Abstract
Hepatitis B virus (HBV) X gene encodes a multifunctional protein that can regulate cellular signaling pathways, interact with cellular transcription factors, and induce hepatocellular oncogenesis. In spite of its diverse activities, the precise role of the X protein in the viral life cycle of HBV remains unclear. To investigate this question, we have produced transgenic mice that carry either the wild-type HBV genome or a mutated HBV genome incapable of expressing the 16.5-kDa X protein. Our results indicate that while the X protein is not absolutely essential for HBV replication or its maturation in transgenic mice, it can enhance viral replication, apparently by activating viral gene expression. These results demonstrate a transactivation role of the X protein in HBV replication in transgenic mice.
Hepatitis B virus (HBV) is a human pathogen that can cause acute and chronic hepatitis and also hepatocellular carcinoma. This virus is a small DNA virus that belongs to the hepadnavirus family. This family contains a group of closely related viruses that infect primarily the livers of their respective animal hosts. The HBV genome is a 3.2-kb DNA molecule that contains four genes named C, S, P, and X (13). The C gene codes for the core protein and the serum e antigen, the S gene codes for three related viral envelope proteins known as surface antigens, the P gene codes for the viral DNA polymerase, and the X gene codes for a 16.5-kDa protein.
The HBV X protein has many activities in vitro (38). It can enhance the expression of RNA polymerase I-, II-, and III-dependent genes through multiple pathways (31, 33, 35, 36). The X protein does not bind to DNA directly. However, it can bind to different transcription factors, including AP1, AP2, ATF-2, CREB, TBP, TFIIB, and TFIIH, to modify their activities (2, 16, 21, 23, 26, 30). It also binds to RBP5, a subunit of all three mammalian RNA polymerases (7, 21), to the proteasome, p53, a DNA repair protein UV-DDB, and a member of the human voltage-dependent ion channel family HVDAC3 (3, 12, 17, 27, 37). The X protein can also activate the ras-raf-mitogen-activated protein kinase signaling pathway and the NF-κB pathway (4, 9, 19, 22, 24, 34). These activities are thought to be important for the X protein to induce oncogenesis and apoptosis in cell cultures and in transgenic mice (8, 10, 18, 25, 32).
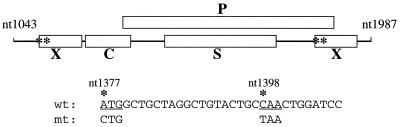
In spite of its diverse activities in cultured cells, the role of the X protein in the HBV life cycle remains a mystery. To investigate its possible functions, we produced transgenic mice that carried either the wild-type HBV genome or a mutated HBV genome that expresses all viral proteins except the X protein. The HBV genomic DNA fragment used for producing the transgenic mice is illustrated in Fig. 1. This DNA fragment starts from nucleotide (nt) 1043, which is located upstream of the ENI enhancer and the X promoter, and terminates at nt 1987, which is located downstream of the unique poly(A) site. This DNA fragment is approximately 1.3 times the size of the HBV genome and has been used to generate transgenic mouse lines that produce high HBV titers (14). To abolish the expression of the X protein, an A-to-C mutation was introduced at nt 1377 to remove the initiation codon of the X protein and a C-to-T mutation was introduced at nt 1398 to introduce a premature termination codon in the X coding sequence. Neither of these two mutations affects the coding sequence of the overlapping polymerase gene. Due to the genomic structure of HBV, the X gene was present in two copies in this DNA fragment, and both copies were mutated to abolish the expression of the X protein. Both the wild-type and the mutant DNA fragments inserted into the pUC19 vector were found to direct the transcription of HBV RNAs and the replication of HBV DNA with no significant difference in Huh7 cells, a well-differentiated human hepatoma cell line (data not shown). These results were consistent with previous reports (5, 40) which indicated that the X gene had no effect on HBV replication in Huh7 cells. These two HBV DNA fragments were then injected into fertilized mouse embryos for the production of transgenic mice.
FIG. 1.
Illustration of the adw2 HBV genome used for the production of transgenic mice. C, S, P, and X indicate the four different HBV genes. The HBV DNA fragment, which starts from the EcoRV site at nt 1043 and ends at the BglII site at nt 1987, contains all of the genomic information of the HBV genome plus a terminal redundancy of nearly 1 kb. This fragment was isolated and inserted into the SmaI/BamHI site of the pUC19 vector. The 4.2-kb PvuII fragment was then isolated from the resulting DNA plasmid for microinjection for the production of the transgenic mice. Asterisks indicate the locations of nt 1377 and 1398. These are the two nucleotides that were mutated to prevent the expression of the 16.5-kDa X protein. The wild-type HBV sequence and the mutated codons are also shown.
The mouse litters produced were analyzed by genomic Southern blotting for the identification of the transgenic mice. The positive mice were further analyzed for the presence of HBV virions in their sera by using the endogenous polymerase assay (EPA). In all of the EPA reactions, the nonionic detergent Nonidet P-40 (NP-40) was included in the reaction mixture to remove the envelope from the HBV virion. Since the omission of NP-40 generated negative EPA results (data not shown), the HBV signals detected by this assay could not be due to the core particles released from injured hepatocytes. Based on these screening procedures, 6 of the 12 litters were found to carry the wild-type HBV genome (1.3HBVwt mice), and among these 6 litters, 3 produced circulating HBV virions. Also, 10 of the 15 litters screened were found to carry the mutated HBV genome (1.3HBVmt mice), and among these 10 litters, 5 were found to produce circulating HBV virions. The ability of five independent 1.3HBVmt F0 mice to produce circulating HBV virions indicates that an intact X gene is not essential for HBV replication and maturation in this mouse model.
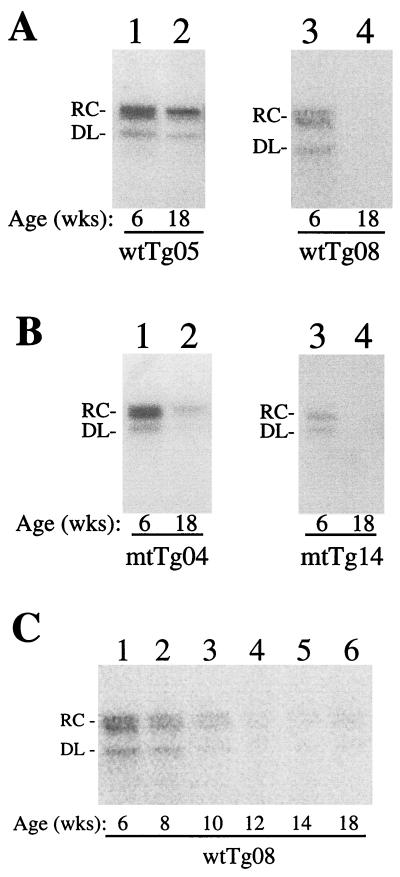
wtTg05 and wtTg08 are two different 1.3HBVwt mouse lines established from two founder mice. The former has a higher circulating HBV level than the latter when age- and sex-matched mice of these two lines were compared. By performing the dot blot analysis with cloned HBV genomic DNA as the standard, the homozygous wtTg05 mouse was found to carry two copies of the HBV genome and the wtTg08 mouse was found to carry six copies (data not shown). mtTg04 and mtTg14 are two different 1.3HBVmt transgenic mouse lines. The former also has a higher circulating HBV level than the latter. Similar dot blot analysis revealed that the homozygous mtTg04 mouse carried eight copies of the HBV genome and the mtTg14 mouse carried four copies. The viral titers of 6- and 18-week-old mice of these representative transgenic mouse lines are compared in Fig. 2A and B, and a more detailed time-point analysis of the viral titers of the wtTg08 mouse line is shown in Fig. 2C. The viral titers in all of the transgenic mice were found to decrease, albeit to different degrees, in an age-dependent manner (Fig. 2). This age-dependent reduction of the viral titers may be due to the methylation of the HBV transgene (1, 29) and/or by the development of anti-HBV antibodies, although the latter possibility appears to be unlikely, based on Northern blotting results (data not shown) (see below). At 6 weeks of age, the circulating viral titers of mtTg04 and wtTg05 male mice were similar and, as determined by the dot blot, were about 3 × 108 genome equivalents per ml of serum. In these and all the subsequent experiments, two to three mice were analyzed per time point to ensure the reproducibility of the results.
FIG. 2.
EPA of circulating HBV virions in transgenic mice. One microliter of mouse serum was mixed with 50 μl of EPA buffer (50 mM Tris-HCl, pH 7.5, 40 mM NH4Cl, 20 mM MgCl2, 1% NP-40, and 0.3% β-mercaptoethanol) containing 50 μCi of [α-32P]dCTP (>3,000 Ci/mmol; ICN) and 0.1 mM (each) dATP, dGTP, and dTTP. The EPA reaction was carried out at 37°C for 2 h and chased with 0.1 mM nonlabeled dCTP at the same temperature for 1 h. The reaction was stopped by the addition of EDTA to a final concentration of 2.5 mM, sodium dodecyl sulfate to 1%, proteinase K to 500 μg/ml, and 50 μg of tRNA carrier. The sample was further incubated at 55°C for 2 h. Afterwards, the HBV DNA was extracted with phenol, ethanol precipitated twice in the presence of 2 M ammonium acetate, resuspended in Tris-EDTA, and electrophoresed on a 1% agarose gel followed by autoradiography, which was analyzed by a PhosphorImager (Molecular Dynamics, Sunnyville, California). (A) wtTg05 and wtTg08 transgenic mouse lines that carried the 1.3HBVwt genome. (B) mtTg04 and mtTg14 transgenic mouse lines that carried the 1.3HBVmt genome. (C) Age-dependent reduction of viral titers in the wtTg08 mouse line. Each set of serum samples was collected from the same mouse at the ages indicated under the gels. Wks, weeks of age; RC, relaxed circular form of the HBV genome; DL, double-stranded linear form of the HBV genome. The DL DNA has an electrophoretic mobility similar to that of the 3.2-kb HBV DNA fragment (compare with Fig. 5B).
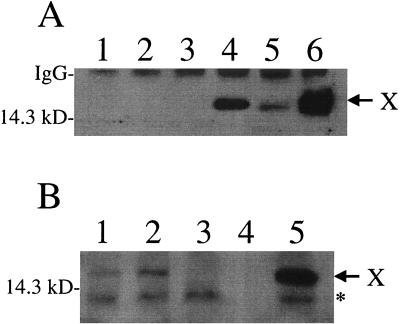
To ensure that the X protein was indeed expressed in wtTg05 and wtTg08 mice and not in mtTg04 and mtTg14 mice, we analyzed the expression of the X protein in the liver of these mice by performing Western blot analysis. As shown in Fig. 3A, the 16.5-kDa X protein band could clearly be detected in the liver homogenates of wtTg05 and wtTg08 mice. This is the first report in which the expression of X protein has been confirmed in the livers of HBV full-genome transgenic mice. In contrast, this X protein could not be detected in the liver homogenates of mtTg04 and mtTg14 mice in the same experiments, indicative of its lack of expression in these mice.
FIG. 3.
Western blot analysis for the expression of the X protein in transgenic mice. Tissue extracts were prepared by homogenizing liver tissue in extraction buffer (50 mM Tris-HCl, pH 8.0; 100 mM NaCl; and 1% NP-40). Immunoprecipitation and Western blot analysis were used to verify expression of the X protein as described previously (30). Briefly, following electrophoresis on 15% sodium dodecyl sulfate-polyacrylamide gels, the separated proteins were transferred to nitrocellulose filters. The presence or absence of the HBV X protein was verified using rabbit anti-HBx polyclonal serum, an avidin-biotin detection kit (Vector Laboratories) and enhanced chemiluminescence (Amersham/Pharmacia). (A) X protein immunoprecipitated from 2 mg of total lysates. Lane 1, nontransgenic control; lane 2, mtTg04; lane 3, mtTg14; lane 4, wtTg05; lane 5, wtTg08; lane 6, transgenic mice harboring the X gene under the control of the human α-1-antitrypsin inhibitor regulatory region (20, 32). All of the mouse liver tissues were analyzed at 6 weeks of age except the tissue in lane 5, which was analyzed at 18 weeks of age. (B) X protein immunoprecipitated from 4 mg of total lysates. Lane 1, T22 X; lane 2, mtTg04/X; lane 3, mtTg04; lane 4, blank; lane 5, wtTg05. The migration of the 14.3-kDa molecular mass marker is shown at the left, and the arrow at the right identifies the migration of the X protein. The asterisk identifies a nonspecific cellular protein seen in all samples in which 4 mg of protein was used for immunoprecipitation.
Overall, there is a large variation of circulating HBV titers among different mouse lines carrying the same transgene, be it the wild-type or the mutated HBV genome. This variation may be caused by the number of HBV DNA copies that integrated into the mouse chromosomes and/or by the locations of their integration sites. Consequently, the effect of the X protein on HBV replication cannot be determined by simply comparing the serum viral titers between different transgenic mouse lines.
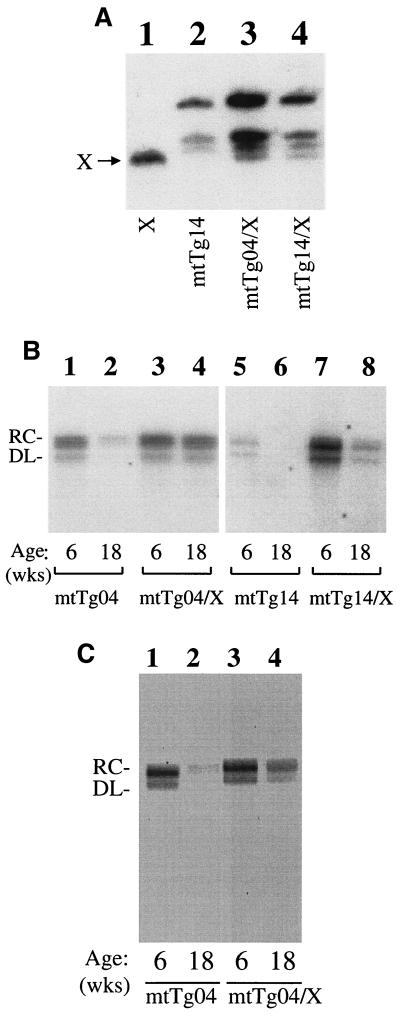
To take a different approach to investigate the possible functions of the X gene in HBV replication, we performed transcomplementation experiments. Kim et al. (18) previously produced a transgenic mouse line that carried only the HBV X gene. The expression of the X gene in their mouse line, named T22, is under the control of its own promoter. This T22 mouse line was crossed to the mtTg04 mouse line to generate crossbred mice that carried both the X gene and the 1.3HBVmt genome. The expression of the X protein in the crossbred mice was again analyzed by Western blotting. As shown in Fig. 3B, although the X protein was not detected in the mtTg04 mice, it was detected in the mtTg04/X crossbred mouse and the T22 X transgenic mouse. We had also performed the genomic Southern blotting experiment. As shown in Fig. 4A, the crossing of the T22 mouse line to the mtTg04 line generated the mtTg04/X crossbred mice that carried the transgenes of both parental mouse lines. Similar results were also obtained when the T22 mouse line was crossed with the mtTg14 mouse line. This successful expression of the X protein in the crossbred mice allowed us to study the possible effects of the X protein on HBV replication. HBV titers in the crossbred mice and their respective age- and sex-matched X-negative mice were then analyzed.
FIG. 4.
Transcomplementation of the 1.3HBVmt genome with the X gene in transgenic mice. (A) Genomic Southern blotting of the transgenic mice. Tails were cut from 6-week-old mice and digested with proteinase K (500 μg/ml) at 55°C overnight in a buffer containing 20 mM Tris-HCl, pH 7.5, 0.5% sodium dodecyl sulfate, 2 mM EDTA, and 400 mM NaCl. The reaction was stopped by phenol extraction, and the genomic DNA was precipitated by ethanol. The DNA was resuspended in Tris-EDTA (10 mM Tris-HCl, pH 7.0, 1 mM EDTA), digested with BamHI, and Southern blotted with the 32P-labeled 3.2-kb HBV genomic DNA as the probe. Lane 1, T22 mouse line that contains the X gene; lane 2, the mtTg14 mouse line; lane 3, the mtTg04/X crossbred mouse; and lane 4, the mtTg14/X crossbred mouse. The arrow denotes the location of the X gene from the T22 mouse line. (B and C) EPA of the circulating HBV virions. (B) Lanes 1 and 2, mtTg04; lanes 3 and 4, mtTg04/X crossbred mice; lanes 5 and 6, mtTg14; lanes 7 and 8, mtTg14/X crossbred mice. (C) Lanes 1 and 2, mtTg04 mice produced by crossing mtTg04 in the B6D2F1 background to CD1 nontransgenic mice; lanes 3 and 4, mtTg04/X mice. The ages of the mice in weeks (wks) are indicated below the gels. The EPA was conducted as described in the legend to Fig. 2.
As shown in Fig. 4B, when mtTg04 and mtTg04/X mice were analyzed, the mtTg04 mice had a slightly lower viral titer than the mtTg04/X mice at the age of 6 weeks. mtTg04 mice had a viral titer that was approximately fourfold lower than that of mtTg04/X mice when the mice were 18 weeks old. This result indicates that the X gene provided in trans can increase the viral titer in mtTg04 mice at 18 weeks of age. As the T22 X transgenic mouse line was derived from the CD1 mouse strain and maintained in this genetic background, the mtTg04/X hybrid mice would contain a partial CD1 genetic background and a partial B6D2F1 background. Thus, it is conceivable that the observed increase in the viral amount in the hybrid mice was caused by the CD1 genetic background and not by the X protein. To rule out this possibility, we crossed nontransgenic CD1 mice to mtTg04 mice and compared the viral titer in the offspring mice with the mtTg04/X mice, which would now have a similar contribution of the CD1 genetic background. As shown in Fig. 4C, the mtTg04/X mice again had a three- to fourfold-higher circulating viral titer than the mtTg04 mice when the mice were 18 weeks old. Thus, the CD1 background did not appear to play any significant role in HBV production in transgenic mice.
When mtTg14 and mtTg14/X mice were analyzed, there was an approximately fivefold increase of the HBV titer in the mtTg14/X crossbred mice when they were 6 weeks old (Fig. 4B). When the mice were 18 weeks old, the HBV titer was undetectable in mtTg14 mice but was clearly detectable in mtTg14/X mice. These results indicate that the X gene provided in trans could also increase the HBV titer in mtTg14 mice whether the mice were 6 or 18 weeks old.
Thus, the results shown in Fig. 4 indicate that the X gene provided in trans could increase the serum viral titer in two independent mouse lines carrying the 1.3HBVmt genome. The lack of a significant induction of the HBV titer in mtTg04 mice by the X gene at 6 weeks of age was probably due to the high basal viral titer in mtTg04 mice at this age.
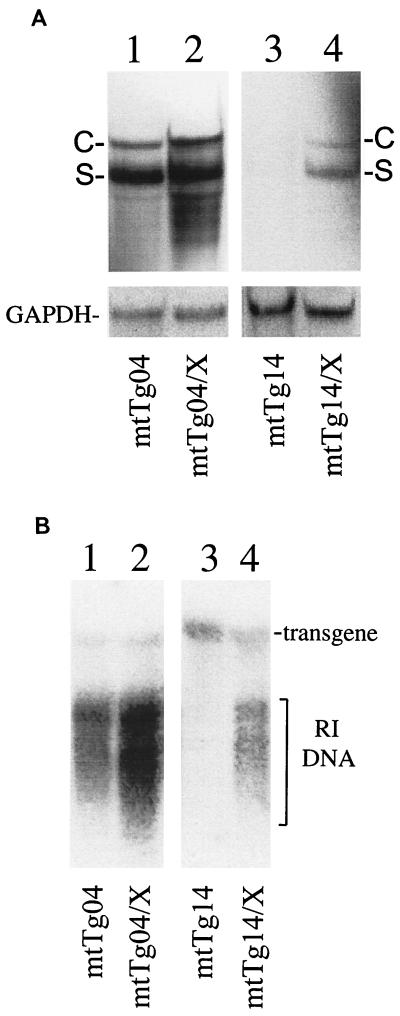
To understand how the X gene provided in trans increased the circulating HBV titer, total liver RNA was isolated from mtTg04 and mtTg14 mice and their respective crossbred progeny and analyzed by Northern blotting. For simplicity, only 18-week-old mice were analyzed because the increase of the viral titer by the X gene was apparent for both mouse lines at this age. As shown in Fig. 5A, the presence of the X gene increases the level of the C gene transcripts approximately threefold in mtTg04 mice. The X gene had no apparent effect on the RNA level of the S gene in this mouse line. This might be due to the higher basal level of the S RNA in this mouse line as the X gene increased significantly both the C RNA and the S RNA levels in the mtTg14 mouse line (Fig. 5A).
FIG. 5.
Enhancement of viral RNA expression and DNA replication in 1.3HBVmt mice by the X gene. (A) Northern blot analysis. To isolate RNA from mouse liver tissues, tissues were homogenized in Trizol (Life Tech), and the RNA was extracted following the manufacturer's instructions. The RNA was then subjected to electrophoresis on a 1% formaldehyde agarose gel, Northern blotted to a nitrocellulose membrane, and hybridized to the 32P-labeled 3.2-kb HBV DNA probe. C and S are HBV C and S gene transcripts, respectively. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) RNA was also analyzed to serve as an internal control. Minor HBV RNA bands smaller than the S gene transcripts were also detected when the mtTg04 mice were analyzed. These minor bands were likely degradation products as they were not always detected in other experiments. (B) Southern blot analysis. The total liver DNA was extracted and analyzed by Southern blotting as described in the legend to Fig. 4, except that the step of BamHI digestion was omitted. The locations of the transgene and the RI DNA are marked. The transgene serves as an internal loading control.
To further investigate the effect of the X gene on viral DNA replication, Southern blotting was performed to analyze the HBV DNA in the livers of the transgenic mice. As shown in Fig. 5B, the X gene increased the level of the HBV replicative intermediate (RI) DNA in mtTg04 mice approximately threefold. This level of increase was close to the level of increase of the viral titers observed in the results shown in Fig. 4. For the mtTg14 mice, the HBV RI DNA was almost undetectable without the X gene but it was clearly detectable in the presence of the X gene, again consistent with the viral titer results shown in Fig. 4.
Thus, the results shown in Fig. 5 indicate that the HBV X gene can increase the HBV RNA and RI DNA levels. The magnitude of this increase is sufficient to explain the observed increase of the circulating viral titers.
In the absence of a convenient animal model system for studying HBV replication, the transgenic mouse provides an alternative choice. By using this system, we found that the X protein is not absolutely essential for HBV replication, but replication is significantly decreased in the absence of X protein. By providing the X gene in trans to the 1.3HBVmt mice, we found that the X protein could increase the viral RNA and DNA levels in liver tissues and the amount of HBV in the serum. Reifenberg et al. (28) found that the X gene provided in trans could stimulate the expression of the C gene when they studied transgenic mice carrying subgenomic HBV DNA. Our results are consistent with these findings and further extend them by showing the activation of HBV gene expression and replication in the context of the entire viral genome. Our results also indicate that, in the context of the whole genome, the X gene can stimulate not only the expression of the C gene but also the expression of the S gene.
The finding that the X protein is not essential for HBV replication in transgenic mice is consistent with the finding that HBV mutants incapable of producing the intact X protein could be isolated from HBV patients (11, 15). However, woodchuck hepatitis virus (WHV) with mutations that abolished the expression of the X gene failed to initiate infection in woodchucks when the DNA was introduced into the animals (6, 40). It is possible that WHV is different from HBV or the mouse provides a factor that may partially replace the function of the X protein. Alternatively, it is tempting to speculate that the apparent inability of the X-negative WHV DNA to initiate infection in woodchucks is due to its lower replication rate. During the natural infection in animals, even a modest difference in the replication rate may be amplified by multiple rounds of infection to generate a profound difference in the amount of progeny viral particles produced. This may cause the X-negative WHV to be eliminated rapidly by the immune system. This speculation is supported by a recent observation of Z. Zhang and colleagues, who found that WHV defective in the X gene may replicate at a low level in vivo (39). Nevertheless, it should be noted that the transgenic mouse model is an imperfect model system and does not allow us to examine the initial infection step of the viral life cycle. Thus, it remains a possibility that the X protein may also be needed for initiating infection.
Acknowledgments
We thank Frank Chisari, Luca Guidotti, and Debbie Johnson for helpful discussions during the course of this research. We are also indebted to Rob Maxson and Frank Sangiorgi at the USC Norris Cancer Center Transgenic Mouse Core Facility for the production of the transgenic mice.
This research was supported by research grants from the National Institutes of Health and Department of Veterans Affairs.
REFERENCES
- 1.Araki, K., K. Akagi, J. Miyazaki, K. Matsubara, and K. Yamamura. 1990. Correlation of tissue-specific methylation with gene inactivity in hepatitis B virus transgenic mice. Jpn. J. Cancer Res. 81:1265-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnabas, S., and O. M. Andrisani. 2000. Different regions of hepatitis B virus X protein are required for enhancement of bZip-mediated transactivation versus transrepression. J. Virol. 74:83-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker, S. A., T. H. Lee, J. S. Butel, and B. L. Slagle. 1998. Hepatitis B virus X protein interferes with cellular DNA repair. J. Virol. 72:266-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benn, J., and R. J. Schneider. 1994. Hepatitis B virus HBx protein activates Ras-GTP complex formation and establishes a Ras, Raf, MAP kinase signaling cascade. Proc. Natl. Acad. Sci. USA 91:10350-10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blum, H. E., Z. S. Zhang, E. Galun, F. von Weizsacker, B. Garner, T. J. Liang, and J. R. Wands. 1992. Hepatitis B virus X protein is not central to the viral life cycle in vitro. J. Virol. 66:1223-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, H. S., S. Kaneko, R. Girones, R. W. Anderson, W. E. Hornbuckle, B. C. Tennant, P. J. Cote, J. L. Gerin, R. H. Purcell, and R. H. Miller. 1993. The woodchuck hepatitis virus X gene is important for establishment of virus infection in woodchucks. J. Virol. 67:1218-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheong, J. H., M. Yi, Y. Lin, and S. Murakami. 1995. Human RPB5, a subunit shared by eukaryotic nuclear RNA polymerases, binds human hepatitis B virus X protein and may play a role in X transactivation. EMBO J. 14:143-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chirillo, P., S. Pagano, G. Natoli, P. L. Puri, V. L. Burgio, C. Balsano, and M. Levrero. 1997. The hepatitis B virus X gene induces p53-mediated programmed cell death. Proc. Natl. Acad. Sci. USA 94:8162-8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doria, M., N. Klein, R. Lucito, and R. J. Schneider. 1995. The hepatitis B virus HBx protein is a dual specificity cytoplasmic activator of Ras and nuclear activator of transcription factors. EMBO J. 14:4747-4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elmore, L. W., A. R. Hancock, S. F. Chang, X. W. Wang, S. Chang, C. P. Callahan, D. A. Geller, H. Will, and C. C. Harris. 1997. Hepatitis B virus X protein and p53 tumor suppressor interactions in the modulation of apoptosis. Proc. Natl. Acad. Sci. USA 94:14707-14712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feitelson, M., L. Lega, J. Guo, M. Resti, M. E. Rossi, C. Azzari, B. S. Blumberg, and A. Vierucci. 1994. Pathogenesis of posttransfusion viral hepatitis in children with beta-thalassemia. Hepatology 19:558-568. [DOI] [PubMed] [Google Scholar]
- 12.Fischer, M., L. Runkel, and H. Schaller. 1995. HBx protein of hepatitis B virus interacts with the C-terminal portion of a novel human proteasome alpha-subunit. Virus Genes 10:99-102. [DOI] [PubMed] [Google Scholar]
- 13.Ganem, D. 1997. Hepadnaviridae: the viruses and their replication, p. 2703-2737. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology. Lippincott-Raven Publishers, Philadelphia, Pa.
- 14.Guidotti, L. G., B. Matzke, H. Schaller, and F. V. Chisari. 1995. High-level hepatitis B virus replication in transgenic mice. J. Virol. 69:6158-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gunther, S., N. Piwon, A. Iwanska, R. Schilling, H. Meisel, and H. Will. 1996. Type, prevalence, and significance of core promoter/enhancer II mutations in hepatitis B viruses from immunosuppressed patients with severe liver disease. J. Virol. 70:8318-8331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haviv, I., M. Shamay, G. Doitsh, and Y. Shaul. 1998. Hepatitis B virus pX targets TFIIB in transcription coactivation. Mol. Cell. Biol. 18:1562-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang, J., J. Kwong, E. C. Sun, and T. J. Liang. 1996. Proteasome complex as a potential cellular target of hepatitis B virus X protein. J. Virol. 70:5582-5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, C. M., K. Koike, I. Saito, T. Miyamura, and G. Jay. 1991. HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature 351:317-320. [DOI] [PubMed] [Google Scholar]
- 19.Klein, N. P., M. J. Bouchard, L. H. Wang, C. Kobarg, and R. J. Schneider. 1999. Src kinases involved in hepatitis B virus replication. EMBO J. 18:5019-5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, T. H., M. J. Finegold, R. F. Shen, J. L. DeMayo, S. L. Woo, and J. S. Butel. 1990. Hepatitis B virus transactivator X protein is not tumorigenic in transgenic mice. J. Virol. 64:5939-5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin, Y., T. Nomura, J. Cheong, D. Dorjsuren, K. Iida, and S. Murakami. 1997. Hepatitis B virus X protein is a transcriptional modulator that communicates with transcription factor IIB and the RNA polymerase II subunit 5. J. Biol. Chem. 272:7132-7139. [DOI] [PubMed] [Google Scholar]
- 22.Lucito, R., and R. J. Schneider. 1992. Hepatitis B virus X protein activates transcription factor NF-κB without a requirement for protein kinase C. J. Virol. 66:983-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maguire, H. F., J. P. Hoeffler, and A. Siddiqui. 1991. HBV X protein alters the DNA binding specificity of CREB and ATF-2 by protein-protein interactions. Science 252:842-844. [DOI] [PubMed] [Google Scholar]
- 24.Natoli, G., M. L. Avantaggiati, P. Chirillo, P. L. Puri, A. Ianni, C. Balsano, and M. Levrero. 1994. Ras- and Raf-dependent activation of c-jun transcriptional activity by the hepatitis B virus transactivator pX. Oncogene 9:2837-2843. [PubMed] [Google Scholar]
- 25.Pollicino, T., O. Terradillos, H. Lecoeur, M. L. Gougeon, and M. A. Buendia. 1998. Pro-apoptotic effect of the hepatitis B virus X gene. Biomed. Pharmacother. 52:363-368. [DOI] [PubMed] [Google Scholar]
- 26.Qadri, I., H. F. Maguire, and A. Siddiqui. 1995. Hepatitis B virus transactivator protein X interacts with the TATA-binding protein. Proc. Natl. Acad. Sci. USA 92:1003-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rahmani, Z., K. W. Huh, R. Lasher, and A. Siddiqui. 2000. Hepatitis B virus X protein colocalizes to mitochondria with a human voltage-dependent anion channel, HVDAC3, and alters its transmembrane potential. J. Virol. 74:2840-2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reifenberg, K., H. Wilts, J. Lohler, P. Nusser, R. Hanano, L. G. Guidotti, F. V. Chisari, and H. J. Schlicht. 1999. The hepatitis B virus X protein transactivates viral core gene expression in vivo. J. Virol. 73:10399-10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schweizer, J., P. Valenza-Schaerly, F. Goret, and C. Pourcel. 1998. Control of expression and methylation of a hepatitis B virus transgene by strain-specific modifiers. DNA Cell Biol. 17:427-435. [DOI] [PubMed] [Google Scholar]
- 30.Seto, E., P. J. Mitchell, and T. S. Yen. 1990. Transactivation by the hepatitis B virus X protein depends on AP-2 and other transcription factors. Nature 344:72-74. [DOI] [PubMed] [Google Scholar]
- 31.Seto, E., T. S. Yen, B. M. Peterlin, and J. H. Ou. 1988. Trans-activation of the human immunodeficiency virus long terminal repeat by the hepatitis B virus X protein. Proc. Natl. Acad. Sci. USA 85:8286-8290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Slagle, B. L., T. H. Lee, D. Medina, M. J. Finegold, and J. S. Butel. 1996. Increased sensitivity to the hepatocarcinogen diethylnitrosamine in transgenic mice carrying the hepatitis B virus X gene. Mol. Carcinog. 15:261-269. [DOI] [PubMed] [Google Scholar]
- 33.Spandau, D. F., and C.-H. Lee. 1988. trans-activation of viral enhancers by the hepatitis B virus X protein. J. Virol. 62:427-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Twu, J. S., C. A. Rosen, W. A. Haseltine, and W. S. Robinson. 1989. Identification of a region within the human immunodeficiency virus type 1 long terminal repeat that is essential for transactivation by the hepatitis B virus gene X. J. Virol. 63:2857-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, H. D., A. Trivedi, and D. L. Johnson. 1997. Hepatitis B virus X protein induces RNA polymerase III-dependent gene transcription and increases cellular TATA-binding protein by activating the Ras signaling pathway. Mol. Cell. Biol. 17:6838-6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, H. D., A. Trivedi, and D. L. Johnson. 1998. Regulation of RNA polymerase I-dependent promoters by the hepatitis B virus X protein via activated Ras and TATA-binding protein. Mol. Cell. Biol. 18:7086-7094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, X. W., K. Forrester, H. Yeh, M. A. Feitelson, J. R. Gu, and C. C. Harris. 1994. Hepatitis B virus X protein inhibits p53 sequence-specific DNA binding, transcriptional activity, and association with transcription factor ERCC3. Proc. Natl. Acad. Sci. USA 91:2230-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yen, T. S. B. 1996. Hepadnaviral X protein: review of recent progress. J. Biomed. Sci. 3:20-30. [DOI] [PubMed] [Google Scholar]
- 39.Zhang, Z., N. Torri, Z. Hu, J. Jacob, and T. J. Liang. 2001. X-deficient woodchuck hepatitis virus mutants behave like attenuated viruses and induce protective immunity in vivo. J. Clin. Investig. 108:1523-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zoulim, F., J. Saputelli, and C. Seeger. 1994. Woodchuck hepatitis virus X protein is required for viral infection in vivo. J. Virol. 68:2026-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]