Abstract
Human cell lines were isolated that express the V protein of either simian virus 5 (SV5) or human parainfluenza virus type 2 (hPIV2); the cell lines were termed 2f/SV5-V and 2f/PIV2-V, respectively. STAT1 was not detectable in 2f/SV5-V cells, and the cells failed to signal in response to either alpha/beta interferons (IFN-α and IFN-β, or IFN-α/β) or gamma interferon (IFN-γ). In contrast, STAT2 was absent from 2f/PIV2-V cells, and IFN-α/β but not IFN-γ signaling was blocked in these cells. Treatment of both 2f/SV5-V and 2f/PIV2-V cells with a proteasome inhibitor allowed the respective STAT levels to accumulate at rates similar to those seen in 2fTGH cells, indicating that the V proteins target the STATs for proteasomal degradation. Infection with SV5 can lead to a complete loss of both phosphorylated and nonphosphorylated forms of STAT1 by 6 h postinfection. Since the turnover of STAT1 in uninfected cells is longer than 24 h, we conclude that degradation of STAT1 is the main mechanism by which SV5 blocks interferon (IFN) signaling. Pretreatment of 2fTGH cells with IFN-α severely inhibited both SV5 and hPIV2 protein synthesis. However, and in marked contrast, pretreatment of 2fTGH cells with IFN-γ had little obvious effect on SV5 protein synthesis but did significantly reduce the replication of hPIV2. Pretreament with IFN-α or IFN-γ did not induce an antiviral state in 2f/SV5-V cells, indicating either that the induction of an antiviral state is completely dependent on STAT signaling or that the V protein interferes with other, STAT-independent cell signaling pathways that may be induced by IFNs. Even though SV5 blocked IFN signaling, the addition of exogenous IFN-α to the culture medium of 2fTGH cells 12 h after a low-multiplicity infection with SV5 significantly reduced the subsequent cell-to-cell spread of virus. The significance of the results in terms of the strategy that these viruses have evolved to circumvent the IFN response is discussed.
Alpha/beta interferons (IFN-α and IFN-β, or IFN-α/β) and gamma interferon (IFN-γ) are induced in vivo following virus infection and are central to the control of virus spread. Interferons (IFNs) induce an antiviral state in uninfected cells and may inhibit virus replication in infected cells as well as enhance the adaptive immune response. IFN-α/β are secreted by cells in response to virus infection, while IFN-γ is produced by subsets of activated T lymphocytes and NK cells. IFN-α/β and IFN-γ bind to independent cell surface receptors and activate distinct but related signal transduction pathways, culminating in the activation of an overlapping set of IFN-stimulated genes. In a cascade of events following the binding of IFN-α/β to their receptors, the inactive cytoplasmic transcription factors STAT1 and STAT2 become phosphorylated, form heterodimers, and migrate to the nucleus, where they become associated with p48 to form the IFN-stimulated gene factor 3 (ISGF3) complex; this complex activates the transcription of IFN-α/β-responsive genes. Following the binding of IFN-γ to its receptor, cytoplasmic STAT1 is phosphorylated and homodimerizes to form the gamma-activated factor (GAF) complex; this complex activates the transcription of IFN-γ-responsive genes. (For reviews on IFN signaling, see references 7 and 35.)
Paramyxoviruses, like most other viruses, have evolved specific molecular mechanisms to circumvent certain IFN-induced antiviral responses (for general reviews of how viruses circumvent IFN responses, see references 6 and 18). Paramyxoviruses have nonsegmented, negative-sense, single-stranded RNA genomes. Their genomes are encapsidated within helical nucleocapsids which are surrounded by pleomorphic lipid-containing envelopes through which protrude the virus glycoproteins. A major characteristic used to help classify paramyxoviruses into different subfamilies and genera is the structure of their P genes. In all paramyxoviruses, the P protein, together with the large (L) protein, forms part of the virus RNA polymerase complexes. The P genes of some paramyxoviruses also encode additional proteins. In the genus Rubulavirus, of which simian virus 5 (SV5), mumps virus, and human parainfluenza virus type 2 (hPIV2) are members, the P gene encodes both the P and the V proteins. The P and V proteins have a common N-terminal domain but unique C-terminal domains. In rubulaviruses, the V mRNA is a faithful transcript of the P gene, while the P mRNA transcript has two additional nontemplate G residues (inserted by a specific polymerase stuttering mechanism during transcription of the gene) that alter the reading frame of the mRNA. In viruses belonging to the Respirovirus (e.g., Sendai virus [SeV]) and Morbillivirus (e.g., measles virus) genera, the P mRNA is a faithful transcript of the gene, and nontemplate residues are inserted to make the V mRNA. In addition, the P genes of respiroviruses and morbilliviruses (but not rubulaviruses) encode a third group of related proteins termed the C proteins. The C proteins are expressed from open reading frames that overlap the 5" portion of the P gene in the +1 reading frame. The P genes of pneumoviruses (e.g., respiratory syncytial virus) are less complex in that they encode only the P protein. However, these viruses have additional genes, homologues of which are not present in the Paramyxovirinae subfamily (for a review on the molecular biology of paramyxoviruses, see reference 16).
It was originally shown that SV5 and SeV at least partially overcome the IFN response by blocking IFN signaling (2). Subsequently, it was demonstrated that many other paramyxoviruses block IFN signaling (5, 8, 14, 15, 25, 39, 40), although they clearly achieve this goal by distinct molecular mechanisms (40). The V protein of SV5 targets STAT1 for proteasome-mediated degradation and is thus responsible for the observed block in IFN signaling (3). The V protein of mumps virus also targets STAT1 for degradation (15). Surprisingly, hPIV2 targets STAT2 rather than STAT1 for degradation and, consequently, while hPIV2 blocks IFN-α/β signaling, it fails to block IFN-γ signaling (24, 25, 40). SV5 and hPIV2 are evolutionarily closely related, showing 43% identity in their HN proteins (27). While the V proteins show a similar level of overall homology (44%), the region of highest homology is found in the conserved C-terminal cysteine-rich domain (34). Although there are extensive areas of homology in the N terminus, there are also regions of very significant divergence. An essential role for the cysteine-rich C-terminal domain of the V protein in circumventing the IFN response was demonstrated for hPIV2 and mumps virus. Thus, recombinant hPIV2 that encoded a truncated version of the V protein that was missing its unique C-terminal domain was sensitive to inhibition by IFN (10). Furthermore, it was reported that the expression of the cysteine-rich C-terminal region of the V protein of mumps virus inhibits the induction of an IFN-induced antiviral state (15). On the other hand, Young et al. also clearly demonstrated a role for the N terminus of the V protein in targeting STAT1 for degradation by demonstrating that a single amino acid substitution at amino acid 100 within the P or V common N terminus can influence the ability of the V protein of SV5 to target STAT1 for degradation in cells from different species (41). Although SeV also encodes a V protein that has a cysteine-rich C-terminal domain, it is the C proteins of SeV, not the V protein, that are responsible for the observed block in IFN signaling (4, 5, 9, 36). It is therefore clear that different paramyxoviruses have solved the problem of how to replicate in the presence of an IFN response in different ways (40). This conclusion is further emphasized by the observation that respiratory syncytial virus does not block IFN signaling but rather has some as-yet-undefined mechanism for circumventing the IFN response (33, 40).
Here we compare the properties of human 2fTGH cells, which stably express the V proteins of SV5 and hPIV2, and examine the replication of SV5 and hPIV2 in these cells in the presence and absence of IFN-α/β and IFN-γ.
MATERIALS AND METHODS
Cells, viruses, and interferons.
Human 2fTGH cells (23, 26) and their derivatives were grown as monolayers in 25- or 75-cm2 tissue culture flasks containing Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (growth medium). All cell lines were negative for mycoplasmas, as screened by 4",6"-diamidino-2-phenylindole (DAPI) staining. Cells were treated with human IFN-α (Intron A; Schering-Plough) or IFN-γ (R & D Systems) at 1,000 IU/ml in medium containing 2% bovine serum (maintenance medium). SV5 (strain W3A) (1) and hPIV2 were grown and titrated under appropriate conditions in Vero cells by using maintenance medium.
Plasmid DNAs and transfection.
The IFN-α/β-responsive plasmid [p(9-27)4tkΔ(−39)lucter] contains four tandem repeat sequences of the IFN-stimulated response elements (ISRE) from the IFN-inducible gene, 9-27, fused to the firefly luciferase gene (13); the IFN-γ-responsive plasmid [p(GAS)2tkΔ(−39)lucter] contains a minimal thymidine kinase promoter and two tandem repeat sequences of the IFN regulatory factor 1 (IRF-1) gamma-activated sequence (GAS) site fused to the luciferase gene (13). pJATlacZ, a plasmid used as a transfection standard, contains a β-galactosidase gene under the control of the rat β-actin promoter (22). The construction of plasmid pEF.SV5-V has been reported elsewhere (3), and the V gene of hPIV2 was cloned into the same vector by using the same strategy, generating plasmid pEF.hPIV2-V. To facilitate ease of isolation of cell clones stably expressing the protein of interest, a vector (pEF.IRES.neo) was constructed that allows the expression of the transgene and the gene for G418 resistance from a single transcript; this transcript is driven by the mammalian elongation factor 1α (EF1α) promoter and has a human β-globin 5" untranslated region for optimal translation of the transgene, with the G418 resistance gene being translated from an internal ribosome entry site (derived from the encephalomyocarditis virus). Furthermore, to encourage selection of high-expression clones, the G418 resistance gene had a glutamic acid-to-aspartic acid conversion at residue 182 that substantially reduced the activity of encoded neomycin phosphotransferase II (38). ApaI/XbaI and ApaI/EcoRI fragments of pEF.SV5-V and pEF.hPIV2-V were introduced into pEF.IRES.neo between the ApaI/Sma and ApaI/EcoRI sites, generating plasmids pEF.SV5-V.IRES.neo and pEF.hPIV2-V.IRES.neo, respectively.
For transfections, monolayers of cells grown in six-well plates to 50 to 70% confluence were transfected with 1 μg of the appropriate DNA and 1.5 μl of Fugene 6 (Roche) according to the manufacturer's instructions. To isolate cells expressing the V protein of SV5 or hPIV2, transfected cells were cultured in the presence of 400 μg of Geneticin/ml, and resistant colonies were isolated. For the IFN signaling assays, at various times posttransfection, cells were or were not induced with 1,000 IU of IFN-α or IFN-γ/ml for 4 h immediately prior to harvesting. Luciferase and β-galactosidase activities were measured as described previously (12). The relative expression levels were calculated by dividing the luciferase values by the β-galactosidase values.
Preparation of radiolabeled antigen extracts, immunoprecipitation, and SDS-PAGE.
Cells were metabolically labeled with l-[35S]methionine (500 Ci/mmol; Amersham International Ltd., Little Chalfont, United Kingdom) for various times in methionine-free tissue culture medium. At the end of the labeling interval, the cells were washed in ice-cold phosphate-buffered saline and lysed in immunoprecipitation buffer (10 mM-Tris-HCl [pH 7.8], 5 mM EDTA, 0.5% Nonidet P-40, 0.65 M NaCl; 4 × 106 to 6 × 106 cells per ml of buffer) by sonication with an ultrasonic probe. Soluble antigen extracts were obtained after pelleting of the particulate material from the total cell antigen extracts by centrifugation at 12,000 × g for 30 min. Immune complexes were formed by incubating (for 2 h at 4°C) 0.2- to 1-ml samples of the soluble antigen extracts with an excess of either anti-SV5 monoclonal antibodies (MAbs) to the HN, F, P, M, and NP proteins (1 μl of concentrated tissue culture fluid of MAbs SV5-HN-4a, -F-1a, -P-e, -M-h, and -NP-a) (32) or anti-hPIV2 MAbs to the HN, NP, and P proteins (1 μl of concentrated tissue culture fluid of MAbs hPIV2-HN-3a, -NP-a, -NP-b, and -P-a) (31). STAT1 and p53 were immunoprecipitated by using either a mixture of anti-N-terminal and anti-C-terminal STAT1 MAbs (Transduction Laboratories) or anti-p53 MAb DO.1 (37). The immune complexes were isolated by using an excess of protein G-Sepharose 4B Fast Flow (Sigma; 20 μl of a 50% [wt/vol] suspension per μl of concentrated tissue culture fluid for 1 h at 4°C). The proteins in the immune complexes were dissociated by heating (100°C for 5 min) in gel electrophoresis sample buffer (0.05 M Tris-HCl [pH 7.0], 0.2% sodium dodecyl sulfate [SDS], 5% 2-mercaptoethanol, 5% glycerol) and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) with 10% gels. After electrophoresis, the gels were fixed, stained, and dried; labeled polypeptides were visualized by phosphorimager analysis.
Immunoblotting.
At the time of harvest, cells were washed twice with phosphate-buffered saline, disrupted in SDS gel loading buffer, sonicated, and boiled for 5 min. Polypeptides were separated by SDS-PAGE with 7% gels and transferred to nitrocellulose membranes, and STAT1 and STAT2 were detected with a polyclonal anti-STAT1 antibody raised against the N-terminal 194 amino acids, a polyclonal antibody to phosphotyrosine (Y701) STAT1, or a MAb to the N-terminal region of STAT2 (Transduction Laboratories). All protein-antibody interactions were detected by enhanced chemiluminescence with horseradish peroxidase-conjugated sheep anti-mouse or donkey anti-rabbit immunoglobulin G (Amersham).
Immunofluorescence.
Cells to be stained for immunofluorescence were grown on 10-mm-diameter coverslips (General Scientific Co. Ltd., Redhill, United Kingdom). The cells were treated and stained with specific MAbs as described previously (30). Antibody binding was detected by indirect immunofluorescence with a secondary Texas Red-conjugated goat anti-mouse immunoglobulin antibody (Seralab, Oxford, United Kingdom; catalogue number SBA 1010-02). The primary antibodies for SV5 were SV5-P-e, -P-k, and -NP-a (32), and those for hPIV2 were hPIV2-NP-a and -P-a (31). Following staining for immunofluorescence, the monolayers of cells were examined by using a Nikon Microphot-FXA immunofluorescence microscope.
RESULTS
Cloning and expression of the V gene of SV5 and hPIV2 in 2fTGH cells.
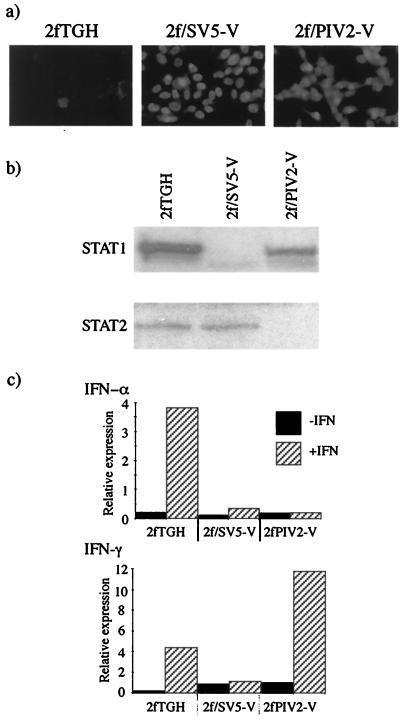
The V genes of SV5 and hPIV2 were cloned into pEF.IRES.neo, a vector that allows expression of the transgene and the G418 resistance gene from a single transcript (see Materials and Methods). 2fTGH cells were transfected with the appropriate plasmids and selected for resistance to G418. Colonies were screened for expression of the V proteins by immunofluorescence. Of 30 colonies screened for expression of the V protein of SV5, 28 were positive, and of 10 colonies screened for the V protein of hPIV2, 8 were positive. In the majority of both SV5 and hPIV2 V protein-positive cell lines, all the cells showed a predominantly nuclear distribution of V protein of similar intensity (Fig. 1a). A cell line expressing the V protein of SV5, termed 2f/SV5-V, and one expressing the V protein of hPIV2, termed 2f/PIV2-V, were selected for further analysis. Immunoblot analysis clearly demonstrated the absence of STAT1 but the presence of STAT2 in 2f/SV5-V cells and the absence of STAT2 but the presence of STAT1 in 2f/PIV2-V cells (Fig. 1b). To determine whether these cells responded to IFN-α/β and IFN-γ, the cells were transiently transfected with either IFN-α/β- or IFN-γ-responsive luciferase reporter plasmids, and the induction of luciferase was assayed following treatment with IFN. Figure 1c demonstrates that while IFN-α/β signaling is blocked in both cell lines, IFN-γ signaling is blocked only in 2f/SV5-V cells.
FIG. 1.
Characterization of 2f/SV5-V and 2f/PIV2-V cells. (a) Photomicrographs showing the intracellular localization of the V proteins of SV5 and hPIV2 in 2f/SV5-V and 2f/PIV2-V cells, respectively. Note that all the cells are positive for V and that V has a primarily nuclear distribution. (b) Immunoblot analysis demonstrating the presence of STAT2 but the absence of STAT1 in 2f/SV5-V cells and the presence of STAT1 but the absence of STAT2 in 2f/PIV2-V cells. Total extracts of 2f/SV5-V and 2f/PIV2-V cells were electrophoresed through 10% polyacrylamide gels and electrophoretically transferred to nitrocellulose, and STAT1 and STAT2 were detected by immunoblot analysis. (c) IFN-α signaling is inhibited in 2f/SV5-V and 2f/PIV2-V cells (top panel), while IFN-γ signaling is inhibited in 2f/SV5-V but not 2f/PIV2-V cells (bottom panel). Cells were transfected with either IFN-α- or IFN-γ-responsive plasmids together with pJATlacZ, and at 46 h posttransfection, cells were either treated with human IFN-α or IFN-γ or left untreated. Four hours later, luciferase and β-galactosidase activities in the cellular lysates were measured. Luciferase activity, expressed in relative light units, was normalized to β-galactosidase activity.
Proteasome-mediated degradation and stability of STAT1 and STAT2.
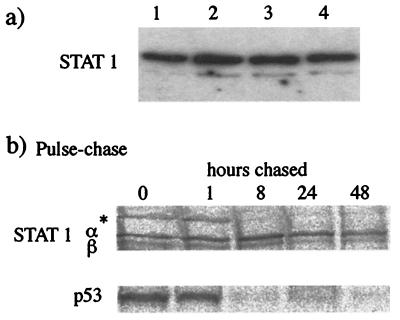
It was previously demonstrated that the proteasome inhibitor MG132 blocks the degradation of STAT1 in cells infected with SV5 (3). It was thus of interest to determine the effect of MG132 on the levels of STAT1 and STAT2 in 2f/SV5-V and 2f/PIV2-V cells, respectively. Surprisingly, following treatment of the cells with MG132 for 8 h, immunoblot analysis revealed very little increase in the levels of either STAT1 or STAT2 in the respective cells (data not shown; see also below). One possible explanation for these results would be if the steady-state synthesis STAT1 and STAT2 was insufficient to restore normal levels of STAT1 in 2fTGH cells in 8 h. To investigate this idea, the half-life of STAT1 was monitored by two methods. First, 2fTGH cells were treated with cycloheximide, and the levels of STAT1 were detected by immunoblot analysis. Under these conditions, there was no detectable reduction in STAT1 levels in 2fTGH cells over 18 h (Fig. 2a). Second, 2fTGH cells were pulsed with 35[S]methionine for 1 h prior to being incubated in the absence of radioactivity (chase) for up to 48 h. STAT1 was immunoprecipitated, and the relative levels were estimated by phosphorimager analysis (Fig. 2b). Quantification of the amount of radioactivity precipitated revealed that only 30 to 40% of STAT1 had been degraded after a 48-h chase. As a control, we demonstrated that p53 was degraded between 1 and 8 h postchase (Fig. 2b).
FIG. 2.
STAT1 has a slow intracellular turnover rate. (a) STAT1 was detected by immunoblot analysis in total extracts of 2fTGH cells that had been treated with cycloheximide (50 μg/ml) for 0, 4, 8, or 18 h (lanes 1 to 4, respectively). (b) 2fTGH cells were radioactively labeled with 35[S]methionine for 1 h and then cultured (chased) for 0, 1, 8, 24, or 48 h in medium that did not contain 35[S]methionine. Soluble antigen extracts were made, and STAT1α and -β and p53 were immunoprecipitated. An asterisk indicates an unidentified host cell protein that is rapidly turned over.
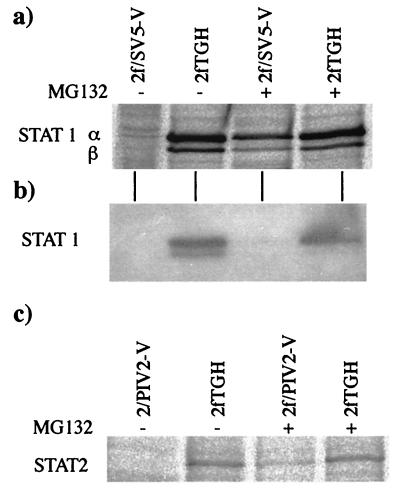
Given the long half-life of STAT1 and the adverse effects of treating cells with the proteasome inhibitor for prolonged periods, we decided to examine whether MG132 could prevent the degradation of newly synthesized STAT1 and STAT2. Cells were pulsed with 35[S]methionine for 8 h in the presence or absence of MG132. STAT1 and STAT2 were immunoprecipitated, and their relative levels were visualized by phosphorimager analysis. The results clearly demonstrated that MG132 blocked the degradation of newly synthesized STAT1 in 2f/SV5-V cells (Fig. 3a) and STAT2 in 2f/PIV2-V cells (Fig. 3c). However, following MG132 treatment, the levels of newly synthesized STAT1 and STAT2 in 2f/SV5-V and 2f/PIV2-V cells, respectively, were slightly lower than those in the control cells. The reason for this result remains unclear, but it may simply have occurred because MG132 does not completely block proteasome-mediated degradation. It should also be noted that for the total cell lysates of 2f/SV5-V cells used in the immunoprecipitation studies, immunoblot analysis failed to demonstrate an obvious increase in the overall levels of STAT1 in cells treated with MG132 (Fig. 3b).
FIG. 3.
The proteasome inhibitor MG132 blocks the degradation of newly synthesized STAT1 in 2f/SV5-V cells and STAT2 in 2f/PIV2-V cells. (a and c) MG132 (10 μM) was or was not added to the culture 1 h before the cells were radioactively labeled with 35[S]methionine for a further 7 h in the presence or absence of the inhibitor. STAT1α and -β were immunoprecipitated from 2f/SV5-V soluble antigen extracts (panel a), and STAT2 was immunoprecipitated from 2f/PIV2-V extracts (panel c). (b) Immunoblot analysis was also used to detect STAT1 in samples of total extracts of the 2f/SV5-V cells used in the immunoprecipitation analysis.
Phosphorylated and nonphosphorylated forms of STAT1 are degraded by SV5.
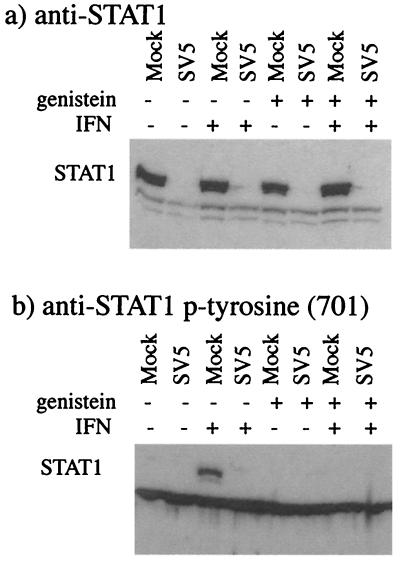
Following stimulation with IFN, activation of STAT1 is usually transient as a result of dephosphorylation. However, it has also been reported that phosphorylated STAT1 is turned over by a mechanism involving proteasome-mediated degradation (11), although there is no evidence that this latter process is important in the regulation of STAT1 function (6). Nevertheless, it was formally possible that following infection with SV5, STAT1 was rapidly phosphorylated, and that only phosphorylated forms of STAT1 were targeted for degradation. To test this idea, 2fTGH cells were infected in the presence or absence of genistein, a general kinase inhibitor that completely inhibited the phosphorylation of STAT1 in response to IFN (Fig. 4). It was clear from these results that both phosphorylated and nonphosphorylated forms of STAT1 were degraded in SV5-infected cells.
FIG. 4.
Both phosphorylated and nonphoshorylated forms of STAT1 are degraded by SV5. 2fTGH cells were or were not pretreated with the kinase inhibitor genistein (100 μM) for 1 h prior to being mock infected or infected with SV5 at an MOI of 10. Cells were or were not cultured in the presence of genistein for a further 6 h. Total cell extracts were made, and the presence of STAT1 was detected by immunoblot analysis with polyclonal anti-STAT1 antibody (a) or anti- phosphotyrosine (Y701) STAT1 [anti-STAT1 p-tyrosine (701)] (b).
Interaction of cellular proteins with the V proteins of SV5 and hPIV2.
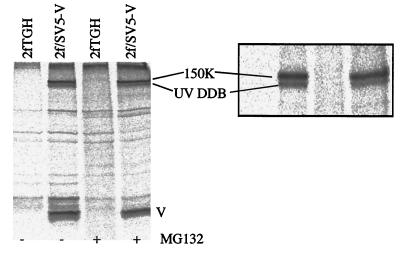
To investigate the interaction of cellular proteins with the V protein of SV5, soluble antigen extracts were made from 35[S]methionine-labeled 2fTGH and 2f/SV5-V cells treated in the presence or absence of MG132. There was no evidence of a protein with a molecular mass similar to that of STAT1 being coimmunoprecipitated with the V protein by anti-V MAbs from 2f/SV5-V cells that had been treated with MG132 (Fig. 5). Similarly, immunoprecipitation with anti-STAT1 antibodies failed to coprecipitate a band with a molecular mass similar to that of the V protein. With these experimental conditions, we also failed to reveal a direct interaction between STAT2 and the V protein of hPIV2 (data not shown). However, two prominent higher-molecular-mass proteins were coprecipitated with the V protein of SV5. Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry confirmed that the lower band was the 127-kDa subunit of the UV-damaged DNA-binding protein (UV DDB). MALDI-TOF analysis of the upper, 150-kDa protein was equivocal, and this protein has yet to be formally identified. Neither of these polypeptides was precipitated with the V protein from 2f/PIV2-V cells. This result was surprising in that it was previously reported that the V proteins of both SV5 and hPIV2 bind UV DDB (19). However, it is possible that the anti-hPIV2 V protein MAb (hPIV2-Pa) used in the immunoprecipitation study binds to a region critical for the interaction of the V protein with UV DDB.
FIG. 5.
Coimmunoprecipitation of host cell proteins with the V protein of SV5. 2fTGH and 2f/SV5-V cells were or were not treated with the proteasome inhibitor MG132 (10 μM) for 6 h, during which time they were also metabolically labeled with 35[S]methionine. Soluble antigen extracts were made, and the V protein was precipitated with MAb SV5-P-k. The precipitated polypeptides were separated on both 12 and 7% (inset) polyacrylamide gels. The lower of the two high-molecular-mass host cell bands that coprecipitated with V was identified by MALDI-TOF analysis as the 127-kDa subunit of UV DDB; the upper, 150-kDa band has yet to be identified (unpublished results).
Cell division and UV sensitivity of 2f/SV5-V and 2f/PIV2-V cells.
Since it has been reported that the V protein of SV5 binds UV DDB (20) and also slows the progression of the cell cycle (19), the rate of proliferation of 2f/SV5-V and 2f/PIV2-V cells and their relative sensitivity to UV light were compared to those of parental 2fTGH cells. However, no significant differences were observed either in the rate of proliferation of the different cells (Table 1) or in their relative sensitivity to UV light (unpublished results).
TABLE 1.
Growth rates for 2fTGH, 2f/SV5-V, and 2f/PIV2-V cells in culturesa
| Cells | Avg no. of cells/flask after the following days in cultures:
|
|||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| 2fTGH | 1.5 × 105 | 3.5 × 105 | 1.2 × 106 | 3.2 × 106 |
| 2f/SV5-V | 1.6 × 105 | 5 × 105 | 1.6 × 106 | 3.6 × 106 |
| 2f/PIV2-V | 1.4 × 105 | 4.0 × 105 | 1.8 × 106 | 4.0 × 106 |
Cells (1.0 × 105) were placed in 25-cm2 flasks and incubated in growth medium at 37°C. After 1, 2, 3, and 4 days, cells from duplicate flasks were trypsinized and counted.
SV5 and hPIV2 protein synthesis in 2f/SV5-V and 2f/PIV2-V cells in the presence and absence of IFNs.
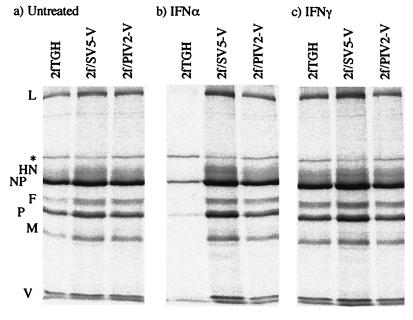
Since IFN-α/β and IFN-γ signaling is blocked in 2f/SV5-V cells but only IFN-α/β signaling is blocked in 2f/PIV2-V cells, we compared the relative levels of SV5 protein synthesis in 2f/SV5-V and 2f/PIV2-V cells that had or had not been pretreated with IFN-α or IFN-γ prior to infection. Figure 6 shows that pretreatment with IFN-α significantly inhibited SV5 protein synthesis in control cells but not in either 2f/SV5-V or 2f/PIV2-V cells. However, unexpectedly, IFN-γ had only marginal effects on the levels of SV5 protein synthesis in control cells; thus, any differential effects of IFN-γ on 2f/SV5-V and 2f/PIV2-V cells could not be convincingly measured.
FIG. 6.
Effect of IFN pretreatment on SV5 protein synthesis in 2fTGH, 2f/SV5-V, and 2f/PIV2-V cells. Cells were not (a) or were pretreated with either IFN-α (b) or IFN-γ (c) (1,000 IU/ml) for 16 h prior to infection with SV5 at an MOI of 0.5 to 1.0 PFU/cell. At 20 h p.i., the cells were metabolically labeled with 35[S]methionine for 1 h. Virus proteins were precipitated from soluble antigen extracts of these cells with a pool of MAbs to the NP, P, M, HN, and F proteins; separated by 10% PAGE; and visualized by phosphorimager analysis. An asterisk indicates an unidentified host cell protein.
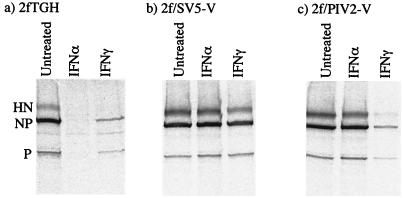
The sensitivity of hPIV2 to IFN-α and IFN-γ was measured in a similar set of experiments. In marked contrast to what was observed with SV5, pretreatment of 2fTGH cells with IFN-γ significantly reduced the levels of hPIV2 protein synthesis, although not as dramatically as pretreatment of cells with IFN-α (Fig. 7a). We also tested whether IFN-α and IFN-γ could induce an antiviral state for hPIV2 in 2f/SV5-V and 2f/PIV2-V cells. As expected, IFN-α failed to induce an antiviral state in both 2f/SV5-V and 2f/PIV2-V cells. In contrast, while IFN-γ failed to induce an antiviral state in 2f/SV5-V cells, it did induce an antiviral state in 2f/PIV2-V cells (Fig. 7b and c).
FIG. 7.
Effect of IFN pretreatment on hPIV2 protein synthesis in 2fTGH (a), 2f/SV5-V (b), and 2f/PIV2-V (c) cells. Cells were or were not pretreated with either IFN-α or IFN-γ (1,000 IU/ml) for 16 h prior to infection with hPIV2 at an MOI of 0.5 to 1.0 PFU/cell. At 20 h p.i., the cells were metabolically labeled with 35[S]methionine for 1 h. Virus proteins were precipitated from soluble antigen extracts of these cells with a pool of MAbs to the NP, P, and HN proteins; separated by 10% PAGE; and visualized by phosphorimager analysis.
SV5 spreads more rapidly in the presence of IFN-α in both 2f/SV5-V and 2f/PIV2-V cells than in 2fTGH cells.
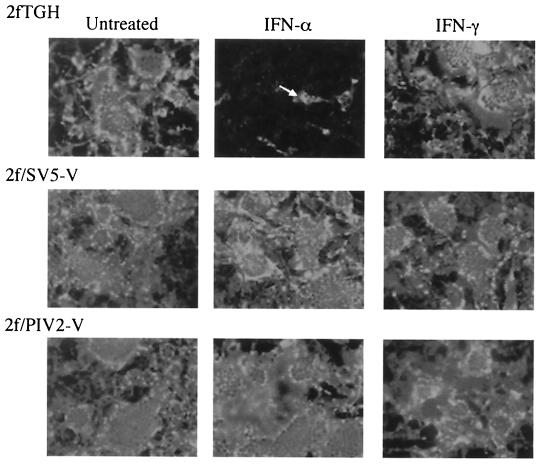
Although SV5 blocks IFN signaling, SV5 protein synthesis was significantly reduced in cells already in an IFN-α-induced antiviral state (Fig. 6). To examine how IFN influences the cell-to-cell spread of SV5, monolayers of 2fTGH, 2f/SV5-V, and 2f/PIV2-V cells were infected with SV5 at 0.01 PFU/cell; at 12 h postinfection (p.i.), exogenous IFN was or was not added to the culture medium. At various times p.i., the monolayers were fixed, and virus-infected cells were visualized by immunofluorescence with a pool of MAbs to the NP and P proteins (Fig. 8). While in the absence of exogenous IFN all of the 2fTGH cells were infected by 72 h p.i., the addition of exogenous IFN-α severely inhibited the spread of SV5 such that only 5 to 10% of the cells were infected. In contrast, IFN-α had no effect on the spread of SV5 in 2f/SV5-V and 2f/PIV2-V cells. In agreement with the above results, IFN-γ had little effect on the cell-to-cell spread of SV5 in control cells; thus, any possible differential effects on 2f/SV5-V and 2f/PIV2-V cells could not be evaluated.
FIG. 8.
Photomicrographs demonstrating the cell-to-cell spread of SV5 in 2fTGH, 2f/SV5-V, and 2f/PIV2-V cells in the presence or absence of exogenous IFN-α or IFN-γ. Cells were infected at 0.01 PFU/cell; 12 h later, IFN-α or IFN-γ (1,000 IU/ml) was or was not added to the culture medium. Monolayers were fixed at various times p.i., and the percentage of infected cells was estimated by immunofluorescence with a pool of MAbs to the NP and P proteins. At 24 h p.i., >5% of the cells in all the monolayers were infected (data not shown). By 72 h p.i., all the cells were infected, except for 2fTGH cells that had been cultured in the presence of IFN-α. For reference, a single cell is highlighted with a white arrow.
STAT1 degradation and the need for virus protein synthesis.
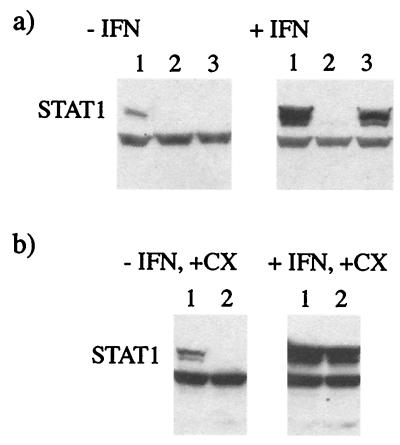
STAT1 is inducible with IFN; thus, the levels of STAT1 are significantly higher in IFN-pretreated cells than in untreated cells. It was previously reported that infectious SV5 can lead to the degradation of STAT1 in untreated and IFN-pretreated cells (3). It was also shown that UV-inactivated SV5 can lead to the degradation of STAT1 in untreated cells; however, the ability of UV-inactivated virus to degrade STAT1 in IFN-pretreated cells was not examined in that study (3). The results shown in Fig. 6 demonstrate that even when cells are in an IFN-induced antiviral state, some SV5 protein synthesis occurs. Consequently, it was possible that some de novo SV5 (V) protein synthesis was required for the complete degradation of STAT1 in IFN-pretreated cells. Two experiments were carried out to investigate whether this was the case. First, cells were pretreated with IFN, and the ability of UV-inactivated SV5 to induce the degradation of STAT1 was examined. Figure 9 shows that while UV-inactivated virus (at the equivalent of 10 PFU/cell) induced the degradation of all of the detectable STAT1 in untreated cells, it did not induce the complete degradation of the much higher levels of STAT1 present in IFN-pretreated cells (irrespective of how long the cells were infected), although there was a partial reduction in the levels. In contrast, “live” virus induced the degradation of STAT1 in both untreated and IFN-α-pretreated cells. Second, to confirm that de novo virus protein synthesis was required to induce the complete degradation of STAT1 in cells already in an IFN-α-induced antiviral state, cells were pretreated with IFN-α for 24 h and then infected with SV5 in the presence or absence of cycloheximide. Although infectious SV5 (at 10 PFU/cell) induced the degradation of STAT1 in untreated cells in the presence of cycloheximide, under the same conditions SV5 did not induce the degradation of STAT1 in IFN-pretreated cells. These results therefore suggest that the degradation of STAT1 is dependent on the concentrations of STAT1 and the V protein. This suggestion is supported by the observation that there was no obvious loss of STAT1 in untreated cells infected at a multiplicity of infection (MOI) of 1 PFU/cell in the presence of cycloheximide (unpublished results). Thus, it is also likely that input virus could degrade STAT1 in IFN-pretreated cells in the absence of virus protein synthesis if the MOI were high enough.
FIG. 9.
De novo SV5 protein synthesis is required for STAT1 degradation in IFN-pretreated cells but not in untreated cells. (a) 2fTGH cells were (+IFN) or were not (−IFN) pretreated with IFN-α (1,000 IU ml) for 18 h prior to being mock infected (lane 1) or infected with “infectious” SV5 (lane 2) or UV-inactivated virus (lane 3; the efficiency of UV inactivation was demonstrated by showing that even after 24 h of infection, no virus protein synthesis had occurred, as judged by immunofluorescence). At 8 h p.i., total cell extracts were made, and STAT1 was detected by immunoblot analysis. (b) 2fTGH cells were (+IFN) or were not (−IFN) pretreated with IFN-α for 18 h prior to being mock infected (lane 1) or infected with “infectious” SV5 (lane 2). From the time of infection, cells were cultured in the presence of cycloheximide (+CX; 50 μg/ml). At 8 h p.i., total cell extracts were made, and STAT1 was detected by immunoblot analysis.
DISCUSSION
The extremely high percentage of cell clones isolated that expressed the V protein of either SV5 or hPIV2 highlights the ease with which STAT1 or STAT2 can be selectively inactivated by using the plasmids constructed. Indeed, we have isolated a number of other cell types, including human liver cells, which express a V protein of SV5 that can no longer respond to either IFN-α/β or IFN-γ (data not shown). All the cells expressed similar levels of the V protein (as judged by immunofluorescence), and expression was stable. Thus, once cells expressing the V protein were isolated, it was not necessary to culture them in the presence of Geneticin to maintain expression. These plasmids should therefore provide valuable reagents with which to selectively degrade STAT1 or STAT2 for further functional analysis of these important cell signaling molecules.
It was previously reported that the proteasome inhibitors MG132 and lactacysteine block the degradation of STAT1 following infection of tissue culture cells with SV5 (3). In the experiments reported here, it was not possible to restore either STAT1 or STAT2 to normal cellular levels in 2f/SV5-V or 2f/PIV2-V cells, respectively, by treating the cells for up to 8 h with MG132. Since STAT1 and STAT2 have been reported to be very stable proteins with half-lives of more than 24 h (17), it is possible that the rates of synthesis were insufficient to allow significant levels of STATs to accumulate during the 8-h MG132 treatment period. We confirmed by pulse-labeling experiments that STAT1 has a half-life of more than 24 h in 2fTGH cells, and we established that MG132 clearly blocks the degradation of newly synthesized STAT1 and STAT2 in 2f/SV5-V and 2f/PIV2-V cells, respectively, thereby allowing STAT levels to accumulate at rates similar to those seen in 2fTGH cells. Given that STAT1 can be completely lost from 2fTGH cells infected with SV5 at 2 to 4 h p.i., we conclude that the degradation of STAT1 by the SV5 V protein is sufficient to account in full for the inhibition of IFN signaling in 2fTGH cells. For similar reasons, specific degradation of STAT2 by hPIV2 appears to be the only mechanism which needs to be invoked to explain the block in IFN signaling in 2fTGH cells. However, it was recently reported that STAT2 may be less stable in HeLa cells, and it was suggested that in these cells, specific translational inhibition of STAT2 mRNA by the PIV2 V protein may also be required to mediate full inhibition of IFN signaling (24).
Following infection with a high MOI of SV5 that prevents cells from entering an IFN-induced antiviral state, even though the majority of cells survive the infection, after 2 to 4 days there is a significant reduction in the rate of virus protein synthesis (unpublished results and reference 41). The reason for the late switching off of virus protein synthesis is unclear, but since inhibition of IFN signaling seems an obvious target for viruses, we speculated that cells may have some other intracellular compensatory mechanism by which they can induce an antiviral state (6, 41). Indeed, one possibility was that the loss of STAT1 or STAT2 may have activated an antiviral default pathway within cells (as a defense against viruses that block IFN signaling). However, given that both SV5 and hPIV2 readily infect 2f/SV5-V and 2f/PIV2-V cells, such a default pathway, if it exists, is clearly not activated simply by the loss of STAT1 or STAT2 from the cells. Indeed, other than a loss in their ability to respond to IFN, there was no obvious effect on the growth or morphology of 2f/SV5-V and 2f/PIV2-V cells compared to 2fTGH cells. A number of recent reports also have demonstrated STAT1-independent regulation of gene expression in response to IFN-γ and the induction of an antiviral state in STAT1 null mice in response to IFN-γ (6, 28). However, the failure of IFN-γ to induce an antiviral state for hPIV2 in 2f/SV5-V cells suggests either that the induction of an IFN-γ-induced antiviral state for hPIV2 is completely dependent on STAT signaling or that the V protein of SV5 also interferes with other STAT-independent signaling cascades that may be induced by IFN-γ.
The inability to induce an antiviral state for SV5 by pretreating 2fTGH cells with IFN-γ is surprising given that pretreatment of cells with IFN-α delays SV5 replication and that IFN-γ inhibits hPIV2 replication, albeit to a lesser extent than IFN-α. Until more is known about the molecular basis of IFN-induced inhibition of SV5 and hPIV2 replication, the reasons for these differences will remain unclear. However, it seems likely from these observations that the different antiviral mechanisms induced by IFN-α/β and IFN-γ in controlling SV5 and hPIV2 vary in relative importance. Furthermore, although many paramyxoviruses block IFN signaling (40), since virus replication is significantly delayed in cells already in an IFN-α/β-induced antiviral state prior to infection, such a mechanism alone will result only in their ability to partially overcome the IFN response. Thus, given the highly pathogenic nature of many paramyxoviruses, it is possible that they have additonal mechanisms, other than blocking of IFN signaling, to help circumvent the IFN response.
It was previously shown that the V proteins of both SV5 and hPIV2 bind to the 127-kDa subunit of UV DDB (19). Further studies have documented that the expression of the V protein of SV5 can slow the progression of the cell cycle, and a role for UV DDB in this process was suggested (20). Furthermore, the hepatitis B virus X protein, which also binds the 127-kDa subunit of UV DDB, also interferes with cell viability (21). It was thus not clear before we started these experiments whether it would be possible to isolate cell lines that express the V protein of SV5 or hPIV2. However, not only was it relatively easy to isolate such cells, but also there was no detectable reduction in the rate of proliferation of 2f/SV5-V (or 2f/PIV2-V) cells compared to parental 2fTGH cells. The reason for this finding remains uncertain, but it may be due to the relatively low levels of V protein expression in 2f/SV5-V cells (at least 20-fold lower than the levels in SV5-infected cells; unpublished results). In transfections with pEFlink plasmids, transient expression levels were more akin to the levels of the V protein observed in SV5-infected cells (data not shown). The studies which documented that the expression of the V protein of SV5 delayed the progression of the cell cycle were performed with transiently transfected cells for which high levels of expression were reported (20). Thus, it cannot be concluded that the results presented here are in conflict with those which demonstrate that the V protein slows the progression of the cell cycle. Indeed, it is possible that cells expressing only low levels of the V protein were selected, as at higher levels, the V protein may have significantly slowed the cell cycle and therefore the rate of cellular proliferation. Nevertheless, the fact that STAT1 was undetectable in 2f/SV5-V cells demonstrates that the ability of the V protein to delay cell cycle progression is not directly linked to the loss of STAT1. Thus, the reason(s) for the large amount of V protein produced in virus-infected cells may relate to its other functions, perhaps in delaying the cell cycle (20) or contributing to the control of virus replication through its interaction with soluble NP (29). Alternatively, the large amount of V protein produced may reflect the need for de novo virus protein synthesis for the degradation of STAT1 in cells already in an antiviral state, i.e., in cells in which there will be significantly reduced initial rates of virus protein synthesis.
The observations that SV5 protein synthesis is not required for input virus to induce the degradation of STAT1 in untreated 2fTGH cells but is required in IFN-pretreated 2fTGH cells (in which there is a significant increase in STAT1 levels) suggest that the degradation of STAT1 is dependent on the relative concentrations of STAT1 and the V protein. Thus, in vivo, during an acute infection, it seems likely that some virus protein synthesis will be required to degrade STAT1 in cells that are in an antiviral state and that have been infected with only small amounts of virus. Furthermore, if the degradation of STAT1 is dependent on the relative concentrations of STAT1 and the V protein, it seems likely that for STAT1 to be degraded, it must form either a direct or an indirect complex with the V protein. Indeed, it has recently been demonstrated that the C protein of SeV is found in high-molecular-weight complexes with STAT1 (30), although the mechanisms of inhibition of IFN signaling for SV5 and SeV appear clearly different. However, even in the presence of proteasome inhbitors and under relatively mild conditions, we failed to show coprecipitation of the V protein of either SV5 or hPIV2 with STAT1 or STAT2, respectively, and the detailed mechanism by which the V protein targets STATs for degradation remains to be elucidated. However, whatever the mechanism for STAT1 degradation by SV5, it is not dependent on IFN signaling, since both phosphorylated and nonphosphorylated forms of STAT1 are degraded. It is also currently unclear which regions of the V protein are responsible for the selective targeting of STAT1 or STAT2 for degradation. Thus, experiments are currently in progress to further define the functional domains of the V protein and their roles in STAT degradation.
Acknowledgments
J. Andrejeva and D. F. Young are supported by grants from the Wellcome Trust and BBSRC, respectively.
REFERENCES
- 1.Choppin, P. W. 1964. Multiplication of a myxovirus (SV5) with minimal cytopathic effects and without interference. Virology 23:224-233. [DOI] [PubMed]
- 2.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. Sendai virus and simian virus 5 block activation of interferon-responsive genes: importance for virus pathogenesis. J. Virol. 73:3125-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. The V protein of simian virus 5 inhibits interferon signaling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 73:9928-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcin, D., P. Latorre, and D. Kolakofsky. 1999. Sendai virus C proteins counteract the interferon-mediated induction of an antiviral state. J. Virol. 73:6559-6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcin, D., J. Curran, and D. Kolakofsky. 2000. Sendai virus C proteins must interact directly with cellular components to interfere with interferon action. J. Virol. 74:8823-8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gil, M. P., E. Bohn, A. K. O'Guin, C. V. Ramana, B. Levine, G. R. Stark, H. W. Virgin, and R. D. Schreiber. 2001. Biological consequences of Stat-1 independent IFN signaling. Proc. Natl. Acad. Sci. USA 98:6680-6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed]
- 8.Gotoh, B., K. Takeuchi, T. Komatsu, J. Yokoo, Y. Kimura, A. Kurotani, A. Kato, and Y. Nagai. 1999. Knockout of the Sendai virus C gene eliminates the viral ability to prevent the interferon-alpha/beta-mediated responses. FEBS Lett. 459:205-210. [DOI] [PubMed] [Google Scholar]
- 9.Kato, A., Y. Ohnishi, M. Kohase, S. Saito, M. Tashiro, and Y. Nagai. 2001. Y2, the smallest of the Sendai virus C proteins, is fully capable of both counteracting the antiviral action of interferons and inhibiting viral RNA synthesis. J. Virol. 75:3802-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawano, M., M. Kato, Y. Kozuka, H. Komada, N. Noda, K. Nanba, M. Trurudome, M. Ito, M. Nishio, and Y. Ito. 2001. Recovery of infectious human parainfluenza virus type 2 from cDNA clones and properties of the defective virus without V-specific cysteine-rich domain. Virology 284:99-112. [DOI] [PubMed] [Google Scholar]
- 11.Kim, T. K., and T. Maniatis. 1996. Regulation of interferon-γ-activated STAT1 by the ubiquitin-proteasome pathway. Science 273:1717-1719. [DOI] [PubMed] [Google Scholar]
- 12.King, P., and S. Goodbourn. 1994. The β-interferon promoter responds to priming through multiple independent regulatory elements. J. Biol. Chem. 269:30609-30615. [PubMed] [Google Scholar]
- 13.King, P., and S. Goodbourn. 1998. STAT1 is inactivated by a caspase. J. Biol. Chem. 273:8699-8704. [DOI] [PubMed] [Google Scholar]
- 14.Komatsu, T., K. Takeuchi, J. Yokoo, Y. Tanaka, and B. Gotoh. 2000. Sendai virus blocks alpha interferon signaling to signal transducers and activators of transcription. J. Virol. 74:2477-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kubota, T., N. Yokosawa, S. Yokota, and N Fujii. 2001. C terminal Cys-RICH region of mumps virus structural V protein correlates with block of interferon alpha and gamma signal transduction pathway through decrease of STAT 1-alpha. Biochem. Biophys. Res. Commun. 283:255-259. [DOI] [PubMed] [Google Scholar]
- 16.Lamb, R. A., and D. Kolakofsky. 2001. Paramyxoviridae: the viruses and their replication, p. 1305-1340. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott-Raven Press, New York, N.Y.
- 17.Lee, C.-K., H. A. R. Bluyssen, and D. E. Levy. 1997. Regulation of interferon-a responsiveness by the duration of Janus kinase activity. J. Biol. Chem. 272:21872-21877. [DOI] [PubMed] [Google Scholar]
- 18.Levy, D. E., and A. Garcia-Sastre. 2001. The virus battles: IFN induction of the antiviral state and the mechanisms of viral evasion. Cytokines Growth Factor Rev. 12:143-156. [DOI] [PubMed] [Google Scholar]
- 19.Lin, G. Y., and R. A. Lamb. 2000. The paramyxovirus simian virus 5 V protein slows progression of the cell cycle. J. Virol. 74:9152-9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin, G. Y., R. G. Paterson, C. D. Richardson, and R. A. Lamb. 1998. The V protein of the paramyxovirus SV5 interacts with damage-specific DNA binding protein. Virology 249:189-200. [DOI] [PubMed] [Google Scholar]
- 21.Lin-Marq, N., S. Bontron, O. Leupin, and M. Strubin. 2001. Hepatitis B virus X protein interferes with cell viability through interactions with the p127-kDA UV-Damaged DNA-Binding Protein. Virology 287:266-274. [DOI] [PubMed] [Google Scholar]
- 22.Masson, N., M. Ellis, S. Goodbourn, and K. A. W. Lee. 1992. Cyclic AMP response-binding protein and the catalytic subunit of protein kinase A are present in F9 embryonal carcinoma cells but are unable to activate the somatostatin promoter. Mol. Cell. Biol. 12:1096-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKendry, R., J. John, D. Flavell, M. Muller, I. M. Kerr, and G. R. Stark. 1991. High-frequency mutagenesis of human cells and characterization of a mutant unresponsive to both alpha and gamma interferons. Proc. Natl. Acad. Sci. USA 88:11455-11459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishio, M., M. Tsurudome, M. Ito, M. Kawano, H. Komada, and Y. Ito. 2001. High resistance of human parainfluenza type 2 virus protein-expressing cells to the antiviral and anti-cell proliferative activities of alpha/beta interferons: cysteine-rich V-specific domain is required for high resistance to the interferons. J. Virol. 75:9165-9176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parisien, J.-P., J. F. Leu, J. J. Rodriguez, B. M. Sullivan, A. Moscona, G. D. Parks, R. A. Lamb, and C. M. Horvath. 2001. The V protein of human parainfluenza virus 2 antagogonizes type I interferon responses by destabilising signal transducer and activator of transcription 2. Virology 283:230-239. [DOI] [PubMed] [Google Scholar]
- 26.Pellegrini, S., J. J. Shearer, I. M. Kerr, and G. R. Stark. 1989. Use of a selectable marker regulated by alpha interferon to obtain mutations in the signaling pathway. Mol. Cell. Biol. 9:4605-4612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Precious, B., J. A. Southern, and R. E. Randall. 1990. Sequence analysis of the HN gene of parainfluenza virus type 2. J. Gen. Virol. 71:1163-1168. [DOI] [PubMed] [Google Scholar]
- 28.Ramana, C. V., M. P. Gil, Y. Han, R. M. Ransohoff, R. D. Schreider, G. R., and Stark. 2001. Stat1-independent regulation of gene expression in response to IFN-γ. Proc. Natl. Acad. Sci. USA 98:6674-6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Randall, R. E., and A. Bermingham. 1996. NP:P and NP:V interactions of the paramyxovirus simian virus 5 examined using a novel protein:protein capture assay. Virology 224:121-138. [DOI] [PubMed] [Google Scholar]
- 30.Randall, R. E., and N. Dinwoodie. l986. Intranuclear localisation of herpes simplex virus immediate-early and delayed-early proteins. Evidence that ICP4 is associated with progeny virus DNA. J. Gen. Virol. 67:2163-2177. [DOI] [PubMed]
- 31.Randall, R. E., and D. F. Young. 1988. Comparison between parainfluenza virus type 2 and simian virus 5: monoclonal antibodies reveal major antigenic differences. J. Gen. Virol. 69:2051-2060. [DOI] [PubMed] [Google Scholar]
- 32.Randall, R. E., D. Young, K. K. A. Goswami, and W. C. Russell. 1987. Isolation and characterisation of monoclonal antibodies to simian virus 5 and their use in revealing antigenic differences between human, canine and simian isolates. J. Gen. Virol. 68:2769-2780. [DOI] [PubMed] [Google Scholar]
- 33.Schlender, J., B. Bossert, U. Buchholz, and K. K. Conzelmann. 2000. Bovine respiratory syncytial virus nonstructural proteins NS1 and NS2 cooperatively antagonize alpha/beta interferon-induced antiviral response. J. Virol. 74:8234-8242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Southern, J. A., B. Precious, and R. E. Randall. 1990. Two nontemplated nucleotide additions are required to generate the P mRNA of parainfluenza type 2 since the RNA genome encodes protein V. Virology 177:388-390. [DOI] [PubMed] [Google Scholar]
- 35.Stark, G. R., I. M. Kerr, B. R. G. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 36.Takeuchi, K., T. Komatsu, J. Yokoo, A. Kato, T. Shioda, Y. Nagai, and B. Gotoh. 2001. Sendai virus C protein physically associates with Stat1. Genes Cells 6:545-557. [DOI] [PubMed] [Google Scholar]
- 37.Vojtesek, B., H. Dolezalova, L. Lauerova, M. Svitakova, P. Havlis, J. Kovarik, C. A. Midgley, and D. P. Lane. 1995. Conformational changes in p53 analysed using new antibodies to the core DNA binding domain of the protein. Oncogene 10:389-393. [PubMed] [Google Scholar]
- 38.Yenofsky, R. L., M. Fine, and J. W. Pellow. 1990. A mutant neomycin phosphotransferase II gene reduces the resistance of transformants to antibiotic selection pressure. Proc. Natl. Acad. Sci. USA 87:3435-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yokosawa, N., T. Kubota, and N. Fujji. 1998. Poor induction of interferon induced 2", 5"-oligoadenylate synthetase (2-5 AS) in cells persistently infected with mumps virus is caused by decrease of STAT-1 alpha. Arch. Virol. 143:1985-1992. [DOI] [PubMed] [Google Scholar]
- 40.Young, D. F., L. Didcock, S. Goodbourn, and R. E. Randall. 2000. Paramyxoviruses utilize different molecular mechanisms to circumvent the interferon response Virology 269:383-390. [DOI] [PubMed] [Google Scholar]
- 41.Young, D. F., N. Chatziandreou, B. He, S. Goodbourn, R. A. Lamb, and R. E. Randall. 2001. A single amino acid substitution in the V protein of simian virus 5 differentiates its ability to block interferon signaling in human and murine cells. J. Virol. 75:3363-3370. [DOI] [PMC free article] [PubMed] [Google Scholar]