. Thus, in addition to the introduction of mutations in the surface glycoproteins, which allows for escape from antibody-mediated immunity, further evidence which shows that influenza viruses can escape from CTL-mediated immunity is accumulating. This would make these viruses masters in disguise, partially explaining their relative success in infecting a large portion of the human population every year for the past several decades.
Acknowledgments
We acknowledge Liane van de Kemp for growing, purifying, and sequencing the influenza A virus samples. Furthermore, we thank Wilfried Levering for performing the HLA typing, Saskia Sakko for organizing the blood collection from the blood donors (Bloodbank Rotterdam), and Ger van der Water for continuous support. We also thank P. van der Bruggen, for FITC-labeled reference peptide, D. Roelen, F. Claas and M. Takiguchi for PBMC cryopreserved in the 1980s, the T2 cell line expressing HLA-B*3501.
Part of this work was supported by the Foundation for Respiratory Virus Infections, Notably Influenza (SRVI), and Numico Research BV. R. A. M. Fouchier is a fellow of the Royal Dutch Academy of Arts and Sciences.
REFERENCES
- 1.Bertoletti, A., A. Costanzo, F. V. Chisari, M. Levrero, M. Artini, A. Sette, A. Penna, T. Giuberti, F. Fiaccadori, and C. Ferrari. 1994. Cytotoxic T lymphocyte response to a wild type hepatitis B virus epitope in patients chronically infected by variant viruses carrying substitutions within the epitope. J. Exp. Med. 180:933-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bertoletti, A., A. Sette, F. V. Chisari, A. Penna, M. Levrero, M. De Carli, F. Fiaccadori, and C. Ferrari. 1994. Natural variants of cytotoxic epitopes are T-cell receptor antagonists for antiviral cytotoxic T cells. Nature 369:407-410. [DOI] [PubMed] [Google Scholar]
- 3.Boon, A. C. M., G. de Mutsert, Y. M. F. Graus, R. A. M. Fouchier, K. Sintnicolaas, A. D. M. E. Osterhaus, and G. F. Rimmelzwaan. 2002. The magnitude and specificity of influenza A virus-specific cytotoxic T-lymphocyte responses in humans is related to HLA-A and -B phenotype. J. Virol. 76:582-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borrow, P., H. Lewicki, X. Wei, M. S. Horwitz, N. Peffer, H. Meyers, J. A. Nelson, J. E. Gairin, B. H. Hahn, M. B. Oldstone, and G. M. Shaw. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3:205-211. [DOI] [PubMed] [Google Scholar]
- 5.Burrows, J. M., S. R. Burrows, L. M. Poulsen, T. B. Sculley, D. J. Moss, and R. Khanna. 1996. Unusually high frequency of Epstein-Barr virus genetic variants in Papua New Guinea that can escape cytotoxic T-cell recognition: implications for virus evolution. J. Virol. 70:2490-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, K. M., B. Rehermann, J. G. McHutchison, C. Pasquinelli, S. Southwood, A. Sette, and F. V. Chisari. 1997. Immunological significance of cytotoxic T lymphocyte epitope variants in patients chronically infected by the hepatitis C virus. J. Clin. Investig. 100:2376-2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Couillin, I., B. Culmann-Penciolelli, E. Gomard, J. Choppin, J. P. Levy, J. G. Guillet, and S. Saragosti. 1994. Impaired cytotoxic T lymphocyte recognition due to genetic variations in the main immunogenic region of the human immunodeficiency virus 1 NEF protein. J. Exp. Med. 180:1129-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Campos-Lima, P. O., R. Gavioli, Q. J. Zhang, L. E. Wallace, R. Dolcetti, M. Rowe, A. B. Rickinson, and M. G. Masucci. 1993. HLA-A11 epitope loss isolates of Epstein-Barr virus from a highly A11+ population. Science 260:98-100. [DOI] [PubMed] [Google Scholar]
- 9.de Campos-Lima, P. O., V. Levitsky, J. Brooks, S. P. Lee, L. F. Hu, A. B. Rickinson, and M. G. Masucci. 1994. T cell responses and virus evolution: loss of HLA A11-restricted CTL epitopes in Epstein-Barr virus isolates from highly A11-positive populations by selective mutation of anchor residues. J. Exp. Med. 179:1297-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong, T., D. Boyd, W. Rosenberg, N. Alp, M. Takiguchi, A. McMichael, and S. Rowland-Jones. 1996. An HLA-B35-restricted epitope modified at an anchor residue results in an antagonist peptide. Eur. J. Immunol. 26:335-339. [DOI] [PubMed] [Google Scholar]
- 11.Elton, D., L. Medcalf, K. Bishop, D. Harrison, and P. Digard. 1999. Identification of amino acid residues of influenza virus nucleoprotein essential for RNA binding. J. Virol. 73:7357-7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gottschalk, S., C. Y. Ng, M. Perez, C. A. Smith, C. Sample, M. K. Brenner, H. E. Heslop, and C. M. Rooney. 2001. An Epstein-Barr virus deletion mutant associated with fatal lymphoproliferative disease unresponsive to therapy with virus-specific CTLs. Blood 97:835-843. [DOI] [PubMed] [Google Scholar]
- 13.Goulder, P. J., C. Brander, Y. Tang, C. Tremblay, R. A. Colbert, M. M. Addo, E. S. Rosenberg, T. Nguyen, R. Allen, A. Trocha, M. Altfeld, S. He, M. Bunce, R. Funkhouser, S. I. Pelton, S. K. Burchett, K. McIntosh, B. T. Korber, and B. D. Walker. 2001. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature 412:334-338. [DOI] [PubMed] [Google Scholar]
- 14.Goulder, P. J., R. E. Phillips, R. A. Colbert, S. McAdam, G. Ogg, M. A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, A. J. McMichael, and S. Rowland-Jones. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212-217. [DOI] [PubMed] [Google Scholar]
- 15.Haanen, J. B., M. C. Wolkers, A. M. Kruisbeek, and T. N. Schumacher. 1999. Selective expansion of cross-reactive CD8+ memory T cells by viral variants. J. Exp. Med. 190:1319-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klenerman, P., U. C. Meier, R. E. Phillips, and A. J. McMichael. 1995. The effects of natural altered peptide ligands on the whole blood cytotoxic T lymphocyte response to human immunodeficiency virus. Eur. J. Immunol. 25:1927-1931. [DOI] [PubMed] [Google Scholar]
- 17.Klenerman, P., S. Rowland-Jones, S. McAdam, J. Edwards, S. Daenke, D. Lalloo, B. Koppe, W. Rosenberg, D. Boyd, A. Edwards, et al. 1994. Cytotoxic T-cell activity antagonized by naturally occurring HIV-1 Gag variants. Nature 369:403-407. [DOI] [PubMed] [Google Scholar]
- 18.Klenerman, P., and R. M. Zinkernagel. 1998. Original antigenic sin impairs cytotoxic T lymphocyte responses to viruses bearing variant epitopes. Nature 394:482-485. [DOI] [PubMed] [Google Scholar]
- 19.Koup, R. A. 1994. Virus escape from CTL recognition. J. Exp. Med. 180:779-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mandruzzato, S., V. Stroobant, N. Demotte, and P. van der Bruggen. 2000. A human CTL recognizes a caspase-8-derived peptide on autologous HLA-B*3503 molecules and two unrelated peptides on allogeneic HLA-B*3501 molecules. J. Immunol. 164:4130-4134. [DOI] [PubMed] [Google Scholar]
- 21.Marsh, S. G. E., P. Parham, and L. D. Barber. 2000. The HLA factsbook, p. 182-186. Academic Press, London, United Kingdom.
- 22.McAdam, S., P. Klenerman, L. Tussey, S. Rowland-Jones, D. Lalloo, R. Phillips, A. Edwards, P. Giangrande, A. L. Brown, F. Gotch, et al. 1995. Immunogenic HIV variant peptides that bind to HLA-B8 can fail to stimulate cytotoxic T lymphocyte responses. J. Immunol. 155:2729-2736. [PubMed] [Google Scholar]
- 23.Oldstone, M. B. 1997. How viruses escape from cytotoxic T lymphocytes: molecular parameters and players. Virology 234:179-185. [DOI] [PubMed] [Google Scholar]
- 24.Parker, C. E., and K. G. Gould. 1996. Influenza A virus—a model for viral antigen presentation to cytotoxic T lymphocytes. Semin. Virol. 7:61-73. [Google Scholar]
- 25.Parker, K. C., M. A. Bednarek, and J. E. Coligan. 1994. Scheme for ranking potential HLA-A2 binding peptides based on independent binding of individual peptide side-chains. J. Immunol. 152:163-175. [PubMed] [Google Scholar]
- 26.Price, D. A., U. C. Meier, P. Klenerman, M. A. Purbhoo, R. E. Phillips, and A. K. Sewell. 1998. The influence of antigenic variation on cytotoxic T lymphocyte responses in HIV-1 infection. J. Mol. Med. 76:699-708. [DOI] [PubMed] [Google Scholar]
- 27.Price, G. E., R. Ou, H. Jiang, L. Huang, and D. Moskophidis. 2000. Viral escape by selection of cytotoxic T cell-resistant variants in influenza A virus pneumonia. J. Exp. Med. 191:1853-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sewell, A. K., D. A. Price, A. Oxenius, A. D. Kelleher, and R. E. Phillips. 2000. Cytotoxic T lymphocyte responses to human immunodeficiency virus: control and escape. Stem Cells (Miamisburg) 18:230-244. [DOI] [PubMed] [Google Scholar]
- 29.Shu, L. L., W. J. Bean, and R. G. Webster. 1993. Analysis of the evolution and variation of the human influenza A virus nucleoprotein gene from 1933 to 1990. J. Virol. 67:2723-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takamiya, Y., C. Schonbach, K. Nokihara, M. Yamaguchi, S. Ferrone, K. Kano, K. Egawa, and M. Takiguchi. 1994. HLA-B*3501-peptide interactions: role of anchor residues of peptides in their binding to HLA-B*3501 molecules. Int. Immunol. 6:255-261. [DOI] [PubMed] [Google Scholar]
- 31.Terajima, M., J. Jameson, J. E. Norman, J. Cruz, and F. A. Ennis. 1999. High-yield reassortant influenza vaccine production virus has a mutation at an HLA-A 2.1-restricted CD8+ CTL epitope on the NS1 protein. Virology 259:135-140. [DOI] [PubMed] [Google Scholar]
- 32.van der Burg, S. H., E. Ras, J. W. Drijfhout, W. E. Benckhuijsen, A. J. Bremers, C. J. Melief, and W. M. Kast. 1995. An HLA class I peptide-binding assay based on competition for binding to class I molecules on intact human B cells. Identification of conserved HIV-1 polymerase peptides binding to HLA-A*0301. Hum. Immunol. 44:189-198. [DOI] [PubMed] [Google Scholar]
- 33.Voeten, J. T., G. F. Rimmelzwaan, N. J. Nieuwkoop, R. A. Fouchier, and A. D. Osterhaus. 2001. Antigen processing for MHC class I restricted presentation of exogenous influenza A virus nucleoprotein by B-lymphoblastoid cells. Clin. Exp. Immunol. 125:423-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Voeten, J. T. M., T. M. Bestebroer, N. J. Nieuwkoop, R. A. M. Fouchier, A. D. M. E. Osterhaus, and G. F. Rimmelzwaan. 2000. Antigenic drift in the influenza A virus (H3N2) nucleoprotein and escape from recognition by cytotoxic T lymphocytes. J. Virol. 74:6800-6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weiner, A., A. L. Erickson, J. Kansopon, K. Crawford, E. Muchmore, A. L. Hughes, M. Houghton, and C. M. Walker. 1995. Persistent hepatitis C virus infection in a chimpanzee is associated with emergence of a cytotoxic T lymphocyte escape variant. Proc. Natl. Acad. Sci. USA 92:2755-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]