Abstract
Enzootic nasal tumor virus (ENTV) induces nasal epithelial cancer in infected sheep, but it is a simple retrovirus lacking a known oncogene. ENTV is closely related to jaagsiekte sheep retrovirus (JSRV), which also causes cancer in sheep but in the epithelial cells of the lower airways and alveoli. Here we show that as with JSRV, the envelope (Env) protein of ENTV can transform cultured cells and thus is likely to be responsible for oncogenesis in animals. In addition, the ENTV Env protein mediates virus entry using the same receptor as does JSRV Env, the candidate tumor suppressor Hyal2. However, ENTV Env mediates entry into cells from a more restricted range of species than does JSRV, and based on this finding we have identified amino acid regions in the Env proteins that are important for virus entry. Also, because ENTV does not efficiently use human Hyal2 as a receptor, we cloned the ovine Hyal2 cDNA and show that the encoded protein functions as an efficient receptor for both ENTV and JSRV. In summary, although ENTV and JSRV use the same cell surface receptor for cell entry and apparently transform cells by the same mechanism, they induce cancer in different tissues of infected sheep, indicating that oncogenesis is regulated at some other level. The transcriptional regulatory elements in these viruses are quite different, indicating that tissue-specific oncogenesis is likely regulated at the level of viral gene expression.
Enzootic nasal tumor virus (ENTV) is a simple retrovirus that is transmitted horizontally and induces nasal adenocarcinoma in sheep (4, 10) and goats (8, 9). ENTV can be found in the nasal fluid of animals with intranasal tumors, which eventually progress and cause severe cranial deformations and respiratory blockage, resulting in death (41). ENTV is closely related to jaagsiekte sheep retrovirus (JSRV) (>95% overall amino acid similarity) (4), which is the causative agent of ovine pulmonary adenocarcinoma (also called sheep pulmonary adenomatosis or jaagsiekte) (30). Unlike tumors caused by ENTV, which arise from nasal epithelial cells, JSRV-induced tumors arise from epithelial cells in the lower airway, including type II alveolar and bronchiolar epithelial cells (27). The mechanism of oncogenesis by ENTV and its relationship to that caused by JSRV are unknown. Both of these viruses are present in many countries worldwide and have a significant economic and animal health impact. In addition, the disease induced by JSRV exhibits histological features similar to those of many human pulmonary adenocarcinomas, including bronchioloalveolar carcinoma (28, 33). Thus, study of adenocarcinoma induced by JSRV and ENTV may provide insights into the etiology of human lung cancer. While ENTV and JSRV do not appear to cause lung cancer in humans having occupational exposure to these viruses, a recent study shows that antiserum directed against the JSRV capsid protein cross-reacts with 30% of human pulmonary adenocarcinoma samples but not with nontumorous lung lesions, normal lung tissue, or many adenocarcinoma samples from other tissues (7) indicating that related viruses may be involved in human lung cancer.
Recent progress in understanding JSRV biology provides clues for the study of ENTV-induced oncogenesis. First, Hyal2 has been identified as the cell surface receptor that mediates JSRV entry (36), and Hyal2 mRNA is widely expressed in different tissues of mice and humans (6, 16, 40). Furthermore, many different sheep cell lines and tissues are permissive to JSRV entry, indicating that the JSRV receptor is expressed in multiple cell types in sheep (14, 29, 31). However, it is not known whether variation in receptor expression in particular cells influences the tissue specificity of JSRV oncogenesis, or whether ENTV uses a different receptor, which could explain the different tissue specificities of oncogenesis by ENTV and JSRV. Second, the JSRV Env protein induces transformation in cultured cells (17, 36), thus identifying it as the oncogenic factor in JSRV pathogenesis. The mechanism of transformation by ENTV is unknown, and differences between the oncogenic mechanisms of ENTV and JSRV could help explain the tissue-specific oncogenesis exhibited by these viruses. Last, productive infection by JSRV is restricted by the preferential activity of the JSRV long terminal repeat (LTR) in type II pneumocytes and Clara cells (26). Sequence analysis of the ENTV LTR shows that ENTV transcriptional regulators differ from those found in JSRV (4), and differences in promoter activity between JSRV and ENTV could explain the tissue specificity of disease induction by the two viruses.
To determine the cell surface receptor used by ENTV, we constructed a retrovirus packaging cell line that produces retrovirus vectors bearing the ENTV envelope (Env) protein to monitor ENTV Env-mediated virus entry. The ENTV vector transduced cells from a restricted range of species compared to that of a JSRV vector, ENTV vector transduction being effectively limited to sheep cells. Interference analysis performed with sheep cells indicated that both vectors used the same receptor for entry, presumably ovine Hyal2 (oHyal2). Indeed, we have cloned the oHyal2 cDNA and show that the encoded protein mediates efficient entry of JSRV and ENTV vectors. We also cloned the closely related bovine Hyal2 (bHyal2) cDNA, but bHyal2 and the human Hyal2 protein (hHyal2) function poorly as receptors for ENTV and must be overexpressed to show receptor function. These differences allowed us to identify residues within Env that are important for virus entry. Last, we show that the env gene of ENTV transforms cultured fibroblasts, thus identifying Env as the likely oncogenic factor in ENTV pathogenesis. These results indicate that the tissue specificity of oncogenesis by ENTV and JSRV is not mediated at the level of virus entry or mechanism of transformation but is likely due to differences in virus expression governed by the divergent enhancer/promoters in these viruses.
MATERIALS AND METHODS
Cell culture.
Mammalian cells including SSF-123 primary sheep skin fibroblast (SSF) cells (a gift from William Osborne, University of Washington, Seattle), HT-1080 human fibrosarcoma cells (ATCC CCL-121), 293 human kidney epithelial cells (ATCC CRL 1573), IB3 human bronchial epithelial cells (44), HeLa cervical carcinoma cells (ATCC CCL-2), NIH 3T3 thymidine kinase-deficient mouse embryo fibroblasts (42), D17 canine osteosarcoma cells (ATCC CRL-6248), 208F rat embryo fibroblasts (34), MDBK bovine kidney epithelial cells (ATCC CCL-22), Vero African green monkey kidney epithelial cells (ATCC CCL-81), and RbTE immortalized rabbit tracheal epithelial cells (a gift from Christine Halbert, Fred Hutchinson Cancer Research Center, Seattle, Wash.) were grown in Dulbecco's modified Eagle medium with 10% fetal bovine serum (HyClone). RbTE cells have been immortalized by transduction with the retrovirus vector LXSN16E6E7, which expresses the human papillomavirus E6 and E7 genes (13). G355 feline embryonic brain cells (11) were grown in McCoy's medium with 15% fetal bovine serum. A23 hamster cells (39) were grown in minimal essential medium-α with 10% fetal bovine serum.
Plasmid expression vectors.
The JSRV Env expression plasmid pSX2.Jenv has been described (35). The JSRV Env coding region in this plasmid was derived from JSRV strain JS7 (GenBank accession no. Y18301). The ENTV Env expression plasmid pSX2.Eenv is similar in structure and was made by inserting an 1881-bp MslI-to-Ecl136 fragment of ENTV that contains the Env coding region into a BsaAI-to-MscI-cut fragment of pSX2 in place of the 10A1 env present in the pSX2 plasmid (20). pSX2.EEJ was made from pSX2.Eenv by replacing the 1,116-bp EcoNI-to-HpaI fragment at the 3" end of the ENTV env gene with that of JSRV. pSX2.JEJ and pSX2.EJJ were made by replacing the 446-bp BsrG1-to-AhdI fragment or the 520-bp AhdI-to-EcoNI fragment, respectively, of ENTV env sequences in pSX2.EEJ with the corresponding fragments from JSRV.
Retrovirus vectors and virus production.
LAPSN is a Moloney murine leukemia virus (MoMLV)-based vector that encodes human placental alkaline phosphatase (AP) and neomycin phosphotransferase (24). A retrovirus vector that expresses the JSRV Env protein, pLJeSN, was constructed by inserting an 1,883-bp MslI-to-Ecl136 fragment of JSRV containing the JSRV Env coding region into the EcoRI site of the retrovirus expression vector LXSN (22).
LAPSN vectors bearing JSRV, ENTV, 10A1, or RD114 Env protein were produced using the PJ4 (35), PN172 (described in this paper), PT67 (20), and FLYRD (3) packaging cell lines, respectively. All of these packaging lines produce vectors with Gag-Pol proteins from MoMLV. LAPSN vectors bearing hybrid ENTV/JSRV Env proteins were made by transient CaPO4-mediated transfection of the Env expression plasmids into NIH 3T3 cells that express MoMLV Gag-Pol proteins (NIH 3T3/LGPS cells) (21) and that contain the LAPSN vector. The cells were fed the day after transfection, and vector-containing medium was collected from confluent dishes 2 days after feeding. All vector preparations were filtered through 0.45-μm-pore-size filters and were stored at −70°C. Vector infections were done in the presence of 4 μg of Polybrene (Sigma)/ml, and transduction was quantitated by fixing cells with 0.5% glutaraldehyde, staining the cell monolayers for AP expression, and counting foci of AP-positive cells as described previously (12) or by exposing cells to G418 and counting G418-resistant colonies.
Cloning of bHyal2 and oHyal2 cDNAs.
Several IMAGE consortium expressed sequence tag (EST) clones (obtained from BACPAC Resources, Oakland, Calif.) were sequenced to obtain the full-length bHyal2 cDNA sequence (GenBank accession no. AF411973). The entire bHyal2 coding region was PCR amplified from an EST clone (GenBank accession no. BE487753) and cloned into the mammalian expression vector pCR3.1 (Invitrogen, Carlsbad, Calif.), and the correct sequence was verified.
The oHyal2 cDNAs were obtained by reverse transcription-PCR from SSF cell mRNA using a cDNA Cycle Kit (Invitrogen). Primers used in the reverse transcription-PCR were designed from bHyal2 cDNA sequences. Primers bLuca2F135 (5"-CCAGCATGTGGACAGGCCTG-3") and bLuca2R600 (5"-TACACATCCTTGTCCTGCCAG-3") produced a 5" fragment of ∼0.4 kb. Primers bLuca2F390 (5"-ACCGGCTGGGCATGTATCCAC-3") and bLuca23UA (5"-CCAGACTCTGGTTTGTCCAC-3") produced a 3" fragment of ∼1.2 kb. The two overlapping fragments (5" and3") contained the complete Hyal2 coding region. Each fragment was then separately cloned into the pCR3.1 vector in the sense orientation. The PmlI/XbaI fragment from the 5" clone was replaced by that of the 3" clone, resulting in a full-length expression construct. The oHyal2 cDNA sequence in this construct was determined (GenBank accession no. AF411974) and was confirmed by PCR sequencing of the genomic region from SSF cell DNA that covered the coding region.
RESULTS
Production of a retrovirus vector with an ENTV pseudotype.
We previously showed that the JSRV Env can be incorporated into virions with a vector and Gag-Pol proteins derived from MoMLV, and we used a similar strategy here to generate vectors bearing the ENTV Env protein. We obtained two uncharacterized molecular clones of the ENTV Env coding region that had been amplified by PCR from tumor tissue of affected sheep (gift from Christina Cousens and Mike Sharp) (4) and inserted these coding regions into the pSX2 expression vector in place of the 10A1 Env coding region present in this plasmid (20). Transient transfection of the resultant plasmids into cells that express MoMLV Gag-Pol proteins and the retrovirus vector LAPSN, followed by an assay for LAPSN vector production using SSF cells as targets for transduction, revealed that transfection of either clone mobilized the LAPSN vector. Tests of several plasmid clones containing the two ENTV env genes showed that one of the genes was clearly more active (>30-fold-higher vector titer), and we used this clone in further studies. We sequenced this clone (GenBank accession no. AF401741) to compare it to the previously determined sequence of cDNAs made from ENTV viral RNA (GenBank accession no. Y16627) and found 55 nucleotide differences in the 1,854-nucleotide Env coding region. However, these resulted in only two conservative amino acid changes. It is not known if the originally sequenced env gene is functional.
We next generated an ENTV packaging cell line by using the same technique as was used for generating JSRV packaging cells (35). Briefly, we transfected the ENTV Env expression plasmid into NIH 3T3/LGPS cells (21), which express MoMLV Gag-Pol proteins, along with a bacterial hygromycin phosphotransferase gene as a selectable marker. The cells were selected in hygromycin, and drug-resistant clones were screened for packaging function. The most active clone was PN172. We introduced the LAPSN vector into these cells to generate a stable cell line for production of ENTV-pseudotype virus for further studies.
The host range of an ENTV vector is restricted compared to that of a JSRV vector.
The ENTV-pseudotype LAPSN vector was used to measure the ability of ENTV Env to promote virus entry into a variety of mammalian cells in comparison to a JSRV-pseudotype LAPSN vector (Table 1). Of the cell types tested, only SSF cells and human 293 cells were transduced by the ENTV vector. The titer of the ENTV vector measured on SSF cells was ∼100-fold lower than that of the JSRV vector. Low-level sporadic transduction of human HT-1080 cells and murine NIH 3T3 cells was detected, but the titer of the ENTV vector was >500-fold lower than that found on SSF cells. In contrast, the JSRV-pseudotype vector transduced cells from many species, including monkey, dog, bovine, rabbit, and cat. All of the cell types that exhibited low to undetectable transduction by either the ENTV- or JSRV-pseudotype vector (<10 focus-forming units[FFU]/ml) could be transduced with a 10A1-pseudotype LAPSN vector at titers of >103 FFU/ml (data not shown). These results indicate that the LAPSN vector and MoMLV Gag-Pol proteins are functional in these cells, as expected, and that the block to transduction by ENTV or JSRV vectors was at the level of virus entry mediated by the ENTV or JSRV Env proteins.
TABLE 1.
The host range of an ENTV vector is restricted compared to that of a JSRV vectora
| Species | Target cells | ENTV vector titer | JSRV vector titer |
|---|---|---|---|
| Sheep | SSF | 5 × 103 | 6 × 105 |
| Human | HT-1080 | <10b | 3 × 104 |
| IB3 | <2 | 4 × 104 | |
| 293 | 5 × 102 | 4 × 104 | |
| HeLa | <2 | 10 | |
| Monkey | Vero | <2 | 1 × 105 |
| Dog | D17 | <2 | 1 × 104 |
| Bovine | MDBK | <2 | 2 × 103 |
| Rabbit | RbTE | <2 | 2 × 103 |
| Cat | G355 | <2 | 20 |
| Rat | 208F | <2 | <2 |
| Hamster | A23 | <2 | <2 |
| Mouse | NIH 3T3 | <10b | <10b |
Target cells were exposed to the LAPSN vector with an ENTV pseudotype (produced from PN172/LAPSN cells) or with a JSRV pseudotype (produced from PJ4/LAPSN cells). Vector titers are expressed in AP+ FFU per except for HeLa cells, for which titers are expressed in G418-resistant CFU per ml due to the high level of endogenous heat-stable AP in HeLa cells, which precludes detection of vector-encoded AP. Results are means of at least two experiments.
Low-level transduction was detected in some assays.
Expression of JSRV Env in SSF cells blocks subsequent transduction by ENTV and JSRV vectors.
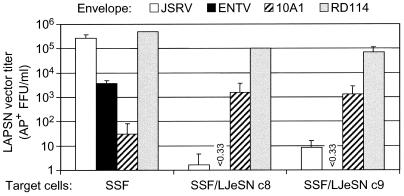
Most retrovirus Env proteins bind their receptors and interfere with entry of viruses that use the same receptor but have no effect on entry by viruses that use other receptors. To test whether ENTV and JSRV Env proteins mediate entry using the same cell surface receptor, we performed interference assays using SSF cells and SSF cells transduced with the retrovirus vector LJeSN that expresses JSRV Env (Fig. 1). Cells were exposed to LAPSN vectors pseudotyped with the JSRV, ENTV, 10A1, or RD114 envelope proteins, and transduction was measured by staining cells for AP+ foci. 10A1 vectors use Pit1 and/or Pit2 (20) and RD114 vectors use RDR (37) as receptors for cell entry; thus, transduction by these vectors should not be affected by JSRV Env expression. The titers of the ENTV and JSRV vectors were >5,000-fold lower on SSF/LJeSN cells than on SSF cells, a strong interference indicating that both viruses use the same receptor. In contrast, there was only a sixfold reduction in transduction by the RD114-pseudotype vector and a 50-fold increase in transduction by the 10A1 vector in SSF cells expressing the JSRV Env in comparison to unmodified SSF cells. Thus, expression of JSRV Env did not significantly interfere with entry of 10A1- or RD114-pseudotyped vectors as expected. We hypothesize that the unexpected increase in 10A1 vector transduction in cells expressing the JSRV Env protein is due to its oncogenic properties, which stimulate metabolism and thus may stimulate phosphate uptake by increasing expression of phosphate transporters, two of which (Pit1 and Pit2) are receptors for 10A1 murine leukemia virus (20).
FIG. 1.
JSRV Env expression blocks transduction by ENTV and JSRV vectors in SSF cells. SSF cells and two clonal SSF cell lines (SSF/LJeSN c8 and c9) which contain the JSRV Env expression vector LJeSN were exposed to LAPSN vectors made with the indicated envelope proteins. Transduction was measured 3 days after vector exposure by staining the cells for AP+ foci. Results are means of two to three independent experiments with duplicate determinations performed in each experiment.
The hHyal2, oHyal2, and bHyal2 proteins function as receptors for JSRV and ENTV.
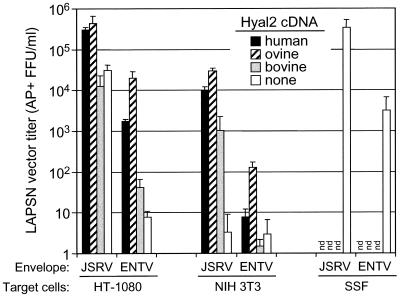
Because many human cell types were not transduced by the ENTV vector, indicating that hHyal2 is a poor receptor for ENTV, we cloned an oHyal2 cDNA and tested whether it encoded a functional receptor for ENTV and JSRV when expressed in HT-1080 human cells and NIH 3T3 mouse cells. In addition, we cloned and tested a bHyal2 cDNA to explore the finding that bovine cells are permissive to JSRV vectors but not to ENTV vectors. oHyal2 functioned best as a receptor for ENTV and JSRV vectors (Fig. 2). hHyal2 also functioned well as a receptor for the JSRV vector but showed a lower level of activity for the ENTV vector (Fig. 2), in agreement with the different host ranges of these viruses (Table 1). bHyal2 functioned poorly as a receptor for either JSRV or ENTV vectors; it showed some activity for the ENTV vector in human cells but not in mouse cells and showed activity for the JSRV vector in mouse cells but not in human cells (Fig. 2). In summary, the oHyal2, hHyal2, and bHyal2 proteins can function as receptors for ENTV and JSRV vectors with activities that parallel the host ranges of these viruses.
FIG. 2.
Evaluation of hHyal2, oHyal2, or bHyal2 proteins as receptors for ENTV and JSRV. HT-1080 and NIH 3T3 cells were seeded at a density of 5 × 105/dish in 60-mm-diameter dishes. After 24 h, the cells were transfected with expression plasmids encoding hHyal2, oHyal2, or bHyal2 or an empty expression plasmid (none). The following day, the transfected cells were trypsinized (1.5 ml of trypsin), and 100-μl aliquots of each preparation were seeded in the wells of six-well dishes. SSF cells were also seeded at a density of 105/well in six-well dishes. After 24 h, the cells were exposed to the ENTV or JSRV vectors. Two days after vector exposure, the cells were stained for AP+ foci. Results are means of data from two to four experiments, each done in duplicate. nd, not done.
Sequence alignments of Hyal2 from five species show that the ovine and bovine receptors are closely related.
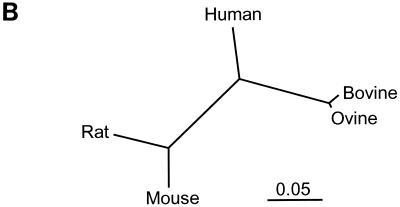
Alignment of ovine, bovine, human, rat, and mouse Hyal2 proteins shows that the ovine and bovine proteins are 99% identical and are more closely related to each other than any of the other Hyal2 proteins (Fig. 3). Although the mature ovine and bovine receptors differ by only seven residues, two of which are conservative changes (note that the divergent C termini of these proteins are removed during glycosylphosphatidylinositol anchor addition), the bovine ortholog functions poorly as a receptor for both JSRV and ENTV. In contrast, the human and ovine receptors share only 86% identity, but the human receptor mediates transduction by JSRV and ENTV vectors at rates that are at least an order of magnitude better than those of the bovine receptor. Mouse Hyal2 and rat Hyal2 proteins exhibit high sequence identity (91%), which in this case correlates with the observations that cells from either species are not transduced by either JSRV or ENTV.
FIG. 3.
Comparison of Hyal2 proteins from different species. (A) Comparison of human (U09577), mouse (AF302843), rat (AF034218), ovine (AF411974), and bovine (AF411973) Hyal2 protein sequences (GenBank accession numbers in parentheses). Amino acid identity (*), strong similarity (:), and weaker similarity (.) and amino acid differences between oHyal2 and bHyal2 (↓) are indicated. (B) Dendrogram of receptor similarity plotted using ClustalW. Scale bar indicates 5% sequence divergence.
Analysis of differences between ENTV and JSRV Env proteins that affect vector titer and species tropism.
The titer of the LAPSN vector produced by the ENTV packaging cell line PN172 is about 100-fold lower than that of the JSRV packaging cell line PJ4 when measured on SSF cells (Table 1). The JSRV and ENTV Env proteins are most divergent within their membrane spanning and cytoplasmic domains (52% identity) (Fig. 4B), and we tested whether the TM subunit of ENTV Env, which contains these domains and which mediates Env incorporation into virions and participates in virus entry following receptor binding, might be responsible for the reduced vector titer (Fig. 4A). Replacement of the TM subunit of the ENTV Env with that of JSRV (construct EEJ) improved the titer (measured on SSF cells) of the LAPSN vector bearing the chimeric Env ∼100-fold compared to that of the LAPSN vector bearing the ENTV Env and was equivalent to that of the LAPSN vector made with the JSRV Env. However, EEJ maintained a strong preference for infection of sheep cells.
FIG. 4.
Transduction of human and sheep cells by vectors bearing chimeric ENTV/JSRV Env proteins. (A) LAPSN vectors bearing the indicated Env proteins were made by transient transfection as described in Materials and Methods. SSF and HT-1080 cells were seeded at 105 cells per well of six-well plates, exposed to the vectors 1 day later, and stained to detect AP+ foci 2 days after vector exposure. Results are means of data from two independent experiments with duplicate determinations performed in each experiment. (B) Sequence alignment of JSRV and ENTV Env proteins. Dots indicate identical amino acids, and the dash indicates a gap. The cleavage sites for restriction enzymes are indicated by vertical lines. The predicted locations of the endoplasmic reticulum signal sequence, the cleavage site between the Env protein surface (SU) and TM subunits, the membrane-spanning domain, and the cytoplasmic tail are shown.
To determine the regions responsible for the species tropism of the JSRV and ENTV Env proteins, we made the exchanges within the surface (SU) region of the EEJ Env using shared restriction sites found in both ENTV and JSRV env genes and tested their functionality (Fig. 4). JEJ and EJJ refer to Env proteins containing amino acids 56 to 204 (10 amino acid differences) or amino acids 205 to 377 (7 amino acid differences) of JSRV Env, respectively. All vector pseudotypes had about the same titer on SSF cells, whereas only the JSRV vector and the JEJ vector were capable of transducing HT-1080 cells well, showing that the 10 residues at positions 59, 62, 63, 75, 174, 177, 180, 191, 195, and 196 of JSRV Env mediate interaction with the human receptor. However, residues 59, 62, 63, and 75 are not expected to be involved in receptor binding because they lie within the predicted endoplasmic reticulum signal sequence (amino acids 1 to 76) (4), and a deletion of the first 71 amino acids of the JSRV envelope protein, which contains most of the signal sequence, did not affect its ability to promote transduction of human or sheep cells (data not shown). Therefore, ENTV Env residues 174, 177, 180, 191, 195, and 196 are responsible for the relatively poor infectivity of human cells by the ENTV vector in comparison to the JSRV vector.
The ENTV env gene can transform rodent fibroblasts.
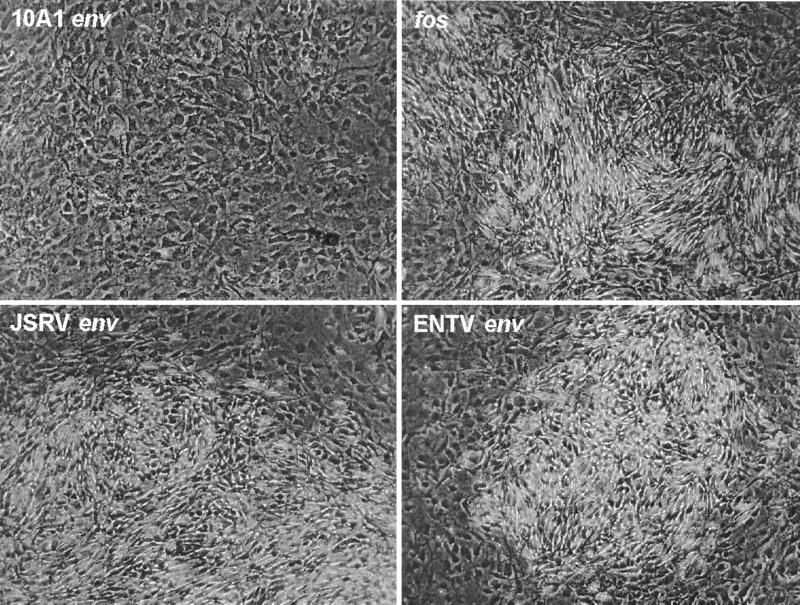
To evaluate whether the ENTV Env protein has oncogenic properties, we tested its ability to transform a morphologically flat variety of NIH 3T3 mouse cells (Fig. 5). ENTV env, JSRV env, and the fos oncogene all produced transformed foci which were similar in size range. The numbers of foci induced by the ENTV and JSRV env genes were similar, while the number produced by the fos gene was about fivefold higher (data not shown). The 10A1 env gene did not induce transformed foci and served as a negative control in this experiment. These results show that the ENTV Env protein can transform cells and has activity similar to that of the JSRV Env protein.
FIG. 5.
The ENTV and JSRV env genes induce transformed foci in cultured mouse cells. A morphologically flat subline of NIH 3T3 mouse cells (originally from Doug Lowy) was seeded at 5 × 105 cells per 6-cm-diameter dish. One day later, the cells were transfected with 10 μg each of the plasmids pSX2.Eenv (ENTV env), pSX2.Jenv (JSRV env), pSX2 (10A1 env) or pFBJ/R (viral fos oncogene) (23) plus 1 μg of pLAPSN (to measure transfection efficiency) by using the CaPO4 coprecipitation method. One day after transfection, the cells were trypsinized and divided in a 1:5 ratio into new dishes. Four days after transfection, the medium was changed to medium containing 5% fetal bovine serum, and the cells were refed with this medium every 3 days thereafter. Eleven days after transfection, the cells were fixed and stained for AP. Representative foci are shown; no foci were observed in the 10A1 env dish. Two AP+ cells are visible at the lower right in the 10A1 env panel.
DISCUSSION
Studies of ENTV and JSRV have been difficult because of the lack of an in vitro cell culture system for growing the viruses. In the case of JSRV, it has been shown that the LTR is inactive in most cell types tested (26). Production of JSRV in culture has been accomplished by transfecting 293T cells with a recombinant JSRV genome in which the U3 region of the upstream JSRV LTR is replaced with a strong human cytomegalovirus promoter (30). However, while virus produced in this manner can infect various ovine cell lines, it replicates poorly and little virus is produced (31). To circumvent these difficulties, we constructed an MoMLV-based packaging line that expresses the JSRV Env protein and showed that JSRV Env is incorporated into virions with MoMLV Gag-Pol and MoMLV-based retroviral vectors (35). We later used this to determine the host range of the JSRV Env (35) and to identify the JSRV cell surface receptor (36). Here we have used the same approach for ENTV and show that the ENTV Env protein can pseudotype MoMLV-based retrovirus vectors, thus allowing us to study the properties of the ENTV Env protein.
We provide strong evidence that JSRV and ENTV use the same cell surface receptor, Hyal2. Nonpermissive cells were rendered susceptible to transduction by both ENTV and JSRV vectors when Hyal2 from ovine, bovine, or human species was expressed in the cells. Furthermore, expression of JSRV Env in sheep cells blocked entry by ENTV and JSRV vectors, showing that ENTV uses a subset of the receptor(s) used by JSRV. Although we cannot entirely rule out the possibility that ENTV and JSRV use multiple receptors, previous studies indicate that at least in human cells, JSRV utilizes only one receptor. Thus, analysis of JSRV entry into radiation hybrid cells shows that only a small region of human chromosome 3 that contains the hyal2 gene can render hamster cells permissive to transduction by a JSRV vector (35, 36). Additionally, while two paralogs of hyal2 that might encode JSRV receptors, hyal1 and hyal3, map to the same region, the proteins they encode do not function as receptors for JSRV, nor do those of the other hyal2 paralogs, hyal4 and spam1, that are located on human chromosome 7 (6, 36). The findings that both ENTV and JSRV use oHyal2 as a receptor and that Hyal2 is ubiquitously expressed in different tissues of other mammals including mice and humans (6, 16, 40) argues that the tissue-specific oncogenesis by these viruses is not determined at the level of receptor-mediated virus entry. Furthermore, JSRV can enter and integrate into cell types from many tissues in culture and in animals, including nasal turbinate cells, the target for ENTV oncogenesis (14, 29, 31), yet JSRV oncogenesis is limited to cells of the lower airway, again indicating that oncogenesis is not regulated at the receptor level.
The ENTV vector titers were lower for all cell types, including sheep cells, compared to the JSRV vector titers. By exchanging the TM domain of the ENTV Env with that of the JSRV Env, we were able to produce a vector that transduced sheep cells as efficiently as a JSRV vector without changing its restricted tropism for cells from other species. This particular clone of the ENTV Env may have a general defect in this region, or perhaps the ENTV Env is less able to interact with MoMLV Gag proteins and pseudotype MoMLV vectors than is the JSRV Env protein.
While both the JSRV and ENTV Env proteins can mediate efficient vector entry using the sheep Hyal2 receptor, they differ in their ability to use the Hyal2 orthologs from other species. This property has been observed previously for other viruses that utilize the same receptor; for example, both MoMLV and the closely related PVC211 retrovirus use the cationic amino acid transporter Cat1 from mouse and rat as a receptor, but only PVC211 can use hamster Cat1 as a receptor (19). Thus, a difference in host range conferred by different Env proteins does not necessarily imply the use of unrelated receptors for Env-mediated cell entry. This apparent paradox can be explained by postulating differences in the abilities of the JSRV and ENTV Env proteins to efficiently bind specific Hyal2 orthologs from different species. Interestingly, while the ENTV vector was unable to transduce bovine cells, it was able to transduce cells transfected with a bHyal2 expression construct, presumably because overexpression of bHyal2 can offset the binding inefficiency of ENTV Env and promote virus entry. Along these lines, we found that ENTV vectors transduce 293 cells but not other human cells and hypothesize that 293 cells express more Hyal2 than do the other cell types. By exchanging a few residues within the SU domain of ENTV with equivalent residues of JSRV, we were able to create a chimeric Env that uses the endogenous human receptor, thus identifying the differences in JSRV Env that allow it to efficiently interact with Hyal2 orthologs.
Mechanisms of retroviral oncogenesis have previously been divided into two major categories: insertional mutagenesis, which typically involves proviral integration in or near a cellular proto-oncogene, such as c-myc (32), leading to increased expression of an oncogenic protein, and virus expression of oncogenes derived from cellular genes, as in the case of v-src or v -sis (2). ENTV provides a new example of a third mechanism of retroviral oncogenesis that involves the presence of oncogenic viral proteins that are not derived from cellular genetic material. JSRV and avian hemangioma virus are recent additions to the latter category, and as we have shown for ENTV, the Env proteins from these viruses all exhibit transforming activity (1, 17, 36). Spleen focus-forming virus is an early example of a retrovirus with an oncogene derived from viral env sequences, but in this case the Env protein is nonfunctional, and spleen focus-forming virus is replication defective (25, 38, 43). Last, human T-cell leukemia virus type 1 and bovine leukemia virus are complex retroviruses that exhibit characteristics of the latter category due to the oncogenic properties of Tax, a viral protein that normally transactivates the viral LTR and which can also interact with cellular transcription factors, leading to the activation of many cellular genes, including those that stimulate growth (5, 15). These results provide increasing evidence that retroviral proteins can evolve oncogenic properties, as have the proteins of viruses in many other virus families.
ENTV and JSRV use the same receptor for cell entry, and both express Env proteins with transforming activity, indicating that some other factor governs their tissue-specific tumorigenesis. JSRV and ENTV have markedly different enhancer regions within their LTRs, suggesting that tissue specificity of disease might be controlled at the transcriptional level. Indeed, the JSRV LTR is specifically active in cells from the lower airway, including type II pneumocytes and Clara cells (26), the targets for JSRV oncogenesis. The sequence of the U3 region of the JSRV LTR reveals potential binding sites for transcription factors hepatocyte nuclear factor 3 (HNF-3), NF-1, and SP-1 (26), which have been shown to function synergistically in regulating expression of lung-specific genes, such as the surfactant genes (18). Biochemical analysis shows that HNF-3 strongly transactivates the JSRV LTR when overexpressed in NIH 3T3 cells, whereas other transcription factors that are expressed in the epithelial cells of the lung did not cause transactivation (26). The U3 region of the ENTV LTR (4) is dramatically different from the JSRV LTR in that it contains only the SP-1 site and lacks many of the other regulatory sequences that are found in JSRV, including the NF-1 site and the two HNF-3 sites. The ENTV LTR contains a binding site for the nuclear transcription factor Y which is not found in the JSRV LTR. Identification of the factors involved in ENTV transcriptional regulation will help to clarify how ENTV and JSRV cause tissue-specific transformation. However, such studies are complicated because it has been difficult to identify candidate transcription factors that transactivate the ENTV LTR and cells which comprise the nasal epithelium are quite diverse, making it difficult to identify the correct lineage of cells for such studies. A good approach to addressing these questions would be to study induction of disease in animals infected with hybrid viruses that contain the structural genes of ENTV or JSRV under the control of the LTR from the other virus. Direct evidence for the transcriptional regulation of ENTV will help to better define the mechanism by which ENTV and JSRV induce tissue-specific cancers.
Acknowledgments
We thank Christina Cousens and Mike Sharp for the gift of ENTV env clones.
This work was supported by grants DK47754 and HL54881 and contracts CO56000 and CO12400 from the National Institutes of Health.
REFERENCES
- 1.Alian, A., D. Sela-Donenfeld, A. Panet, and A. Eldor. 2000. Avian hemangioma retrovirus induces cell proliferation via the envelope (env) gene. Virology 276:161-168. [DOI] [PubMed] [Google Scholar]
- 2.Coffin, J. M. 1996. Retroviridae: the viruses and their replication, p. 1767-1847. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology. Lippincott-Raven, Philadelphia, Pa.
- 3.Cosset, F. L., Y. Takeuchi, J. L. Battini, R. A. Weiss, and M. K. Collins. 1995. High-titer packaging cells producing recombinant retroviruses resistant to human serum. J. Virol. 69:7430-7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cousens, C., E. Minguijon, R. G. Dalziel, A. Ortin, M. Garcia, J. Park, L. Gonzalez, J. M. Sharp, and M. de las Heras. 1999. Complete sequence of enzootic nasal tumor virus, a retrovirus associated with transmissible intranasal tumors of sheep. J. Virol. 73:3986-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cross, S. L., M. B. Feinberg, J. B. Wolf, N. J. Holbrook, F. Wong-Staal, and W. J. Leonard. 1987. Regulation of the human interleukin-2 receptor alpha chain promoter: activation of a nonfunctional promoter by the transactivator gene of HTLV-I. Cell 49:47-56. [DOI] [PubMed] [Google Scholar]
- 6.Csoka, A. B., S. W. Scherer, and R. Stern. 1999. Expression analysis of six paralogous human hyaluronidase genes clustered on chromosomes 3p21 and 7q31. Genomics 60:356-361. [DOI] [PubMed] [Google Scholar]
- 7.de las Heras, M., S. H. Barsky, P. Hasleton, M. Wagner, E. Larson, J. Egan, A. Ortin, J. A. Gimenez-Mas, M. Palmarini, and J. M. Sharp. 2000. Evidence for a protein related immunologically to the jaagsiekte sheep retrovirus in some human lung tumours. Eur. Respir. J. 16:330-332. [DOI] [PubMed] [Google Scholar]
- 8.de las Heras, M., J. A. Garcia de Jalon, E. Minguijon, E. W. Gray, P. Dewar, and J. M. Sharp. 1995. Experimental transmission of enzootic intranasal tumors of goats. Vet. Pathol. 32:19-23. [DOI] [PubMed] [Google Scholar]
- 9.de las Heras, M., J. A. Garcia de Jalon, and J. M. Sharp. 1991. Pathology of enzootic intranasal tumor in thirty-eight goats. Vet. Pathol. 28:474-481. [DOI] [PubMed] [Google Scholar]
- 10.de las Heras, M., J. M. Sharp, L. M. Ferrer, J. A. Garcia de Jalon, and L. M. Cebrian. 1993. Evidence for a type D-like retrovirus in enzootic nasal tumour of sheep. Vet. Rec. 132:441. [DOI] [PubMed]
- 11.Dunn, K. J., C. C. Yuan, and D. G. Blair. 1993. A phenotypic host range alteration determines RD114 virus restriction in feline embryonic cells. J. Virol. 67:4704-4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fields-Berry, S. C., A. L. Halliday, and C. L. Cepko. 1992. A recombinant retrovirus encoding alkaline phosphatase confirms clonal boundary assignment in lineage analysis of murine retina. Proc. Natl. Acad. Sci. USA 89:693-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halbert, C. L., G. W. Demers, and D. A. Galloway. 1991. The E7 gene of human papillomavirus type 16 is sufficient for immortalization of human epithelial cells. J. Virol. 65:473-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holland, M. J., M. Palmarini, M. Garcia-Goti, L. Gonzalez, I. McKendrick, M. de las Heras, and J. M. Sharp. 1999. Jaagsiekte retrovirus is widely distributed both in T and B lymphocytes and in mononuclear phagocytes of sheep with naturally and experimentally acquired pulmonary adenomatosis. J. Virol. 73:4004-4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inoue, J., M. Seiki, T. Taniguchi, S. Tsuru, and M. Yoshida. 1986. Induction of interleukin 2 receptor gene expression by p40x encoded by human T-cell leukemia virus type 1. EMBO J. 5:2883-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lepperdinger, G., B. Strobl, and G. Kreil. 1998. HYAL2, a human gene expressed in many cells, encodes a lysosomal hyaluronidase with a novel type of specificity. J. Biol. Chem. 273:22466-22470. [DOI] [PubMed] [Google Scholar]
- 17.Maeda, N., M. Palmarini, C. Murgia, and H. Fan. 2001. Direct transformation of rodent fibroblasts by jaagsiekte sheep retrovirus DNA. Proc. Natl. Acad. Sci. USA 98:4449-4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Margana, R. K., and V. Boggaram. 1997. Functional analysis of surfactant protein B (SP-B) promoter. Sp1, Sp3, TTF-1, and HNF-3α transcription factors are necessary for lung cell-specific activation of SP-B gene transcription. J. Biol. Chem. 272:3083-3090. [DOI] [PubMed] [Google Scholar]
- 19.Masuda, M., C. A. Hanson, P. M. Hoffman, and S. K. Ruscetti. 1996. Analysis of the unique hamster cell tropism of ecotropic murine leukemia virus PVC-211. J. Virol. 70:8534-8539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller, A. D., and F. Chen. 1996. Retrovirus packaging cells based on 10A1 murine leukemia virus for production of vectors that use multiple receptors for cell entry. J. Virol. 70:5564-5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller, A. D., J. V. Garcia, N. von Suhr, C. M. Lynch, C. Wilson, and M. V. Eiden. 1991. Construction and properties of retrovirus packaging cells based on gibbon ape leukemia virus. J. Virol. 65:2220-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller, A. D., and G. J. Rosman. 1989. Improved retroviral vectors for gene transfer and expression. BioTechniques 7:980-982. [PMC free article] [PubMed] [Google Scholar]
- 23.Miller, A. D., I. Verma, and T. Curran. 1985. Deletion of the gag region from FBR murine osteosarcoma virus does not affect its enhanced transforming activity. J. Virol. 55:521-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller, D. G., R. H. Edwards, and A. D. Miller. 1994. Cloning of the cellular receptor for amphotropic murine retroviruses reveals homology to that for gibbon ape leukemia virus. Proc. Natl. Acad. Sci. USA 91:78-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishigaki, K., D. Thompson, C. Hanson, T. Yugawa, and S. Ruscetti. 2001. The envelope glycoprotein of Friend spleen focus-forming virus covalently interacts with and constitutively activates a truncated form of the receptor tyrosine kinase Stk. J. Virol. 75:7893-7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palmarini, M., S. Datta, R. Omid, C. Murgia, and H. Fan. 2000. The long terminal repeat of jaagsiekte sheep retrovirus is preferentially active in differentiated epithelial cells of the lungs. J. Virol. 74:5776-5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palmarini, M., P. Dewar, M. De las Heras, N. F. Inglis, R. G. Dalziel, and J. M. Sharp. 1995. Epithelial tumour cells in the lungs of sheep with pulmonary adenomatosis are major sites of replication for jaagsiekte retrovirus. J. Gen. Virol. 76:2731-2737. [DOI] [PubMed] [Google Scholar]
- 28.Palmarini, M., H. Fan, and J. M. Sharp. 1997. Sheep pulmonary adenomatosis: a unique model of retrovirus-associated lung cancer. Trends Microbiol. 5:478-483. [DOI] [PubMed] [Google Scholar]
- 29.Palmarini, M., M. J. Holland, C. Cousens, R. G. Dalziel, and J. M. Sharp. 1996. jaagsiekte retrovirus establishes a disseminated infection of the lymphoid tissues of sheep affected by pulmonary adenomatosis. J. Gen. Virol. 77:2991-2998. [DOI] [PubMed] [Google Scholar]
- 30.Palmarini, M., J. M. Sharp, M. de las Heras, and H. Fan. 1999. Jaagsiekte sheep retrovirus is necessary and sufficient to induce a contagious lung cancer in sheep. J. Virol. 73:6964-6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palmarini, M., J. M. Sharp, C. Lee, and H. Fan. 1999. In vitro infection of ovine cell lines by jaagsiekte sheep retrovirus. J. Virol. 73:10070-10078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Payne, G. S., J. M. Bishop, and H. E. Varmus. 1982. Multiple arrangements of viral DNA and an activated host oncogene in bursal lymphomas. Nature 295:209-214. [DOI] [PubMed] [Google Scholar]
- 33.Perk, K., and I. Hod. 1982. Sheep lung carcinoma: an endemic analogue of a sporadic human neoplasm. J. Natl. Cancer. Inst. 69:747-749. [PubMed] [Google Scholar]
- 34.Quade, K. 1979. Transformation of mammalian cells by avian myelocytomatosis virus and avian erythroblastosis virus. Virology 98:461-465. [DOI] [PubMed] [Google Scholar]
- 35.Rai, S. K., J. C. DeMartini, and A. D. Miller. 2000. Retrovirus vectors bearing jaagsiekte sheep retrovirus Env transduce human cells by using a new receptor localized to chromosome 3p21.3. J. Virol. 74:4698-4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rai, S. K., F. M. Duh, V. Vigdorovich, A. Danilkovitch-Miagkova, M. I. Lerman, and A. D. Miller. 2001. Candidate tumor suppressor HYAL2 is a glycosylphosphatidylinositol (GPI)-anchored cell-surface receptor for jaagsiekte sheep retrovirus, the envelope protein of which mediates oncogenic transformation. Proc. Natl. Acad. Sci. USA 98:4443-4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rasko, J. E., J. L. Battini, R. J. Gottschalk, I. Mazo, and A. D. Miller. 1999. The RD114/simian type D retrovirus receptor is a neutral amino acid transporter. Proc. Natl. Acad. Sci. USA 96:2129-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruscetti, S. K. 1999. Deregulation of erythropoiesis by the Friend spleen focus-forming virus. Int. J. Biochem. Cell. Biol. 31:1089-1109. [DOI] [PubMed] [Google Scholar]
- 39.Stewart, E. A., K. B. McKusick, A. Aggarwal, E. Bajorek, S. Brady, A. Chu, N. Fang, D. Hadley, M. Harris, S. Hussain, R. Lee, A. Maratukulam, K. O'Connor, S. Perkins, M. Piercy, F. Qin, T. Reif, C. Sanders, X. She, W. L. Sun, P. Tabar, S. Voyticky, S. Cowles, J. B. Fan, and D. R. Cox. 1997. An STS-based radiation hybrid map of the human genome. Genome Res. 7:422-433. [DOI] [PubMed] [Google Scholar]
- 40.Strobl, B., C. Wechselberger, D. R. Beier, and G. Lepperdinger. 1998. Structural organization and chromosomal localization of Hyal2, a gene encoding a lysosomal hyaluronidase. Genomics 53:214-219. [DOI] [PubMed] [Google Scholar]
- 41.Vitellozzi, G., L. Mughetti, M. Palmarini, M. T. Mandara, L. Mechelli, J. M. Sharp, and I. Manocchio. 1993. Enzootic intranasal tumour of goats in Italy. Zentbl. Vetmed. B 40:459-468. [DOI] [PubMed] [Google Scholar]
- 42.Wei, C. M., M. Gibson, P. G. Spear, and E. M. Scolnick. 1981. Construction and isolation of a transmissible retrovirus containing the src gene of Harvey murine sarcoma virus and the thymidine kinase gene of herpes simplex virus type 1. J. Virol. 39:935-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamamura, Y., H. Senda, Y. Kageyama, T. Matsuzaki, M. Noda, and Y. Ikawa. 1998. Erythropoietin and Friend virus gp55 activate different JAK/STAT pathways through the erythropoietin receptor in erythroid cells. Mol. Cell. Biol. 18:1172-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeitlin, P. L., L. Lu, J. Rhim, G. Cutting, G. Stetten, K. A. Kieffer, R. Craig, and W. B. Guggino. 1991. A cystic fibrosis bronchial epithelial cell line: immortalization by adeno-12-SV40 infection. Am. J. Respir. Cell Mol. Biol. 4:313-319. [DOI] [PubMed] [Google Scholar]