Abstract
The full-length genome of human cytomegalovirus strain AD169 was cloned as an infectious bacterial artificial chromosome (BAC) plasmid, pAD/Cre. The BAC vector, flanked by LoxP sites, was inserted immediately after the Us28 open reading frame without deletion of any viral sequences. The BAC vector contained the Cre recombinase-encoding gene disrupted by an intron under control of the simian virus 40 early promoter. When pAD/Cre was transfected into primary human foreskin fibroblast cells, Cre was expressed and mediated site-specific recombination between the two LoxP sites, excising the BAC DNA backbone. This gave rise to progeny virus that was wild type with the exception of an inserted 34-bp LoxP site. We performed site-directed mutagenesis on pAD/Cre to generate a series of viruses in which the TRL/IRL13 diploid genes were disrupted and subsequently repaired. The mutants reach the same titer as the wild-type virus, indicating that the TRL/IRL13 open reading frames are not required for virus growth in cell culture. The sequence of the TRL13 open reading frame in the low-passage Toledo strain of human cytomegalovirus is quite different from the corresponding region in the AD169 strain. One of multiple changes is a frameshift mutation. As a consequence, strain Toledo encodes a putative TRL13 protein whose C-terminal domain is larger (extending through the TRL14 coding region) and encodes in a reading frame different from that of strain AD169. We speculate that the strain AD169 coding region has drifted during passage in the laboratory. We propose that TRL13 has been truncated in strain AD169 and that the partially overlapping TRL14 open reading frame is not functional. This view is consistent with the presence of both TRL13 and -14 on all mRNAs that we have mapped from this region, an organization that would include the much longer strain Toledo TRL13 open reading frame on the mRNAs.
Human cytomegalovirus (HCMV) is one of the eight herpesviruses known to infect humans. It is a ubiquitous pathogen that can cause a variety of diseases. Although HCMV infection is asymptomatic in healthy individuals, the virus can cause serious disease in immunocompromised individuals and unborn children (5).
The molecular functions of most HCMV genes are not well understood, and to a great extent, this results from the difficulty in constructing HCMV mutants. Recently, several herpesviruses, including HCMV (3, 14), mouse CMV (16, 28), pig CMV (15), mouse gammaherpesvirus 68 (1), pseudorabies virus (PRV) (22, 23), Epstein-Barr virus (10), and herpes simplex virus type 1 (13, 21), have been cloned into an F plasmid as infectious bacterial artificial chromosomes (BACs). In each case, infectious virus is generated upon delivery of the BAC plasmid into mammalian cells that support viral replication. In the earlier HCMV BACs, portions of the viral genome, Us2 to Us6 (3) or Us1 to Us12 (14), were deleted to accommodate insertion of the BAC vector. Although this region is not required for HCMV replication in cultured cells, the recombinant viruses generated from these BACs are not genotypically full length. Recently, a novel approach was described for construction of a BAC plasmid containing the PRV genome (23). It does not contain a deletion of the viral sequence, and the BAC vector DNA is excised from the viral genome when the BAC plasmid enters mammalian cells.
Employing a strategy similar to that used to create the PRV BAC, we cloned the full-length genome of HCMV strain AD169 into a BAC vector to generate pAD/Cre. The BAC sequence is inserted following the Us28 open reading frame (ORF) with no viral sequence deletion. The BAC includes an intron containing Cre-coding regions. Cre recombinase is produced, and the BAC vector DNA is excised by site-specific Cre/LoxP recombination upon introduction of the BAC DNA into human cells, generating a wild-type virus with only a 34-bp LoxP site following the Us28 gene.
We used this BAC clone to construct HCMV mutants lacking one or both copies of a diploid viral gene. The AD169 genome contains identical TRL13 and IRL13 ORFs that reside within a repeated region of the viral genome (TRL, terminal repeat long; IRL, internal repeat long). The TRL/IRL13 mRNA, whose synthesis is first detected during the early phase of infection and is expressed at high levels during the late phase of infection (7), was recently shown to be one of a set of five virus-encoded mRNAs that are packaged into HCMV particles (4). Although the presence of this mRNA in virions implies an important role for the RNA or its encoded 147-amino-acid protein (9) early during infection, no function has been described for this ORF. A mutant virus lacking one copy of the ORF within a larger deletion (TRL4-14) is viable in cultured cells (24, 25), demonstrating that one copy of the gene is sufficient to support viral replication. In this study, we deleted both TRL13 and IRL13 and demonstrated that they are not essential for virus growth in cell culture.
To complete our initial characterization of this diploid locus, we mapped the transcripts encoding TRL13 and IRL13 and determined that TRL/IRL13 encodes a protein. Finally, we observed that the TRL13 sequence is quite different in the low-passage Toledo strain compared to that in the AD169 laboratory strain of HCMV.
MATERIALS AND METHODS
Cells and viruses.
Primary human foreskin fibroblast (HFF) cells at passages 8 to 15 were propagated in Dulbecco's modified Eagle medium with 10% fetal calf serum at 37°C with a 5% CO2 atmosphere. HCMV strain AD169 was used as the wild-type virus and the parental virus for all of the recombinant viruses constructed in this study. Strain AD169 and its recombinant derivatives were propagated in HFF cells. Viral titers were determined in duplicate by plaque assay with HFF cells.
Plasmids.
Recombination plasmid YD-C29 was derived from pMBO131, a mini-F plasmid that has been described previously (18). In YD-C29, pMBO131 vector DNA, together with the GFP/Puro expression cassette, was flanked by two LoxP sites and inserted right after the Us28 ORF (Fig. 1). YD-C29 was constructed as follows. The 1.4-kb Us29 sequence was PCR amplified by using primers YD-Pri13 (5"-GGCTCGAGGCGCGCCTTAATTAATAACTTCGTATAGCATACATTATACGAAGTTAT GCTTTTCGCGTCGGAGCT-3"; including sites for XhoI, AscI, and PacI, LoxP, and the sense Us29 sequence, in that order, to the 3" side of the Us28 ORF) and YD-Pri15 (5"-GGGTTTAAACCCGGGGGTTCCCCATAAG-3"; including a PmeI site and the antisense Us29 sequence) and cloned into pGEM-T (Promega), resulting in YD-C15. The GFP/Puro expression cassette was released from pGET-015 (26) by PacI digestion and subcloned into YD-C15 to make YD-C25. The 1.4-kb Us28 sequence was PCR amplified by primers YD-Pri14 (5"-GGCTCGAGATAACTTCGTATAATGTATGCTATACGAAGTTAT TCATGCTGTGGTACCAGG-3"; including sites for XhoI, LoxP, and antisense sequence to the stop codon region of the Us28 ORF) and YD-Pri16 (5"-GGGTTTAAAC TACGTTCGCGCTTGTCTG-3"; including a PmeI site and sense sequence to the Us28 region) and cloned into p-GEMT. The Us28 sequence was then released by PmeI/NdeI digestion and cloned into the PmeI/NdeI sites of YD-C25 to construct YD-C27. Finally, the pGEM-T vector backbone DNA in YD-C27 was removed by XhoI digestion and replaced with SalI-linearized pMB131 to produce YD-C29.
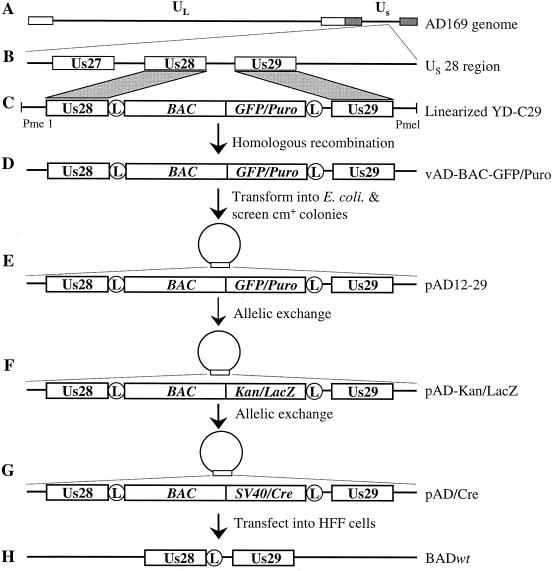
FIG. 1.
Strategy used to clone the genome of HCMV strain AD169 into a BAC. The HCMV genome (A) and a detail showing the ORFs present in the Us28 region (B) are diagrammed. AD169 viral DNA and PmeI-linearized recombination plasmid pYD-C29 (C) were cotransfected into HFF cells to generate the recombinant virus vAD-BAC-GFP/Puro (D). Circular vAD-BAC-GFP/Puro DNA was electroporated into E. coli DH10B cells to generate the BAC clone pAD12-29 (E). pAD12-29 was electroporated into the recA+ derivative of DH10B cells, GS500, where allelic exchange replaced the GFP/Puro cassette in the BAC plasmid with the kan/lacZ cassette (F) and subsequently with the SV40/Cre cassette to construct self-excisable BAC plasmid pAD/Cre (G). Transfection of pAD/Cre alone into HFF cells generated BAD wt (H), a phenotypically wild-type AD169 virus with a single LoxP site remaining that is located after the Us28 ORF. The circle enclosing an L represents the LoxP site.
pGS403 contains a gene that encodes Cre under the control of the HCMV major immediate-early promoter. The Cre-encoding gene contained a synthetic intron that prevented expression of the functional protein in Escherichia coli (23). The delivery vector used for allelic exchange, pGS284, is a derivative of pCVD422 that is based on the RP4oriT and the R6K replication origin of pGP704, which allowed replication of the plasmid only in the presence of the π protein. In allelic exchange, the π protein was provided by the S17-1γpir host cells (22). pGS284 also contained a β-lactamase gene (Ampr) and the sacB gene. The sacB gene rendered E. coli cells harboring pGS284 sensitive to sucrose. pGS403, pGS284, and the pp71 expression plasmid pCMV71 were previously described (2, 22, 23).
Two mutagenic cassettes were used to facilitate allelic exchange in this study. YD-C54 contained a kanamycin resistance-encoding gene (kan) and a lacZ gene cloned into p-GEMT. Both kan and lacZ were expressed in E. coli. The 1.8-kb kan/lacZ cassette could be released by PacI digestion. YD-C33 contained kan and green fluorescent protein (GFP)-encoding genes in the pGET-007 background (26). kan was expressed in E. coli, while GFP was under control of the simian virus 40 (SV40) promoter. The 2.6-kb kan/GFP cassette could be released by XbaI digestion.
YD-C60 was the delivery plasmid used in the construction of pAD-kan/lacZ from pAD12-29 by allelic exchange and was constructed as follows. The GFP/Puro cassette was removed from YD-C29 by PacI digestion, resulting in YD-C48. The 3-kb sequence flanking the PacI site in YD-C48 was PCR amplified and cloned into p-GEMT, resulting in YD-C56. The PacI fragment of the kan/lacZ cassette was released from YD-C54 and ligated into the PacI site of YD-C56 to make YD-C58. Finally, the BglII fragment of the pGEM-T vector backbone in YD-C58 was replaced with BglII-linearized pGS284, resulting in YD-C60. YD-C70 was the delivery plasmid used in allelic change to construct pAD/Cre from pAD-kan/lacZ. It was constructed as follows. YD-C66 contained the Cre-encoding gene under the control of the SV40 promoter that could be released by PacI digestion. The PacI fragment of SV40/Cre from YD-C66 was ligated into the PacI site of YD-C56 to make YD-C68. Finally, the EagI fragment of the pGEM-T vector sequence in YD-C68 was replaced with NotI-linearized pGS284, resulting in YD-C70.
The following delivery plasmids were pGS284 derivatives and used for construction of various TRL13 and IRL13 recombinant BAC plasmids by allelic exchange. (i) YD-C72 was used in construction of the TRL13 knockout BAC plasmid. It contained the C-terminal part of the TRL13 ORF and its flanking sequence (strain AD169 sequence nucleotides [nt] 9791 to 10795 and 10994 to 11998) generated by PCR while the 66-amino-acid N terminus of the TRL13 ORF was replaced with the 2.6-kb XbaI fragment of the GFP/kan cassette released from YD-C33. (ii) YD-C80 was used in construction of the IRL13 knockout BAC plasmid. It contained the C terminus of the IRL13 ORF and its flanking sequence (nt 177469 to 178473 and 178672 to 179676) generated by PCR while the 66-amino-acid N terminus of the IRL13 ORF was replaced with the 1.1-kb zeo/lacZ cassette. (iii) YD-C87 was used in construction of the recombinant BAC plasmid in which the GFP ORF was fused to the C terminus of the IRL13 putative ORF. It contained the 1.9-kb PCR-generated IRL13 upstream flanking sequence including the IRL13 putative ORF without the stop codon (nt 178231 to 180116) and the 726-bp GFP ORF followed by the 1.4-kb IRL13 downstream flanking sequence (nt 176812 to 178227). (iv) YD-C85 was used in construction of the TRL13 revertant BAC plasmid. It contained the PCR-generated 2.2-kb viral sequence spanning the TRL13 gene and its flanking sequences (nt 9791 to 11998).
All of the PCR fragments used in plasmid construction were verified by sequencing. All PCRs amplifying viral sequences used AD169 DNA isolated from virions as the template.
RACE.
A 10-cm-diameter plate of confluent HFF cells was infected with virus at a multiplicity of 5 PFU/cell for 72 h. Total RNA was extracted from cells with Trizol reagent (Gibco BRL), and polyadenylated RNA was subsequently purified by using RNeasy columns (Qiagen). The purified transcripts were subjected to 3" and 5" SMART (switching mechanism at 5" end of RNA transcript) RACE (rapid amplification of cDNA ends) (Clontech). The two gene-specific primers (GSPs) used were located inside the putative RL13 ORF. 3" RACE GSP2 (5"-CCTGTTCCTGTCAATGTCCCTGTAG-3") was sense to the RL13 ORF, and 5" RACE GSP1 (5"-CTACATTGTGCCATTTCTCAGTC-3") was antisense to the RL13 ORF. The specific RACE fragments generated by PCR were T/A cloned into pGEM-T easy vectors and sequenced.
DNA isolation, transformation, and transfection.
Linear viral DNA for cotransfection was isolated from virions, and total DNA for Southern blot assays was isolated from cell lysates as previously described (2, 19). Circular viral DNA was isolated from infected HFF cells by the Hirt method (12, 16). In brief, 106 HFF cells in a 100-mm-diameter plate were infected with virus at a multiplicity of 5 PFU/cell for 72 h. Cells were scraped off the plate, collected by centrifugation, washed with 5 ml of solution I (10 mM Tris, 10 mM EDTA, pH 8.0), and resuspended in 0.5 ml of solution I with 0.25 mg of proteinase K/ml. Sodium dodecyl sulfate was added to a final concentration of 0.6%, and NaCl was added to a final concentration of 1 M. The cell lysate was incubated at 4°C overnight, and cell debris was cleared by centrifugation. The supernatant was phenol-chloroform extracted twice, and DNA was precipitated with ethanol. The circular DNA was transformed into E. coli Genehogs (Research Genetics) by electroporation in an E. coli Pulser with 1-mm gap cuvettes at 1.8 kV (Bio-Rad). Transformed bacteria were grown on Luria-Bertani (LB) plates containing chloramphenicol at 15 mg/ml. To isolate the BAC plasmid, the bacteria harboring the BAC plasmid were grown in 100 ml of LB medium with chloramphenicol at 15 mg/ml at 37°C overnight and the BAC plasmid DNA was purified with Nucleobond AX100 columns (Clontech) and resuspended in 50 μl of TE (10 mM Tris, 1 mM EDTA, pH 8.0) buffer.
To generate virus from the BAC plasmid, 15 μl of purified BAC plasmid, 1 μg of HCMV pp71-expressing plasmid (2), and 1 μg of Cre-expressing plasmid (23) were mixed with 5 × 106 HFF cells in a total volume of 265 μl, electroporated in a Gene Pulser with 4-mm-gap cuvettes (Bio-Rad) with settings of 260 mV and 960 μF and seeded on a 10-cm-diameter plate. Cells were supplied with fresh medium on the next day, and fresh HFF cells were supplemented to 50% confluency if necessary. Cells were incubated at 37°C for 2 weeks or until a 100% cytopathic effect was reached.
Pulsed-field gel electrophoresis.
Linear viral DNA and linearized BAC plasmid DNA were resolved on 1.0% agarose pulsed-field gels with 0.5× TBE buffer (44.5 mM Tris, 44.5 mM boric acid, 1 mM EDTA, pH 8.0) run in a CHEF-DRIII pulsed-field gel electrophoresis system (Bio-Rad). The gel was run at 15°C and 4.5 V/cm, and switch times were ramped from 5 to 120 s for 48 h.
Growth curves.
For single-step growth curves, 2 × 105 HFF cells/well in a 12-well dish were infected with virus at a multiplicity of 5 PFU/cell. The infected-cell supernatants containing extracellular virus were harvested at 24-h intervals for up to 7 days postinfection. For multistep growth curves, 2 × 105 HFF cells/well in a 12-well dish were infected with virus at a multiplicity of 0.01 PFU/cell. Medium from infected cells was harvested at 3- to 4-day intervals for 21 days. The infect-cell supernatants were stored at −70°C until the end of the experiment. The samples of medium were thawed, cleared of cell debris, and sonicated, and the virus titer was determined by plaque assay. Assays were done in triplicate at each time point.
Allelic exchange.
Allelic exchange was carried out as previously described (22) with modifications. A blue-white screening step was incorporated to facilitate the identification of BAC clones undergoing successful allelic exchange. To introduce a nonselectable modification into an allele in the BAC plasmid, we first substituted a kan/lacZ cassette into the allele. The resulting colonies containing the BAC plasmid with kan/lacZ inserted were identified as blue colonies on LB plates containing 5% sucrose, chloramphenicol at 15 μg/ml, and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) and isopropyl-β-d-thiogalactopyranoside (IPTG) at 30 μg/ml. We subsequently replaced the kan/lacZ insertion with the desired modification by a second round of allelic exchange and identified the resulting colonies harboring the BAC plasmid with kan/lacZ replaced with the desired modification as white colonies on Suc+ Cm+ X-Gal+ IPTG+ plates. The candidate clones were confirmed by phenotype (Cmr, Carbs, and Kanr or Kans), depending on the presence or absence of the kan/lacZ cassette.
Southern and Northern blot assays.
For Southern blot assays, viral or plasmid DNA was digested with appropriate restriction enzymes and separated by 0.8% agarose gel electrophoresis. For Northern blot assays, total RNA was isolated from HFF cells with Trizol reagent and separated by 1% formaldehyde denaturing agarose gel electrophoresis. DNA or RNA was transferred onto a Nytran SuPerCharge membrane by using the TURBOBLOTTER Rapid Downward Transfer System (Schleicher & Schuell), hybridized with 32P-labeled double-stranded DNA probes, and visualized by autoradiography. Double-stranded DNA probes were prepared by labeling gel-purified restriction fragments or PCR fragments with a Random Primed DNA Labeling Kit (Boehringer Mannheim).
Nucleotide sequence accession number.
The GenBank accession number of the RL13/14-encoding gene of HCMV strain Toledo is AY064173.
RESULTS
A BAC containing the full-length HCMV genome.
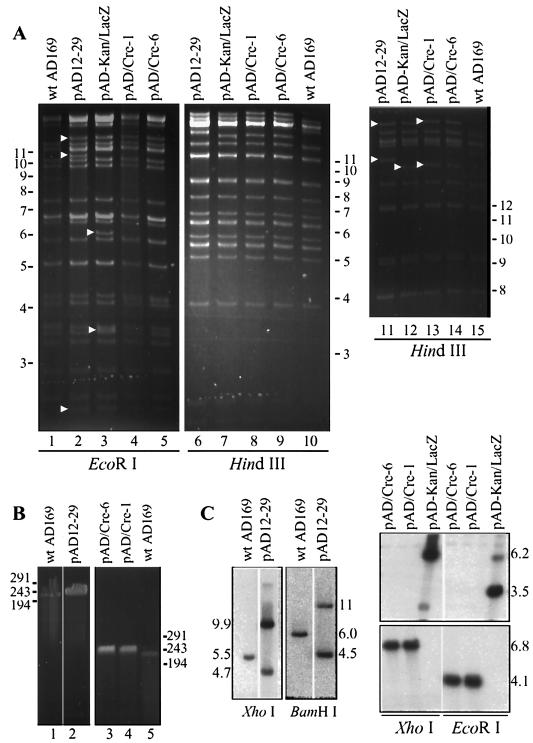
We cloned the full-length HCMV genome (strain AD169) into a BAC. The modified BAC vector was inserted immediately after the HCMV Us28 ORF without deletion of any viral sequences. Since the Us28 region is dispensable for viral growth in HFF cells (11, 27), we reasoned that insertion of the BAC vector after the Us28 ORF would have no effect on virus replication in cell culture if the insertion affected expression of the nearest gene. The cloning strategy is summarized in Fig. 1. Recombination plasmid pYD-C29 contains the mini-F plasmid and markers that function in human cells bracketed by LoxP sites inserted between Us28 and Us29 sequences. A unique restriction site was engineered in YD-C29 so that the plasmid could be linearized by PmeI. Linearized YD-C29 and linear strain AD169 viral DNA were cotransfected into HFF cells. The recombinant virus vAD-BAC-GFP/Puro was enriched by puromycin selection, and plaques showing green fluorescence were purified. After one round of plaque purification, circular viral DNA was isolated from infected HFF cells and electroporated into E. coli. Clones that were Cmr (the BAC contains a chloramphenicol resistance marker) were screened for the presence of the Us28 and Us29 regions by colony hybridization and PCR analysis (data not shown). Candidate clones were examined by restriction digestion and Southern blot analyses. The results for one clone, pAD12-29, are shown in Fig. 2. Both EcoRI and HindIII digestions of pAD12-29 yielded complex patterns similar to those of AD169 viral DNA, with the following predicted exceptions. HindIII digestion of pAD12-29 produced a unique fragment of 16.3 kb due to the BAC insertion and a fragment of 27.4 kb due to circularization of the termini of the viral genome (Fig. 2A, lane 11). EcoRI digestion produced unique fragments of 2.3, 10.4, and 11.3 kb as a result of the same alterations (Fig. 2A, lane 2). PacI digestion of pAD12-29 was expected to generate two fragments of 235 and 2.7 kb. Consistent with this prediction, the large fragment released by PacI digestion migrated no faster than 229-kb strain AD169 viral DNA when analyzed by pulsed-field gel electrophoresis (Fig. 2B, left). Taken together, these results argue that pAD12-29 contains the complete sequence of the HCMV genome. Correct insertion of the BAC vector following the Us28 ORF was confirmed by Southern blot assay (Fig. 2C, left). As expected, the strain AD169 DNA contained only one Us28/29-specific XhoI fragment (5.5 kb) and one BamHI fragment (6.0 kb). In contrast, pAD12-29 had two Us28/29-specific XhoI fragments (4.7 and 9.9 kb) and two BamHI fragments (4.5 and 11 kb). Thus, we successfully cloned the full-length strain AD169 genome as a BAC plasmid.
FIG. 2.
Structural analysis HCMV BAC plasmids pAD12-29, pAD-kan/lacZ, and pAD/Cre. (A) HindIII and EcoRI digestion of wild-type (wt) AD169 viral DNA and the cloned HCMV BAC plasmid DNA. The fragments identified by arrowheads are unique to the BAC plasmids. Two independent isolates, pAD/Cre-1 and pAD/Cre-6, are shown. Lanes 11 to 15 exhibit an experiment in which the digested fragments were subjected to extended electrophoresis to resolve the larger HindIII fragments. Molecular size markers, in kilobases, are indicated. (B) Pulsed-field gel electrophoresis of BACs linearized with PacI and linear wild-type AD169 viral DNA. Molecular size markers, in kilobases, are indicated. (C) Southern blot analyses of viral and BAC DNAs. The left half was probed with a 32P-labeled Us28-specific DNA fragment. The top of the right half received a kan/lacZ-specific probe, and the bottom half was hybridized with a Cre-specific probe.
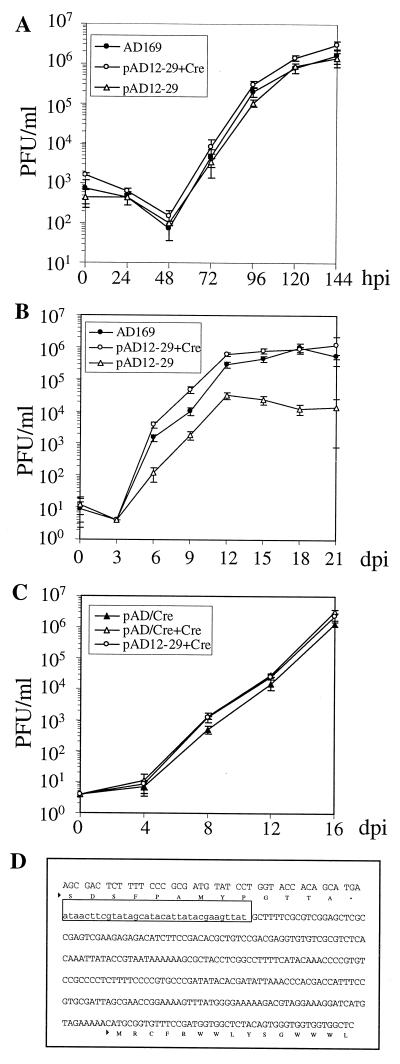
To test whether pAD12-29 was infectious, we transfected it into HFF cells with or without cotransfection of the Cre expression plasmid. Coexpression of Cre was expected to remove the foreign DNA sequence between the two LoxP sites from the viral genome in HFF cells. Plaques appeared in 2 weeks, and a 100% cytopathic effect was reached in 3 weeks, indicating that pAD12-29 was able to produce infectious virus (vAD12-29). To test if vAD12-29 had wild-type growth kinetics, single-step (Fig. 3A) and multistep (Fig. 3B) growth curves were plotted. In the single-step growth curve, there was no notable difference in growth kinetics among the viruses generated by transfection with pAD12-29 alone, pAD12-29 plus the Cre expression plasmid, and the wild-type strain AD169 virus. In the multistep growth curve, however, the virus derived by transfection with pAD12-29 alone grew to an ∼100-fold reduced yield in comparison with the wild-type virus, whereas the virus derived from pAD12-29 plus the Cre expression plasmid grew as well as the wild-type virus. Sequence analysis of the Us28 region confirmed that the 9-kb BAC vector DNA was retained in the genome of the virus generated by transfection of pAD12-29 alone (data not shown). In contrast, the BAC sequence between two LoxP sites was successfully removed from the genome of the virus generated by pAD12-29 plus the Cre expression plasmid.
FIG. 3.
Growth kinetics of viruses generated from pAD12-29 or pAD/Cre and analysis of the residual Lox site after excision of the BAC. pAD12-29 or pAD/Cre was transfected into HFF cells in the presence or absence of Cre-expressing plasmid GS403. The titers of viruses generated from transfected cells were determined, and growth curves were plotted. The suffix +Cre indicates viruses generated by cotransfection with GS403. AD169 was used as a wild-type control. (A) Single-step growth curve of the viruses generated by pAD12-29 transfection. HFF cells were infected at a multiplicity of 5 PFU/cell. hpi, hour postinfection. (B) Multistep growth curve of the viruses generated by pAD12-29 transfection. HFF cells were infected at a multiplicity of 0.01 PFU/cell. dpi, days postinfection. (C) Multistep growth curve of the viruses generated from pAD/Cre transfection. HFF cells were infected at a multiplicity of 0.01 PFU/cell. (D) DNA sequence of the Us28 region in the viruses generated by cotransfection of pAD12-29 plus GS403 or by transfection of pAD/Cre. The C terminus of the Us28 ORF, the 34-bp LoxP site (framed), and the N terminus of the Us29 ORF are indicated.
To simplify the use of pAD12-29, we incorporated the Cre expression cartridge into pAD12-29 to construct self-excisable pAD/Cre, bypassing the need for cotransfection with a separate Cre-expressing plasmid. The cartridge contained the Cre-encoding gene interrupted by an intron (23). As a consequence, Cre is expressed in human cells but not in E. coli. The modified BAC was produced by two rounds of allelic exchange facilitated by blue-white screening (Fig. 1). In the first exchange, a kan/lacZ cassette was used to replace the GFP/Puro cassette to construct pAD-kan/lacZ. E. coli colonies harboring pAD-kan/lacZ were readily identified by their blue color when growing on LB plates containing X-Gal and IPTG. The pAD-kan/lacZ clones were confirmed by their kanamycin-resistant phenotype. In the second round of allelic exchange, the Cre expression cassette was used to replace the kan/lacZ cassette to construct pAD/Cre. The resulting pAD/Cre clones, in which the kan/lacZ cassette was lost, were easily identified as white colonies on LB plates containing X-Gal and IPTG. As expected, all of the white colonies examined were kanamycin sensitive. EcoRI and HindIII cleavage showed that pAD-kan/lacZ and pAD/Cre clones retained complex digestion patterns characteristic of pAD12-29 (Fig. 2A). Compared to pAD12-29, EcoRI digestion of pAD-kan/lacZ produced unique fragments of 3.5 and 6.0 kb and lacked a fragment of 10.4 kb. HindIII digestion produced a unique fragment of 15.2 kb and lacked the 16.3-kb fragment. Compared to pAD-kan/lacZ, EcoRI digestion of pAD/Cre lacked the fragments of 3.5 and 6.0 kb. HindIII digestion produced unique fragments of 15.4 and 28.3 kb but not the fragments of 15.2 and 27.2 kb. All of these results were consistent with predictions. Pulsed-field gel electrophoresis with the two PacI-linearized pAD/Cre clones showed that pAD/Cre had a size slightly larger than that of the wild-type strain AD169 genome (Fig. 2B, right). These results indicate that pAD-kan/lacZ and pAD/Cre contained the full-length viral genome. pAD/Cre and pAD-kan/lacZ were also subjected to Southern blot analysis (Fig. 2C, right). pAD-kan/lacZ contained XhoI (6.2-kb) and EcoRI (3.5-kb) fragments specific for the kan/lacZ cassette. pAD/Cre, however, lost the kan/lacZ-specific fragments and gained the Cre-specific XhoI (6.8-kb) and EcoRI (4.1-kb) fragments, as expected. Taken together, these results indicate that allelic exchange replaced the GFP/Puro cartridge with the SV40/Cre cassette without affecting the overall integrity of the cloned viral genome.
We next examined the infectivity of the virus generated by transfection with pAD/Cre by plotting a multistep growth curve (Fig. 3C). Virus from either cotransfection with the Cre expression plasmid (pAD/Cre+Cre) or transfection without the Cre expression plasmid (pAD/Cre) was examined. Virus from pAD12-29 plus the Cre expression plasmid was used as the wild-type control (pAD12-29+Cre). We found that both pAD/Cre and pAD/Cre+Cre grew as well as pAD12-29+Cre, suggesting that the Cre-encoding gene in pAD/Cre was expressed from the BAC vector and mediated site-specific recombination efficiently to excise the BAC vector DNA from pAD/Cre. Therefore, transfection of pAD/Cre alone was sufficient to generate wild-type virus. We further sequenced the Us28 region of the virus from pAD/Cre transfection and confirmed that only one LoxP site remained following the Us28 ORF, as shown in Fig. 3D. Thus, we have constructed a self-excisable BAC that produces wild-type virus upon transfection into HFF cells.
BADsubTRL13, a BAC-derived mutant lacking TRL13.
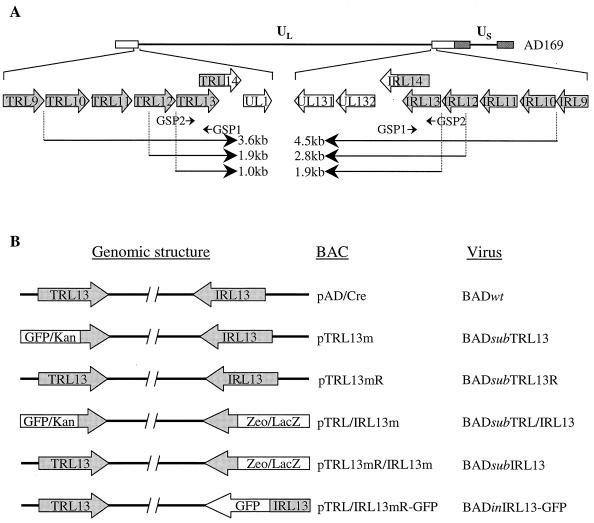
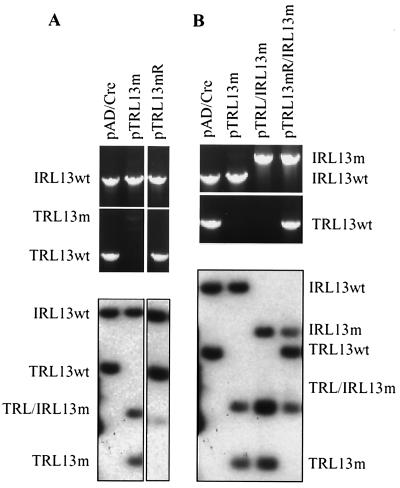
TRL13 and IRL13 (Fig. 4A) are diploid ORFs located in the inverted repeats flanking the unique long region of the HCMV genome with one copy in the TRL region and the other one in the IRL region. As noted above, the recent finding that transcripts containing TRL/IRL13 were packaged within the virions suggested that they might play an important role early in infection (4). Previously, it was reported that a mutant with the large deletion including TRL13 was viable in cell culture, although it was not clear whether the kinetics of virus growth was impaired in this mutant (24, 25). Further, it is likely that the second copy in the IRL region would compensate for the loss of TRL13. To fully understand the functions of TRL/IRL13 during virus infection in cultured cells, we constructed a double mutant in which both TRL13 and IRL13 were disrupted. A series of recombinant BACs were generated from pAD/Cre in E. coli by using a two-step homologous recombination approach (Fig. 4B). First, we constructed the single mutant pTRL13m and its revertant pTRL13mR. In pTRL13m, we replaced the N-terminal half of TRL13 with the GFP/kan cassette. In pTRL13mR, we repaired the mutated TRL13 allele to generate a wild-type revertant. pTRL13m and pTRL13mR were examined by cleavage with HindIII and EcoRI (data not shown), and the digestion patterns indicated that allelic exchange in recombination-competent cells was not detrimental to the overall integrity of the cloned viral genome and that the sizes of several fragments were altered by the mutations. To further confirm that TRL13 was specifically targeted, the TRL/IRL13 loci were examined by PCR amplification and Southern blot analysis (Fig. 5A). A wild-type TRL13 PCR-generated fragment of 1.6 kb was present in wild-type pAD/Cre and revertant pTRL13mR, while only the mutant TRL13 PCR fragment of 4.0 kb containing the GFP/kan cassette appeared in pTRL13m. A 2.2-kb IRL13-specific fragment, however, was amplified from wild-type pAD/Cre, mutant pAD-TRL13m, and revertant pAD-TRL13mR. These results were confirmed by Southern blot analysis. While pAD/Cre and pTRL13mR contained wild-type PstI fragments generated from TRL13 and IRL13, pTRL13m contained PstI fragments corresponding to both wild-type IRL13 and mutant TRL13 loci. These results indicated that we were able to preserve the wild-type allele of IRL13 and the overall structure of the cloned HCMV genome while specifically targeting TRL13 for mutagenesis and that TRL13 was successfully mutated in pTRL13m and subsequently repaired in pTRL13mR.
FIG. 4.
Diagrams of the TRL13/IRL13 locus, its encoded mRNAs, and the BACs in which the locus is altered. (A) Diagram of the genomic TRL13 and IRL13 regions. The top line represents the HCMV strain AD169 genome. Below is a detail displaying the region spanning TRL13 and IRL13. Shadowed arrows represent ORFs in the repeated sequences. Whereas the N termini of TRL14 and IRL14 are identical, their C-terminal portions, which extend into the unique long region, are distinct. The three TRL13-specific transcripts of ∼1.0, ∼1.9, and ∼3.6 kb and the three IRL13-specifc transcripts of ∼1.9, ∼2.8, and ∼4.5 kb are shown as lines, and their direction of transcription is indicated by arrowheads. Also shown are the TRL/IRL13 GSPs used for SMART RACE. GSP1 and GSP2 lie within the TRL/IRL13 ORFs. (B) Diagram of BACs containing alterations in the TRL/IRL13 ORFs. In the single mutant pTRL13m, the N-terminal portion of the TRL13 ORF was replaced with the GFP/kan cassette. In the double mutant pTRL/IRL13m, the N-terminal portions of the TRL13 and IRL13 ORFs were replaced with the GFP/kan and zeo/lacZ cassettes, respectively. The revertant pTRL13mR was based on the pTRL13m background, except that the mutated TRL13 sequence was repaired to the wild-type sequence. The revertant pTRL13mR/IRL13m was identical to pTRL/IRL13m, except that the mutated TRL13 sequence was repaired to the wild-type sequence. In pTRL/IRL13mR-GFP, the IRL13 ORF was tagged at its C terminus with the GFP ORF. Also listed are the recombinant viruses generated from the corresponding BAC plasmids.
FIG. 5.
Structural analysis of BACs containing mutations in the TRL/IRL loci. (A) Analysis of the single mutant pTRL13m and its revertant pTRL13mR. The PCR assay (top) employed IRL13- and TRL13-specific primers. For the Southern blot (bottom), DNA was digested with PstI and probed with 32P-labeled TRL/IRL13 DNA. (B) Analysis of the double mutant pTRL/IRL13m and its revertant pTRL13mR/IRL13m. The PCR analysis (top) amplified the TRL13 and IRL13 loci, and the Southern blot analysis (bottom) displays DNA that was digested with PstI and probed with 32P-labeled TRL/IRL13 DNA.
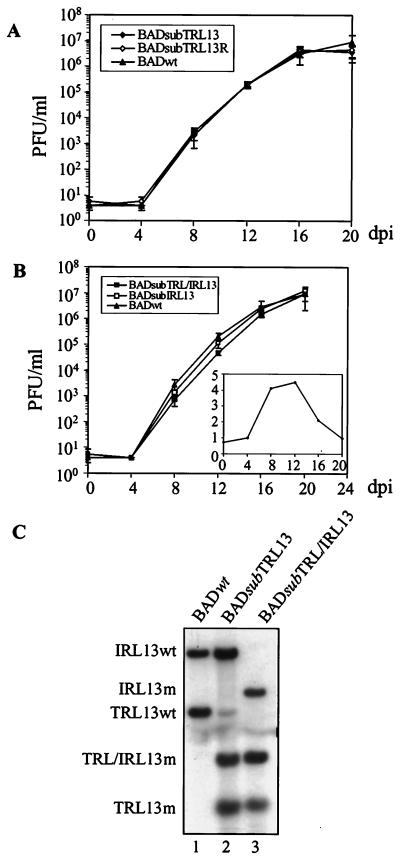
Recombinant viruses BADsubTRL13 and BADsubTRL13R were generated by transfection of HFF cells with pTRL13m and pTRL13mR, respectively. To determine if the kinetics of virus growth was impaired by loss of one copy of TRL13, we plotted a multistep growth curve for the mutant and revertant viruses (Fig. 6A). BADsubTRL13 and BADsubTRL13R grew as well as the pAD/Cre-derived wild-type virus (BADwt). Since both mutant and wild-type copies of TRL/IRL13 were present in pTRL13m, it was possible that homologous recombination could occur between two alleles of IRL/TRL13 when the virus was propagated in cell culture. We examined the stability of the introduced mutation by Southern blot assay (Fig. 6C). In BADsubTRL13, the major fragments hybridizing with the TRL/IRL13 probe were the mutant TRL13 and wild-type IRL13 fragments. A faint fragment representing the wild-type TRL13 locus appeared after one passage in cell culture, indicating that TRL13 and IRL13 underwent spontaneous recombination to repair the mutant TRL13 locus to the wild-type sequence at low frequency. Nevertheless, the blot argues that the identical growth kinetics of the mutant and wild-type viruses was not caused by extensive contamination of the mutant stock with spontaneously generated wild-type virus. The loss of one copy of TRL13 did not measurably affect virus growth in cell culture.
FIG. 6.
Growth properties and DNA analysis of viruses with mutations in the TRL/IRL loci. Wild-type and mutant viruses were generated by transfection of HFF cells with BACs. (A) Multistep growth curve of the single mutant BADsubTRL13 and its revertant BAD subTRL13R. HFF cells were infected at a multiplicity of 0.01 PFU/cell. (B) Multistep growth curve of double mutant BADsubTRL/IRL13 and its revertant BADsubIRL13. The insert shows the ratio of the titer of BADwt to that of BADsubTRL/IRL13 at the time points indicated. (C) Southern blot analysis of DNAs of wild-type and mutant viruses. DNA was digested with PstI and probed with 32P-labeled TRL/IRL13 DNA. dpi, days postinfection.
BADsubTRL/IRL13, a BAC-derived mutant lacking TRL13 and IRL13.
In addition to the single mutant pTRL13m, we also constructed a double mutant BAC, pTRL/IRL13m, in which TRL13 and IRL13 were disrupted by GFP /kan and zeo/lacZ cassettes, respectively (Fig. 4). We also repaired the mutant TRL13 allele in pTRL/IRL13m to regenerate the wild-type TRL13 sequence, producing the single revertant pTRL13mR/IRL13m. We examined the DNA structures of constructed recombinant BAC plasmids by EcoRI and HindIII digestion (data not shown), and the patterns of fragments were as expected. The TRL/IRL13 alleles were further examined by PCR and Southern blot analyses (Fig. 5B). Both assays demonstrated that pTRL/IRL13m had the mutant IRL13 fragment and lacked the wild-type TRL13 fragment, whereas pTRL13mR/IRL13m had mutant IRL13 and wild-type TRL13 fragments. Taken together, these results demonstrate that we successfully generated a double TRL13/IRL13 mutant BAC and subsequently repaired the TRL13 allele.
The recombinant BACs were transfected into HFF cells, and plaques appeared in 2 weeks. Viral DNAs were purified from infected-cell lysates and analyzed by Southern blot assay (Fig. 6C). BADsubTRL/IRL13 contained only the mutant TRL13 and IRL13 fragments and lacked the wild-type fragments, indicating that the two mutations were stably maintained in the recombinant virus. To determine if the loss of both alleles altered the kinetics of virus growth, we plotted a multistep growth curve (Fig. 6B). BADsubTRL/IRL13 grew almost as well as BADsubIRL13 and BADwt. The difference in yield between BADsubTRL/IRL13 and BADwt was less than fivefold at any time measured (Fig. 6B, insert), and at day 20, their yields were identical. We concluded that both copies of TRL/IRL13 are dispensable for virus growth in HFF cells.
Altered TRL13 and IRL13 RNAs in mutant virus-infected cells.
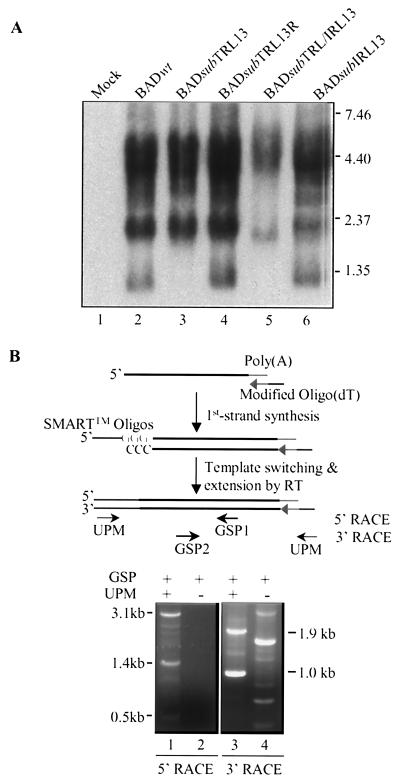
Several transcripts from the TRL-UL junction region have been reported (8, 20). It was unclear, however, exactly where these transcripts were located and whether they were spliced. Accordingly, we fine-mapped the TRL13 and IRL13 transcripts. We first performed Northern blot analyses to assess the sizes of TRL/IRL13 transcripts expressed late in infection by mutant and wild-type viruses. HFF cells were either mock infected or infected with wild-type or mutant virus for 72 h, and total RNA was isolated and subjected to Northern blot analysis with a TRL/IRL13-specific probe (Fig. 7A). Several TRL/IRL13 transcripts were evident in BADwt-infected cells, including a transcript of ∼1.0 kb, a cluster of transcripts of ∼1.9 kb, and transcripts of 3.0 to 5.0 kb. Comparison of the transcripts in mutant virus-infected cells with those in wild-type virus-infected cells allowed the following assignments. (i) The ∼1.0-kb transcript was TRL13 specific, as evidenced by its absence after BADsubTRL13 infection and its presence after infection with the revertant, BADsubTRL13R. (ii) The cluster of ∼1.9-kb transcripts was lost after infection with the double mutant BADsubTRL/IRL13, and the cluster reappeared in the single revertant BADsubIRL13, indicating that an ∼1.9-kb TRL13-specific transcript comigrated with an IRL13-specific transcript of the same size; a transcript in BADsubTRL/IRL13-infected cells migrated below the ∼1.9-kb band seen in BADwt infection (Fig. 7A, lanes 2 and 5), which might result from expression of a truncated TRL13 transcript from the SV40 promoter of the inserted GFP/kan cassette (Fig. 5). (iii) The substantial reduction in the amount of the 3.0- to 5.0-kb transcript cluster in the double mutant (BADsubTRL/IRL13) infection suggested that the majority of these transcripts were TRL/IRL13 specific, and the presence of a 3.0- to 5.0-kb species after infection with BADsubIRL13 indicated that at least one transcript from this cluster was derived from TRL13. Taken together, these results suggested that TRL13 encodes ∼1.0-, ∼1.9-, and 3.0- to 5.0-kb transcripts and that IRL13 encodes at least one ∼1.9-kb transcript and some of the 3.0- to 5.0-kb transcripts.
FIG. 7.
Map of the TRL/IRL13 transcripts. (A) Northern blot analysis of RNA from HFF cells infected with wild-type or mutant viruses. The blot was hybridized with a 32P-labeled TRL13-specific DNA. (B) RACE to amplify the 5" and 3" ends of TRL13- and IRL13-specific transcripts. At the top is a simplified diagram of the SMART RACE reaction (Clontech) with the GSPs and the universal primer mix (UPM) that anneals to the SMART oligonucleotides (Oligos) and modified oligo(dT) sequences. At the bottom are the bands amplified in the RACE PCRs; their size, relative to those of marker DNAs, are indicated.
To more precisely map the TRL13- and IRL13-specific transcripts, we performed SMART RACE on RNA from BAD wt-infected cells by using two overlapping primers located inside the TRL/IRL13 putative ORFs (Fig. 7B). The 5" RACE assay identified three fragments (0.5, 1.4, and 3.1 kb). Two fragments of 1.0 and 1.9 kb were identified in the 3" RACE assay. These RACE PCR-generated fragments were cloned and sequenced. By comparing the RACE sequence data and the published genomic sequence of HCMV strain AD169 (9), we deduced the complete sequences of the TRL13 and IRL13 transcripts. All of the transcripts were unspliced. Since the transcripts started within the inverted repeat sequences, the N termini of the TRL13 and IRL13 transcripts are identical. Three transcription start sites were identified, which were at nt 8101 to 181366 (upstream of TRL/IRL10), nt 9827 to 179640 (in the TRL/IRL12 ORF), and nt 10736 to 178731 (upstream of TRL/IRL13) for the TRL13 and IRL13 transcripts (Fig. 4A). Each start site resided downstream of a TATA motif. The TRL13 and IRL13 transcripts terminated at different positions since the 3" ends of the transcripts extended into the UL region. The TRL13 transcripts terminated at nt 11736, 17 nucleotides downstream of an AATAAA (nt 11714) signal residing upstream of the UL1 gene, and the IRL13 transcripts terminated at nt 176829, 19 nucleotides downstream of an AATAAA (nt 176853) signal within the UL131 gene. Thus, the three TRL13-specific transcripts were 1.0, 1.9, and 3.6 kb, while the IRL13-specific transcripts were 1.9, 2.8, and 4.5 kb. The sizes of these transcripts correlate with those identified in Fig. 7A. All of the transcripts that were identified contain the TRL13/14 or the IRL13/14 ORFs, and several of the transcripts contain additional ORFs as well.
Marked variation in the TRL13 ORF from the Toledo strain compared to that of the AD169 strain of HCMV.
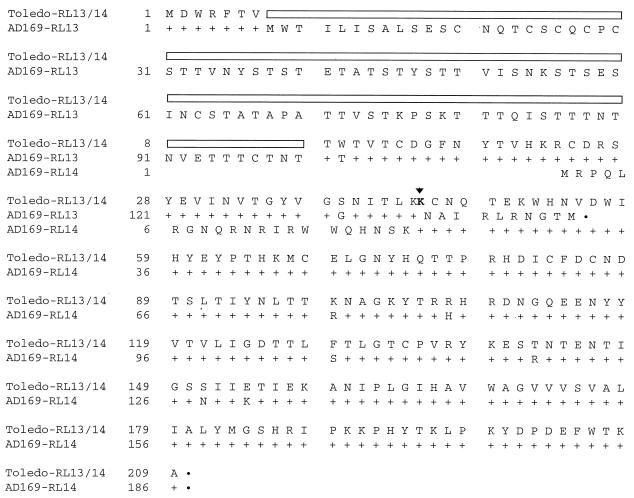
Since the IRL/TRL13 ORFs are not required for the production of normal HCMV yields in HFF cells, we reasoned that their sequence might have drifted during the continued passage of the AD169 laboratory strain in cultured cells. To investigate this possibility, we compared the sequence of this locus in AD169 to the less extensively passaged Toledo strain of the virus (Fig. 8). The strain AD169 sequence was identical to the published sequence (9) but markedly different from the strain Toledo sequence. In comparison to the Toledo strain, the AD169 strain has a 279-bp insertion near the 5" end of TRL/IRL13, a deletion of 1 bp near the C terminus of TRL/IRL13, and several single-base-pair missense mutations. The Toledo strain should produce a protein lacking 93 amino acids that are present in the N-terminal domain of the strain AD169 protein and extending into a different reading frame through the region assigned to IRL/TRL14 in strain AD169. As a result, the Toledo strain encodes a single protein extending through the region corresponding to the TRL13 and TRL14 regions of strain AD169, and if it were to use the internal AUG assigned to the TRL/IRL14 ORF, it would produce a protein of only 15 amino acids.
FIG. 8.
Sequence comparison of the TRL13 and -14 domains in two strains of HCMV. The TRL13 and -14 regions were amplified from strain AD169 and Toledo DNAs by PCR. Both strands of the DNA fragments produced in two independent PCRs were sequenced for each strain. Putative proteins encoded by the region are shown. The AD169 sequence is identical to the published sequence (9). The strain Toledo amino acid sequence is shown in full, identical matches of the strain AD169 and Toledo sequences are shown by plus signs, and amino acid changes in strain AD169 relative to strain Toledo are indicated by the single-letter code. The box identifies a segment of the strain AD169 sequence that is absent in strain Toledo. The arrow designates the location of an extra A residue in strain Toledo that causes a frame shift relative to strain AD169.
TRL/IRL13 encode a protein.
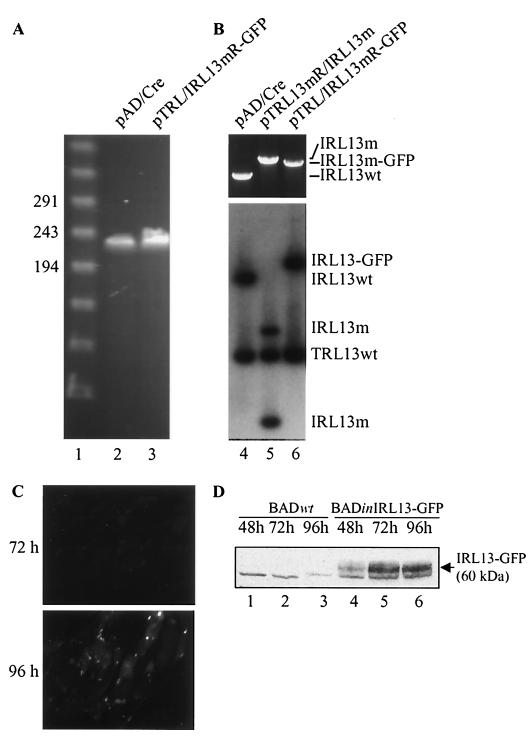
Since the TRL/IRL13 protein of strain AD169 is quite different from that encoded by the Toledo strain, we confirmed that the ORF encoded a protein that accumulated in strain AD169-infected cells. We constructed a recombinant BAC plasmid, pTRL/IRL13mR-GFP, in which the GFP ORF, without its start codon, was fused in frame to the C terminus of the IRL13 ORF. If the putative IRL13 ORF indeed encoded a protein, the IRL13-GFP fusion protein would be expressed and easily detected by fluorescence microscopy or Western blot analysis. Pulsed-field gel electrophoresis showed that pTRL/IRL13mR-GFP migrated at the predicted size (Fig. 8A), and EcoRI and HindIII digestions produced restriction patterns as predicted (data not shown). Generation of the IRL13-GFP fusion in pTRL/IRL13mR-GFP was also examined by PCR and Southern blot assay (Fig. 8B). pTRL/IRL13mR-GFP contained a IRL13-GFP-specific PCR fragment and lacked the wild-type IRL13 fragment. A blot probed with the TRL/IRL13-specific probe revealed that while pAD/Cre contained wild-type TRL13 and IRL13 PstI fragments, pTRL/IRL13mR-GFP contained wild-type TRL13 and an IRL13-GFP fusion fragment (Fig. 8B, lanes 4 and 6).
pTRL/IRL13mR-GFP was transfected into HFF cells, and the virus BADinIRL13-GFP was produced. HFF cells were infected with the variant at a multiplicity of 5 PFU/cell, and expression of the IRL13-GFP protein was monitored by fluorescence microscopy. GFP fluorescence was first detected at 72 h, and a maximal signal was evident at 96 h postinfection (Fig. 8C). The GFP signal was located mostly in the cytoplasm. Expression of the IRL13-GFP fusion protein was confirmed by Western blot assay (Fig. 8D). A band was recognized by a GFP-specific polyclonal antibody beginning at 48 h after infection with BADinIRL13-GFP but not after infection with BAD wt. Its ∼60-kDa size was substantially larger than that of the ∼30-kDa GFP protein, consistent with the expression of a fusion protein. However, an IRL13-GFP fusion protein would be predicted to have a molecular mass of 48 kDa. The IRL13 sequence contains several N-glycosylation and phosphorylation sites (data not shown), so the increased size might be due to posttranslational modification. Our results indicate that the TRL/IRL13 ORF in strain AD169 encodes a protein that accumulates during the late phase of infection.
DISCUSSION
Infectious BAC clones have been described previously for the AD169 (3) and Towne (14) strains of HCMV. In both cases, a portion of the US region that is not essential for viral replication in cultured cells was deleted to accommodate insertion of the BAC replicon, and the BAC sequences remain in the HCMV genome after the virus has been regenerated in human cells. The infectious BAC that we describe here for strain AD169 differs from the earlier clones in that the BAC vector sequence is rapidly excised when virus is regenerated in human cells, and consequently, it was not necessary to delete viral sequences. This was achieved by flanking the BAC sequences with LoxP sites and adding an intron-containing Cre-encoding gene within the BAC sequences. The intron prevents expression of Cre protein in bacterial cells, but when splicing occurs in mammalian cells, the recombinase is produced and the BAC sequences are excised. This strategy was first used to produce an infectious PRV BAC (23).
Excision of the 9-kb BAC from the strain AD169 genome within human cells was required to produce a virus with wild-type growth characteristics. When it was not excised from the viral genome, the virus exhibited delayed growth kinetics and produced a reduced yield (Fig. 3B). The reduced yield was likely caused by inefficient packaging of the oversized genome carrying the inserted BAC. Alternatively, the poor growth may have resulted from an effect of the inserted BAC element on expression of a neighboring gene, although its nearest neighbor, Us28, has been shown to be dispensable for growth in cultured cells (11, 27).
A two-step recombination process (allelic exchange) was used to insert a modification into the BAC plasmid in E. coli. In the first step, the delivery plasmid, harboring the ampicillin resistance marker, was integrated into the BAC plasmid by homologous recombination. The recombinants were selected through growth in the presence of carbenicillin (an analog of ampicillin). In the second step, the delivery plasmid was excised from the BAC plasmid by a second homologous recombination. Since the delivery plasmid contained the sacB gene that rendered the host cell sucrose sensitive, colonies that grew on sucrose-containing medium potentially contained the desired recombinant. The approach has worked well with the PRV BAC (22). When applying allelic exchange to the HCMV BAC, we found that the efficiency with which the correct recombinant BAC clones were obtained was lower than that previously observed with the PRV BAC. Ninety-five percent of the clones that resulted from allelic exchange retained the phenotype of the parental BAC. There are several potential reasons for this problem: (i) inefficient homologous recombination due to the larger size of the HCMV BAC compared to the PRV BAC, (ii) incomplete selection for integration of the delivery plasmid into the BAC during the first step, and (iii) spontaneous mutation resulting in loss of function of the sacB gene. To more efficiently identify clones that underwent successful allelic exchange, we modified the target allele by replacement of a kan/lacZ cassette in the first round of allelic exchange. Correctly resolved exconjugants were identified as blue colonies on LB plates containing sucrose, chloramphenicol, and X-Gal-IPTG and confirmed by their kanamycin-resistant phenotype. In the second round of allelic exchange, the desired modification was introduced into the BAC plasmid replacing the kan/lacZ cassette and confirmed by identification of white colonies on LB plates containing sucrose-chloramphenicol-X-Gal-IPTG. All of the white colonies proved to exhibit the appropriate kanamycin-sensitive phenotype. The incorporation of blue-white screening made identification of the desired products of allelic exchange reactions in the HCMV BAC much easier.
We employed the infectious BAC initially to construct mutant viruses lacking either one or both copies of the diploid TRL/IRL13 ORFs (Fig. 4B). Although there have been reports of mutant viruses lacking single copies of ORFs within the strain AD169 TRL/IRL region (24, 25), to the best of our knowledge, this is the first example of a double mutation in which both copies of an ORF within the repeated region have been altered. Mutants lacking one copy of the ORF, BAD subTRL13 and BADsubIRL13, grew indistinguishably from their wild-type parent, BADwt (Fig. 6A). The double mutant, BADsubTRL/IRL13, displayed a modest but transient delay in the accumulation of infectious virus, eventually producing a wild-type yield (Fig. 6B). Conceivably, BADsubTRL/IRL13 will display a more substantial growth defect in a cell type other than the primary diploid fibroblasts that we have employed in our study. Many, and probably all, of the HCMV genes that have been designated nonessential for growth in cell culture contribute importantly in the infected human host by optimizing growth and dissemination, modulating tissue tropism, performing immunoevasion functions, etc. (17). It is likely that the TRL/IRL13 product has such a role in vivo.
The Toledo strain of HCMV has a shortened IRL relative to that of strain AD169 (6) and appears to contain only the TRL13 copy of the pair of ORFs that are duplicated in strain AD169. Comparison of the TRL13 ORF sequence in the AD169 strain with that in the Toledo strain (Fig. 8) revealed that the two virus strains encode markedly different proteins. The protein predicted to be encoded by the Toledo strain lacks 93 amino acids present in the N-terminal domain of the protein encoded by strain AD169, and the C-terminal domain of the strain Toledo protein extends through the region assigned to TRL14 in strain AD169. This extension results from the presence of an additional A residue in the strain Toledo sequence that shifts the reading frame of the strain Toledo protein compared to that of the strain AD169 protein. Although we cannot be certain, we suspect that the strain Toledo arrangement represents the wild-type organization of the TRL13 ORF since strain Toledo has been passaged to a more limited extent in the laboratory. If the TRL13 ORF is not essential for HCMV growth in HFF cells, its sequence could have drifted during its extensive propagation in the laboratory. The structures of the mRNAs carrying the TRL/IRL13 and -14 ORFs in strain AD169-infected cells (Fig. 7) are consistent with this conclusion. Each of the mRNAs includes both ORFs and, in the case of the Toledo strain, would be expected to encode the large TRL13 protein with its extension through the TRL14 domain. The mRNAs encoded by strain AD169 should express the shortened TRL/IRL13 ORF of that strain, and we employed a fusion protein to confirm this prediction (Fig. 9). However, it appears unlikely that the TRL/IRL14 ORF would be expressed from these mRNAs. Its AUG appears third in the mRNA, and it resides in a suboptimal sequence context, suggesting that it cannot efficiently serve as an initiator codon.
FIG. 9.
The AD169 IRL13 ORF encodes a protein. (A) Pulsed-field gel electrophoresis of PacI-linearized pTRL/IRL13mR-GFP and pAD/Cre DNA. (B) Analysis of pTRL/IRL13mR. At the top is a PCR performed with a pair of IRL13-specific primers. This pair of primers could not amplify the TRL13 allele. At the bottom is a Southern blot in which DNA was digested with PstI and probed with 32P-labeled TRL/IRL13 DNA. (C) GFP fluorescent image of HFF cells infected with BADinIRL13-GFP for 72 and 96 h. (D) Western blot analysis of IRL13-GFP expression. HFF cells were infected with BADwt, BAD inIRL13-GFP, or BADsubTRL13 for 48, 72, and 96 h, and lysates were prepared and analyzed by Western blot assay using a GFP-specific antibody.
It is conceivable that the Toledo strain-specific TRL13 protein can influence the growth of the virus in cultured cells. Experiments are in progress to substitute the Toledo strain TRL13 domain for the corresponding AD169 strain TRL/IRL13 and -14 sequences.
Acknowledgments
We thank Felicia Goodrum, Robert Kalejta, and Bret Wing for thoughtful comments on the manuscripts.
This work was supported by grants from the National Cancer Institute (CA 85786 and CA 87661). D. Yu is a Fellow of The Leukemia & Lymphoma Society.
REFERENCES
- 1.Adler, H., M. Messerle, M. Wagner, and U. H. Koszinowski. 2000. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J. Virol. 74:6964-6974. [DOI] [PMC free article] [PubMed]
- 2.Baldick, C. J., Jr., A. Marchini, C. E. Patterson, and T. Shenk. 1997. Human cytomegalovirus tegument protein pp71 (ppUL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J. Virol. 71:4400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borst, E. M., G. Hahn, U. H. Koszinowski, and M. Messerle. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bresnahan, W. A., and T. Shenk. 2000. A subset of viral transcripts packaged within human cytomegalovirus particles. Science 288:2373-2376. [DOI] [PubMed] [Google Scholar]
- 5.Britt, W. J., and C. A. Alford (ed.). 1996. Cytomegalovirus, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 6.Cha, T. A., E. Tom, G. W. Kemble, G. M. Duke, E. S. Mocarski, and R. R. Spaete. 1996. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J. Virol. 70:78-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chambers, J., A. Angulo, D. Amaratunga, H. Guo, Y. Jiang, J. S. Wan, A. Bittner, K. Frueh, M. R. Jackson, P. A. Peterson, M. G. Erlander, and P. Ghazal. 1999. DNA microarrays of the complex human cytomegalovirus genome: profiling kinetic class with drug sensitivity of viral gene expression. J. Virol. 73:5757-5766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, C. P., C. L. Malone, and M. F. Stinski. 1989. A human cytomegalovirus early gene has three inducible promoters that are regulated differentially at various times after infection. J. Virol. 63:281-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison 3rd, T. Kouzarides, J. A. Martignetti, et al. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 10.Delecluse, H. J., T. Hilsendegen, D. Pich, R. Zeidler, and W. Hammerschmidt. 1998. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc. Natl. Acad. Sci. USA 95:8245-8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirsch, A. J., and T. Shenk. 1999. Human cytomegalovirus inhibits transcription of the CC chemokine MCP-1 gene. J. Virol. 73:404-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 13.Horsburgh, B. C., M. M. Hubinette, D. Qiang, M. L. MacDonald, and F. Tufaro. 1999. Allele replacement: an application that permits rapid manipulation of herpes simplex virus type 1 genomes. Gene Ther. 6:922-930. [DOI] [PubMed] [Google Scholar]
- 14.Marchini, A., H. Liu, and H. Zhu. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J. Virol. 75:1870-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGregor, A., and M. R. Schleiss. 2001. Molecular cloning of the guinea pig cytomegalovirus (GPCMV) genome as an infectious bacterial artificial chromosome (BAC) in Escherichia coli. Mol. Genet. Metab. 72:15-26. [DOI] [PubMed] [Google Scholar]
- 16.Messerle, M., I. Crnkovic, W. Hammerschmidt, H. Ziegler, and U. H. Koszinowski. 1997. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 94:14759-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mocarski, E. S. (ed.). 1996. Cytomegalovirus and their replication, 3rd ed. Lippincott-Raven Publishers, Philadelphia. Pa.
- 18.O'Connor, M., M. Peifer, and W. Bender. 1989. Construction of large DNA segments in Escherichia coli. Science 244:1307-1312. [DOI] [PubMed] [Google Scholar]
- 19.Oliveira, S. A., and T. E. Shenk. 2001. Murine cytomegalovirus M78 protein, a G protein-coupled receptor homologue, is a constituent of the virion and facilitates accumulation of immediate-early viral mRNA. Proc. Natl. Acad. Sci. USA 98:3237-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rawlinson, W. D., and B. G. Barrell. 1993. Spliced transcripts of human cytomegalovirus. J. Virol. 67:5502-5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saeki, Y., T. Ichikawa, A. Saeki, E. A. Chiocca, K. Tobler, M. Ackermann, X. O. Breakefield, and C. Fraefel. 1998. Herpes simplex virus type 1 DNA amplified as bacterial artificial chromosome in Escherichia coli: rescue of replication-competent virus progeny and packaging of amplicon vectors. Hum. Gene Ther. 9:2787-2794. [DOI] [PubMed] [Google Scholar]
- 22.Smith, G. A., and L. W. Enquist. 1999. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J. Virol. 73:6405-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith, G. A., and L. W. Enquist. 2000. A self-recombining bacterial artificial chromosome and its application for analysis of herpesvirus pathogenesis. Proc. Natl. Acad. Sci. USA 97:4873-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spaete, R. R., and E. S. Mocarski. 1987. Insertion and deletion mutagenesis of the human cytomegalovirus genome. Proc. Natl. Acad. Sci. USA 84:7213-7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takekoshi, M., F. Maeda-Takekoshi, S. Ihara, S. Sakuma, and Y. Watanabe. 1991. Site-specific stable insertion into the human cytomegalovirus genome of a foreign gene under control of the SV40 promoter. Gene 101:209-213. [DOI] [PubMed] [Google Scholar]
- 26.Tullis, G. E., and T. Shenk. 2000. Efficient replication of adeno-associated virus type 2 vectors: a cis-acting element outside of the terminal repeats and a minimal size. J. Virol. 74:11511-11521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vieira, J., T. J. Schall, L. Corey, and A. P. Geballe. 1998. Functional analysis of the human cytomegalovirus US28 gene by insertion mutagenesis with the green fluorescent protein gene. J. Virol. 72:8158-8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wagner, M., S. Jonjić, U. H. Koszinowski, and M. Messerle. 1999. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J. Virol. 73:7056-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]