Abstract
We have used a virus overlay assay to detect cellular proteins associated with human cytomegalovirus (HCMV) particles. The radiolabeled HCMV particles specifically bound to two host proteins with molecular sizes of 150 and 180 kDa. By a micro-amino-acid sequencing technique, the 180-kDa protein was identified as a human homologue of the ES130/p180 ribosome receptor (p180), which is an integral endoplasmic reticulum (ER) membrane protein possessing a very unique tandem repeat domain at its N-terminal region. The virus overlay assay using truncated p180 polypeptides revealed that HCMV binding to human p180 occurred through the N-terminal region. In HCMV-permissive cells the high level of expression of the human p180 protein was clearly observed regardless of cell type. Furthermore, we showed that p180 binds to the UL48 gene product, which is one of the predominant tegument proteins of HCMV and which is considered to be tightly associated with the capsid. The interaction between the two proteins was assumed to be specific and was observed both in vitro and in vivo. During the late phase of infection, the unique relocation of human p180 was observed, that is, to the juxtanuclear region, which appeared to be in the vicinity of the area where naked virions were frequently observed in an electron-microscopic study. Thus our data suggest that p180 interacts with the HCMV tegument, at least through pUL48, during the HCMV replication process. We discuss the possible role of the interaction between p180 and pUL48 in the intracellular transport of HCMV virions.
Human cytomegalovirus (HCMV) is a member of the beta herpesvirus family containing a double-stranded 230-kbp DNA genome that is expected to contain over 200 open reading frames (ORFs) (20). It comprises four structural elements: a DNA-containing core, an icosahedral capsid, a layer that surrounds the capsid termed the tegument, and a lipid envelope containing viral glycoproteins (14).
The infectious cycle of HCMV begins with virus binding to the cell surface heparan sulfate (10, 17, 21). The binding process is assumed to consist of multiple steps, which involve subsequent high-affinity receptors responsible for fusion and penetration. Although several candidates for cellular receptors, including CD13 and annexin II, have been reported (34, 35, 42), intrinsic receptors have not been completely elucidated. After release of the capsids into the cytoplasm and dissociation of some tegument components, the capsids are transported to the nuclear membrane. Then uncoating at the nuclear pore results in release of the genome into the nucleus, where transcription and genomic replication occur. Progeny capsids are assembled inside the nucleus and bud into the perinuclear space between the inner and outer nuclear membranes after acquiring a primary envelope. The subsequent maturation is a complex and poorly understood process. Recently, however, accumulated reports showed that naked nucleocapsids are released into the cytoplasm and then acquire final envelopment by budding into a cytoplasmic compartment (24, 26). This maturation route is consistent with electron-microscopic studies which have shown cytoplasmic nonenveloped particles apparently acquiring the envelope (29, 30). Among the many unresolved processes, the mechanism of HCMV capsid transport in the cytoplasm is entirely unknown. Since movement of virus-sized particles through the cytosol is not likely to occur by free diffusion but rather most likely by the cellular transport system (32), it is conceivable that interaction of viral tegument proteins with the host machinery involved in the cellular transport system permits an active process.
In vivo replication of HCMV can be observed in a wide variety of cell types including epithelial cells, endothelial cells, fibroblasts, and monocytes/macrophages (7, 31), while efficient replication in cultured cells often appears to be limited to human diploid fibroblasts such as MRC-5 cells (20). A limited number of epithelial cell lines have been shown to support productive HCMV infection; these include epithelial cell lines originating from human retinal pigment epithelial cells (2, 38) and colon carcinoma cells (16). Little is known, however, about the control of cellular susceptibility to HCMV infection. Although immediate-early (IE) gene expression could be frequently detected in a variety of cell lines including those of nonhuman origin, in most cases fully productive replication was not supported (22). This observation suggests that critical factors may work at postinternalization stages. From the complexity of the virion structure and its replication process, multiple host factors are assumed to regulate the HCMV replication process.
The aim of our study is to explore and identify host factors that play a crucial role in the HCMV replication process and to investigate molecular mechanisms of virus-host cell interaction. In the present study, we used a virus overlay assay to detect cellular proteins associated with HCMV particles. This assay allowed us to define two host proteins with molecular sizes of 150 and 180 kDa. We identified the latter protein as a human homologue of ES130/p180, which was reported to be a member of the ES130/kinectin family (25). ES/130 was originally reported as a morphogenesis inducer in the chicken embryonic heart (25), while canine p180 was first identified as a ribosome receptor in MDCK cells (28). The latter protein possesses a unique tandem repeat domain with a recently proposed function related to membrane proliferation and secretion (5). In HCMV-permissive cells the high-level expression of the human p180 protein was clearly observed. We further demonstrate that the binding of p180 to HCMV occurs at least partially through interaction with the tegument protein encoded by UL48. It is the largest HCMV polypeptide and is also referred to as high-molecular-weight protein (6, 14). The HCMV UL48 gene is conserved among herpesviruses (9); its precise function remains to be elucidated. It has been reported that the herpes simplex virus type 1 (HSV-1) homologue of UL48, UL36, is required for the uncoating process at the nuclear membrane at early times after infection (4). Recently it was proposed that the HSV-1 homologue also functions in targeting cytoplasmic capsids to the proper compartment during the late phase of HSV-1 infection (12). We discuss the possible role of the interaction between p180 and the UL48 protein in the intracellular transport of viral particles.
MATERIALS AND METHODS
Cells and virus.
Human embryonic lung fibroblasts (MRC-5) between passages 20 and 35 (Riken Cell Bank, Tsukuba, Japan) were used throughout the study. The cells were grown in Eagle's minimum essential medium (MEM) supplemented with 10% fetal bovine serum (FBS) (Gibco BRL) and were maintained in MEM supplemented with 2% FBS. HCMV (strain AD169 or Towne) was purified from the culture medium of infected-cell monolayers as previously described (23). THP-1, U937, HSB-2, and Ramos 3 cells were cultured in RPMI 1640 supplemented with 10% FBS. Other cell lines including HeLa, 293T, CHO, Vero, NIH 3T3, and Caco-2 cells were cultured in Dulbecco MEM supplemented with 10% FBS. In some experiments, THP-1 cells were treated with 10 ng of 12-O-tetradecanoylphorbol-13-acetate (TPA)/ml (final concentration) for 24 h.
Biotinylation and radiolabeling of HCMV.
The HCMV probe was prepared as described previously (23). In brief, confluent cell monolayers were infected with HCMV at a multiplicity of infection of 1 to 2 PFU/cell at 37°C. For purification of unlabeled virions, the infected cells were incubated until the maximum cyopathic effects (CPE) were observed. After removal of cell debris by low-speed centrifugation (4,000 × g, 20 min), virus particles in the culture medium were collected as pellets by high-speed centrifugation (32,000 × g, 1 h). The virus particles resuspended in MEM were further purified by a d-sorbitol gradient (30 to 70%) centrifugation at 90,000 × g for 1 h, pelleted by centrifugation at 85,000 × g for 30 min, and resuspended in an appropriate volume of 0.01 M phosphate-buffered saline (PBS), pH 7.4. Biotinylation of HCMV was carried out by incubating the purified virion suspension (about 1 mg of protein/ml) with biotin-3-sulfo-N-hydroxysuccinimide ester (final concentration, 90 μg/ml) (Sigma, St. Louis, Mo.), and then the suspension was dialyzed against PBS at 4°C. To obtain radiolabeled virions, the culture medium of the infected cells was replaced with Dulbecco MEM containing a reduced level of methionine, 2% FBS, and 10 μCi of [35S]methionine/ml (Trans35S-labeling mixture; ICN, Irvine, Calif.) when 30 to 50% of cells in the culture exhibited CPE. After further incubation for 4 days, viral particles were purified as described above. Specific activities of the virus particles ranged from 0.03 to 0.07 cpm per PFU.
Detection of HCMV-binding proteins by virus overlay assay.
The membrane fractions of MRC-5 cells were prepared as described previously (13), and 10 μg of proteins was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and electroblotted onto polyvinylidene difluoride (PVDF) membranes (Immobilon-P; Millipore, Bedford, Mass.). To achieve an efficient electrotransfer of the 180-kDa protein, strong blotting conditions were required; these included an aqueous blotting buffer (50 mM Tris-glycine buffer [pH 8.4] without methanol) and a prolonged blotting time. Under these conditions, the proteins that were approximately less than 80 kDa were not retained on the membrane. After being blocked with 5% bovine serum albumin (BSA) for 2 h at 37°C, the membrane was incubated with a solution containing 35S-HCMV (about 1 × 105 to 3 × 105 cpm/ml) at 4°C overnight. After extensive washing with buffer A (150 mM NaCl, 1 mM EDTA, 1% gelatin, 10 mM Tris-HCl, pH 7.0), radioactive bands were detected with an image analyzer (BAS 1000; Fuji Photofilm, Kanagawa, Japan). Alternatively, the membrane was incubated with biotinylated virions in place of radiolabeled virions and then with streptavidin conjugated with horseradish peroxidase (Vector Laboratory, Burlingame, Calif.). The positive bands were detected by an ECL system (Amersham Pharmacia Biotech, Little Chalfont, Buckinghamshire, United Kingdom). In some experiments, either heparin (100 μg/ml; Sigma) or myelin basic protein (500 μg/ml; Sigma) was added to the probe solution.
Preparative SDS-PAGE and micro-amino-acid sequencing.
The 180-kDa protein of MRC-5 cells was concentrated from spots excised from the first preparative SDS-5% PAGE gels using the concentration gel system (11). Electroblotting onto a PVDF membrane (Immobilon-P) was carried out under the strong transfer condition described above, and the membrane was stained with Ponceau-S. The spot corresponding to the 180-kDa protein was excised and digested with lysylendopeptidase AP-1 (0.2 μg; Wako Pure Chemical Industries, Osaka, Japan) in 100 μl of 20 mM Tris-HCl (pH 8.5) with 8% acetonitrile at 37°C overnight (1). The digested peptides were extracted with 8% acetonitrile and purified using a high-performance liquid chromatography system (model 130A; Applied Biosystems) equipped with a reverse-phase C8 column (2.1 by 30 mm; RP-300; Applied Biosystems). Rechromatography using a C18 column was carried out when necessary. The amino acid sequences of the recovered peptides were determined by a pulse liquid phase sequencer (model 477A; Applied Biosystems). The sequence homology was analyzed using the GENETYX computer program (Software Development, Tokyo, Japan).
PCR cloning, sequencing, and subcloning of human p180-expressing plasmids.
Fragments of human ES130/p180 cDNA were amplified by PCR with appropriate primers and cDNA of MRC-5 cells as the template. Briefly, total RNA was extracted from cultured MRC-5 cells using guanidine isothiocyanate-phenol. Single-stranded cDNA was synthesized using avian myeloblastosis virus reverse transcriptase with oligo-p(dT)15 primers (Roche Diagnostics, Mannheim, Germany). The PCR products were cloned into pUC118 or pUC19 using appropriate restriction enzyme sites. The resulting 3.8-kbp cDNA was excised by BamHI and SpeI digestion and subcloned into vector pTSX-1, which was derived from vector pTrc99A (Amersham Pharmacia Biotech) by insertion of synthetic oligonucleotides encoding additional SpeI, XhoI, and NotI cloning sites. Finally, we generated pTSX3.8F containing a 3.8-kbp cDNA for the entire p180 isoform, which includes a 0.8-kbp tandem-repeat region. Because a molecular size estimated from the deduced amino acid sequence of the coding region of pTSX3.8F was apparently smaller than 180 kDa, a 1.8-kbp DNA encoding 54 decapeptide repeats, which corresponded to the full-length tandem-repeat region, was amplified from genomic DNA as reported previously (19). A 1.6-kbp fragment excised from the 1.8-kbp clone by EcoRI and SacI digestion was inserted into a corresponding region of pTSX3.8F to obtain pTSX4.8F, encoding the full-length p180. Sequencing was performed by an ABI 373 sequencer (Applied Biosystems Japan) using a dye terminator ready reaction kit (Perkin-Elmer) according to the manufacturer's instructions.
Generation of GST fusion polypeptides.
The cDNA fragments encoding various regions of human ES130/p180 were subcloned into pGEX vectors (pGEX5X-3 and pGEX4T-3; Pharmacia Biotech) using convenient restriction enzyme sites to generate glutathione S-transferase (GST) fusion polypeptides. These polypeptides, designated C1, C2, N1, N2, and N3, contain amino acids 553 to 1043, 505 to 1242, 342 to 644, 158 to 373, and 25 to 157, respectively, of the human p180 protein. The plasmids were verified by nucleotide sequencing. All the polypeptides were expressed in Escherichia coli XL1-blue, solubilized in 50 mM Tris-HCl (pH 8.0), purified by glutathione-Sepharose column chromatography (Amersham Pharmacia Biotech), and dialyzed against PBS.
In vitro overlay assay using biotinylated polypeptides.
Biotinylation of the recombinant GST fusion polypeptides was carried out by incubating the purified polypeptides (about 0.3 mg of protein/ml) with biotin-3-sulfo-N-hydroxysuccinimide ester (final concentration, 200 μg/ml; Sigma). After a 30-min incubation at room temperature, 20 μl of 1 M Tris-HCl (pH 7.4) was added and then the mixture was dialyzed against PBS at 4°C. For in vitro overlay assays, proteins were separated by SDS-PAGE and transferred to a PVDF membrane. After being blocked with Block Ace (Snowbrand Milk Products, Tokyo, Japan) at 4°C overnight, the membrane was incubated with a diluted biotinylated probe (1:100) in 50% Block Ace in PBS containing 0.05% Tween-20 (PBS-T) at 25°C for 2 h. The membrane was washed with PBS-T extensively and then was incubated with streptavidin conjugated with horseradish peroxidase (Vector Laboratory) at 25°C for 1 h. The positive bands were detected by nitroblue tetrazolium-BCIP (5-bromo-4-chloro-3-indolylphosphate)-NADH (Wako) or an ECL system.
Anti-p180 antibody production and affinity purification.
Antibodies against C1 or N1 fragments of human p180 recombinant polypeptides were produced in New Zealand White rabbits. To remove cross-reactivity due to an overlapping between the C1 and N1 polypeptides corresponding to amino acids 553 to 644, the anti-C1 antiserum was preabsorbed with mixtures of N1 polypeptides and glutathione-Sepharose beads and the anti-N1 antiserum was preabsorbed with a C1 polypeptide-glutathione-Sepharose bead mixture. The antibody against C1 or N1 polypeptide in the preabsorbed serum was purified by immunoaffinity chromatography using an antigen-coupled Sepharose gel as follows. First, residual anti-GST antibodies were separated and purified by a GST-coupled Sepharose gel, and then anti-C1 or -N1 antibodies were purified with the antigen-coupled Sepharose. Western blotting analysis revealed no cross-reactivity between the affinity-purified anti-N1 and -C1 antibodies (data not shown). The purified anti-GST antibody was used as the control in the immunofluorescence (IF) and immunoprecipitation experiments discussed below.
Antibodies and IF analysis.
The monoclonal antibodies (MAbs) against calnexin, the Xpress epitope, and CD13 (WM-15) were obtained from Transduction (Lexington, Ky.), Invitrogen (Carlsbad, Calif.), and Silenus Lab (Hawthorn, Australia), respectively. The goat polyclonal antibody against calreticulin was from Santa Cruz Biotechnology (Santa Cruz, Calif.). The rabbit polyclonal antibody against HCMV UL48 was a generous gift from W. Gibson (Johns Hopkins University). The MAb against the HCMV IE protein (MAB 810) was purchased from Chemicon (Temecula, Calif.).
The cells grown on coverslips were fixed with 4% paraformaldehyde in PBS for 1 h and then permeabilized with 0.1% Triton X-100 in PBS for 2 min. After being blocked with 10% FBS in PBS, the cells were incubated with primary antibodies for 1 h in PBS containing 0.5% gelatin, 10 mM glycine, and 10 mM EDTA and then were incubated with the secondary antibodies (1 h at room temperature), which were anti-rabbit or anti-mouse immunoglobulin G (IgG) antibodies labeled with fluorescein isothiocyanate (FITC) or Alexa Fluor 568 (Molecular Probes). For triple staining, cells were incubated with a FITC-labeled swine anti-goat IgG antibody first, followed by a Cy5-labeled goat anti-rabbit IgG antibody (Chemicon) and an Alexa Fluor 568-labeled goat anti-mouse IgG antibody. Cells were examined under a confocal laser-scanning microscope (LSM 410; Carl Zeiss, Oberkochen, Germany).
PCR cloning and construction of UL48-expressing plasmids and antibody production.
HCMV genomic DNA was extracted from the purified HCMV particles. DNA fragments of the UL48 gene were amplified by PCR. The PCR products were cloned into pTSX-1 to generate pTSX6.8F, which covers 6.8 kbp of DNA encoding UL48. A mammalian expression vector encoding UL48 tagged with the Xpress epitope was generated by insertion of a BamHI and XbaI digestion fragment of pTSX6.8F in frame into a pcDNA4 HisMax vector (Invitrogen, Groningen, The Netherlands), designated pcUL48. Orientation and sequence were verified by nucleotide sequencing. An 0.8-kbp DNA fragment of the UL48 gene excised by SphI and SmaI digestion was subcloned into pQE32 (Qiagen) to generate bacterial expression vector pHisUL48/S1. The histidine-tagged polypeptide (UL48/S1), which contains amino acids 457 to 738 of the UL48 protein, were expressed in E. coli XL1-blue, solubilized in 50 mM Tris-HCl (pH 8.0) containing 0.3 M NaCl and 8 M urea, purified by TALON column chromatography (Clontech) according to the manufacturer's instruction, and dialyzed against PBS containing 5 mM EDTA. Antibodies against UL48/S1 were produced in rabbits and affinity purified as described above.
Transfection, immunoprecipitation, and Western blotting analysis.
293T cells were transiently transfected with 1.5 μg of pcUL48 or an HCMV IE1 expression plasmid (18) using Fugene-6 (Roche Diagnostics) according to the manufacturer's instructions. After 48 h, cells were lysed in 1 ml of lysis buffer (20 mM Tris-HCl [pH 7.4], 5 mM EDTA, 300 mM NaCl, 1% Nonidet P-40, 1% sodium deoxycholate) containing proteinase inhibitor mixtures (Complete; Roche Diagnostics) for 30 min on ice. Insoluble debris was removed by centrifugation. After cell lysates were precleared with protein A-Sepharose (Amersham Pharmacia Biotech), the precipitating antibody was added, and the antibody-antigen complex was recovered with protein A-Sepharose. The immune complexes were separated by SDS-PAGE and subjected to either Western blotting or a virus overlay assay. In the Western blotting analyses of human p180 in various cell lines, 2 × 106 to 4 × 106 cells were lysed with SDS sample buffer (0.1 M Tris-HCl [pH 6.8], 1% SDS) and then centrifuged for 15 min at 10,000 × g. Protein concentration was measured by a BCA kit (Pierce, Rockford, Ill.). Equal amounts of lysates (20 μg of proteins) were separated by SDS-PAGE and transferred to a PVDF membrane as described above. The filters were probed with the affinity-purified antibodies against human p180 protein, and the proteins were detected by an ECL system.
Transmission electron microscopy.
HCMV-infected or mock-infected cells were harvested by scraping at various times postinoculation (p.i.) after washing the monolayers with PBS. The sedimented cell pellet was fixed with 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.2) for 1 h at 4°C, postfixed in OsO4, and embedded in Epon. Ultrathin sections were prepared on a grid and stained with uranyl acetate and lead citrate. The sections were examined with a JEOL 1200 EX (Tokyo, Japan) at 80 kV.
RESULTS
Detection of specific binding of 35S-HCMV to MRC-5 proteins.
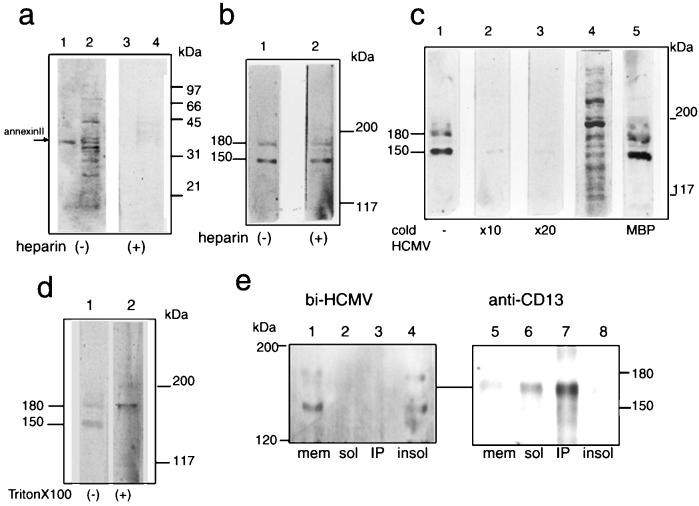
HCMV-permissive cells were used for the initial screening of 35S-HCMV binding proteins of host cells. The membrane fraction of MRC-5 was isolated and subjected to the virus overlay assay. While the assay detected several positive bands including approximately 30- to 35-, 45-, and 70-kDa proteins after separation in SDS-12% PAGE gels (Fig. 1a, lane 2) most of them turned out to be negative when tested in the presence of heparin (lane 4). 35S-HCMV binding to authentic annexin II was also observed only in the absence of heparin (compare lanes 1 and 3). However, more-specific positive signals were obtained for proteins of more than 120 kDa when proteins were separated in SDS-5% PAGE gels and electrotransferred under strong blotting conditions as described in Materials and Methods. Figure 1c represents a typical staining pattern under these conditions. Two proteins, the molecular sizes of which were 150 and 180 kDa, have high abilities to bind with the 35S-HCMV probe (Fig. 1c, lane 1). Unlike binding to the 30- to 70-kDa proteins, 35S-HCMV binding to the 150- and 180-kDa proteins was not affected by the addition of 100 μg of heparin/ml (Fig. 1b, lane 2). The binding was assumed to be specific since cold-virion competition studies showed that the positive bands became faint following the addition of excess amounts of cold virion (Fig. 1c, lane 2 and 3), while it was not affected by the addition of myelin basic protein (lane 5). Similar binding patterns were observed with the biotinylated HCMV probe (Fig. 1e, lane 1) but not with biotinylated BSA (data not shown).
FIG. 1.
The radiolabeled HCMV probe specifically bound to two host proteins with molecular masses of 150 and 180 kDa. (a) The membrane fractions of MRC-5 (lanes 2 and 4) and authentic annexin II (lanes 1 and 3) were separated in SDS-12% PAGE gels and subjected to the virus overlay assay as described in Materials and Methods in the absence (lanes 1 and 2) and the presence (lanes 3 and 4) of heparin (100 μg/ml). (b to e) The membrane fraction was separated in SDS-5% PAGE gels, transferred to a PVDF membrane under strong conditions, and a virus overlay assay was carried out as for panel a. (b) Assay in the absence (lane 1) and the presence (lane 2) of heparin. (c) Assay in the absence (lane 1) and the presence of cold virions at a 10-fold excess (lane 2) and a 20-fold excess (lane 3) or in the presence of myelin basic protein (MBP; lane 5). Lane 4, protein pattern of the membrane fraction stained by amido black. (d) Virus overlay assay performed in the absence (lane 1) and the presence (lane 2) of Triton X-100 (0.5%, final concentration). (e) The membrane fraction (mem) was solubilized with 1% Triton X-100 in PBS for 30 min on ice and centrifuged to separate the soluble (sol) and insoluble (insol) fractions. Then immunoprecipitation with a MAb against CD13 (WM15) was carried out, followed by either a virus overlay assay (lanes 1 to 4) or immunostaining by WM15 (lanes 5 to 8). Shown are the membrane fraction of MRC-5 cells prior to solubilization (lanes 1 and 5), the Triton X-100-soluble fraction (lanes 2 and 6), the immunoprecipitates (IP) with WM15 (lanes 3 and 7), and the Triton X-100-insoluble fraction (lanes 4 and 8). bi-HCMV, biotinylated HCMV.
CD13/aminopeptidase N, whose molecular size ranged approximately from 150 to 165 kDa, has been reported to bind and associate with HCMV (34). To determine whether an anti-CD13 antibody recognizes the virus-binding 150-kDa protein, immunoprecipitation by a MAb against CD13 (WM15) was performed, followed by either Western blotting analysis or the virus overlay assay (Fig. 1e). The WM15 immunoprecipitates were not bound by the HCMV (lane 3) and showed a mobility different from that of the 150-kDa protein (compare lanes 4 and 7). In contrast, the virus-binding 150-kDa protein was found exclusively in the insoluble fraction by Triton X-100 treatment (lane 4). Therefore the 150-kDa protein was distinguished from CD13. The 180-kDa protein was also detected in the insoluble fraction under these conditions. Addition of detergent to the probe solution resulted in increased binding ability to the 180-kDa protein but, in contrast, markedly reduced binding to the 150-kDa protein (Fig. 1d).
Identification of the 180-kDa protein by partial amino acid sequencing.
Our preliminary study using the HCMV overlay assay revealed that the HCMV-binding activity was well correlated with the presence of the 180-kDa protein, showing a marked metachromasia in Coomassie brilliant blue staining (data not shown). We isolated the metachromatic 180-kDa protein by preparative SDS-PAGE as described in Materials and Methods; this permitted us to analyze the internal amino acid sequence. Protein sequencing of the isolated fragments derived from the 180-kDa band provided four sequences, (i) (S)HVE(D)(G)(D), (ii) HPPAPAEPSSD(L)A(S)(K), (iii) (S)VEEEQVX(R), and (iv) KTESASVQ(G)RN (residues that are uncertain from protein sequence analysis are in parentheses). A homology search of the EMBL database revealed that internal amino acid sequences i, ii, and iii matched the human homologue of the ES130/p180 (p180) protein (19), which has been reported to be a member of the ES130/kinectin family. Sequence iv was found to be a new sequence in the database, but our cDNA cloning study revealed that the human homologue of ES130/p180 has sequence identical to sequence iv, as described below. Based on these results, the 180-kDa protein appeared to be the human homologue of the p180 protein (19, 40).
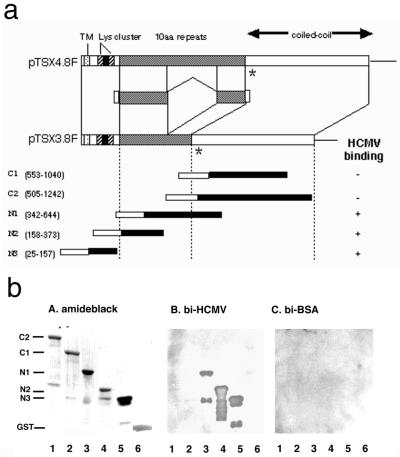
Human cDNA of p180 was amplified by PCR as described in Materials and Methods, and we isolated a 3.8-kbp clone (pTSX3.8F) containing a 0.8-kbp tandem repeat region. Multiple splicing isoforms of human p180, whose repeat numbers varied, have been reported (19). Sequencing analyses revealed that the 3.8-kbp clone was basically identical to the reported sequence of the human p180 except for the length of the tandem repeats and other minor differences such as the presence of a three amino-acid insertion. The product of the 3.8-kbp clone was found to be a new isoform possessing 24 decapeptide repeats. Figure 2a shows the domain structures deduced from the primary amino acid sequences and the relationship between the human p180 isoforms possessing 24 and 54 decapeptide repeats (encoded by pTSX3.8F and pTSX4.8F, respectively). They each have a predicted transmembrane domain close to the N terminus and a large cytoplasmic domain consisting of two distinct regions, namely, a highly basic N-terminal domain containing a basic tandem repeat and a C-terminal acidic coiled-coil domain (19). The cDNA sequence encoding human p180 submitted to the database was divided into two sequences, namely, that encoding a tandem repeat-lacking form and that encoding a 54-repeat-only domain. The internal amino acid sequence (sequence iv) provided above was assigned to a junction site between the tandem repeat and C-terminal domains: this was the reason why we could not find sequence iv in the database. Therefore, sequence iv is direct evidence that the p180 protein in human cells is expressed as a tandem repeat-possessing form.
FIG. 2.
The domain structure of human p180 isoforms and HCMV binding to recombinant p180 polypeptides containing various regions. (a) Schematic representation of the domain structure of the human p180 protein and the relationship between isoforms possessing 54 and 24 tandem repeats, which were deduced from the primary amino acid sequences encoded by pTSX3.8F and pTSX4.8F. They each have a predicted transmembrane domain (TM) close to the N terminus, a highly basic N-terminal domain consisting of lysine clusters, a proline-rich domain (solid), a basic tandem repeat domain (10aa repeats), and a C-terminal acidic coiled-coil domain (19). ∗, insertion of three amino acids in this clone at amino acid 488. Various truncated human p180-GST fusion polypeptides were designed to address the domain responsible for the binding to HCMV. Open portion of the bar for each truncated mutant, N-terminal GST. Numbers in parentheses denote the amino acid residues encoded by pTSX3.8F. A summary of the binding experiments in the virus overlay assay is shown on the right. (b) Various truncated human p180/GST-fusion polypeptides were purified and used for the overlay assay. (A) Protein staining with amido black; (B) overlay assay with a biotinylated HCMV (bi-HCMV) probe; (C) overlay assay with biotinylated BSA.
HCMV binding to recombinant and cellular p180 polypeptides.
Bacterially expressed polypeptides encoded in various regions of the cDNA were used to address HCMV binding to human p180. The virus overlay assay clearly showed that N1 and N2 fragments (Fig. 2b, lanes 3 and 4), which contain the tandem repeat domain of human p180 polypeptides, bound to the biotinylated HCMV probe but neither to C1 nor to C2 (lanes 1 and 2). The N3 polypeptide, consisting of the basic N-terminal domain, was also bound by the probe (lane 5). Other basic proteins including GST (lane 6) and myelin basic protein (data not shown) were not bound by the probe, suggesting specific binding to the human p180 polypeptides. These results suggest that HCMV binding to human p180 occurred through the N-terminal domain.
We next examined the binding of the HCMV probe to the endogenous p180 protein. Antibodies were raised against C1 and N1 recombinant polypeptides and affinity purified. Cell lysates from MRC-5 cells were subjected to immunoprecipitation using either the affinity-purified anti-p180 antibody or the anti-GST antibody as the control. The binding of the biotinylated HCMV probe was detected in immunoprecipitates by the anti-p180 antibody with an apparent molecular size of 180 kDa (Fig. 3a, lane 2), but not by the anti-GST antibody (Fig. 3a, lane 1). These results verified that it was human p180 to which the HCMV probe specifically bound.
FIG. 3.
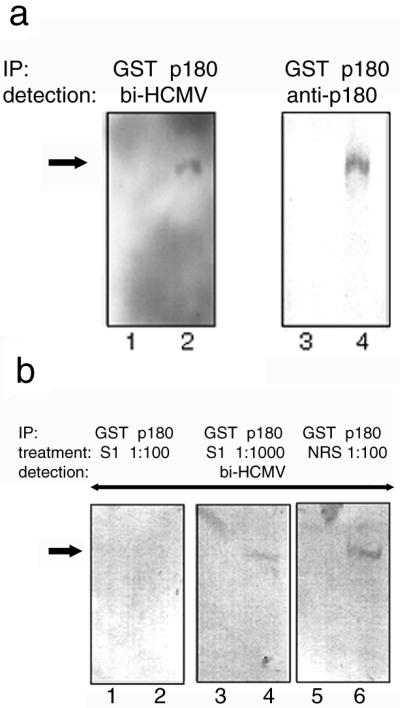
HCMV binding to the cellular p180 protein. The binding of the HCMV probe to the cellular p180 protein was tested using immunoprecipitated (IP) antigens. MRC-5 cell lysates were subjected to immunoprecipitation and either to a virus overlay assay or Western blotting. (a) Proteins immunoprecipitated by the anti-GST antibody (lanes 1 and 3) or the anti-p180 antibody (lanes 2 and 4) were used for either virus overlay assays (left) or Western blotting (right). In lane 2, a positive band with an apparent molecular size of 180 kDa was detected (arrow). bi-HCMV, biotinylated HCMV. (b) The biotinylated HCMV probe was pretreated with antiserum against UL48/S1 at dilution of 1:100 (left) or 1:1,000 (middle) or normal rabbit serum at a 1:100 dilution (right). After 2 h of incubation at 4°C, a virus overlay assay was carried out for panel a. Proteins immunoprecipitated by the anti-GST antibody (lanes 1, 3, and 5) or the anti-p180 antibody (lanes 2, 4, and 6) were used as antigens. p180 is shown by an arrow.
Human p180 proteins were highly expressed in HCMV-permissive cells.
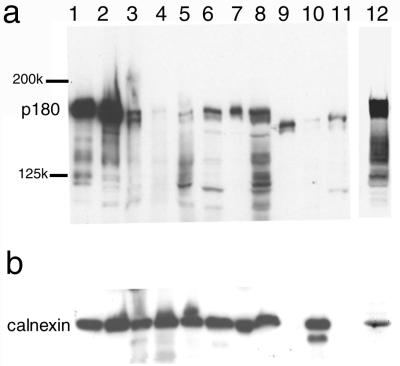
It is well known that HCMV can replicate only in a restricted range of human-derived cells, including MRC-5, K-1034, and Caco-2 cells. We examined the levels of expression of human p180 in various cell lines by Western blotting analysis (Fig. 4). Multiple positive bands were detected in most human cells, and the levels of expression of the 180-kDa protein varied considerably among the cells, whereas those of calnexin were similar. The highest levels of p180 expression were evidently observed in MRC-5, K-1034, and Caco-2 cells (Fig. 4, lanes 1, 2, and 12, respectively). Several positive bands corresponding to molecular sizes between 110 and 140 kDa are assumed to be isoforms of the human p180 protein having shorter tandem repeats. In nonhuman cell lines such as Vero, NIH 3T3, and CHO, ca.160-kDa proteins were primarily detected instead of the 180-kDa form, although their identity remains unclear.
FIG. 4.
Human p180 and related proteins were highly expressed in the HCMV-permissive cell lines. Total cell lysates from various cell lines were subjected to Western blotting and analyzed with the affinity-purified anti-p180 C1 antibody (a) or a MAb against calnexin, an ER resident membrane protein (b). p180 was most highly expressed in MRC-5, K-1034, and Caco-2 cells, which were known as fully permissive for HCMV infection. Lanes: 1, MRC-5; 2, K-1034; 3, U937; 4, THP-1; 5, HSB-2; 6, Ramos 3; 7, HeLa; 8, 293T; 9, NIH 3T3; 10, Vero; 11, CHO; 12, Caco-2.
Among the above cell lines tested, monocytic cell line THP-1 contained a trace amount of human p180. Because THP-1 has been reported to become susceptible to HCMV infection after treatment with phorbol ester (41), the effect of TPA treatment on p180 expression was studied. Interestingly, p180 protein expression in THP-1 cells was induced by TPA treatment (Fig. 5a), while that of calnexin was not (Fig. 5b). Immunofluorescence analysis also confirmed the induction of human p180 expression in the TPA-treated THP-1 cells, which showed the typical multinucleated differentiated morphology (Fig. 5c). Thus, a close correlation between levels of human p180 expression and HCMV permissiveness was observed.
FIG. 5.
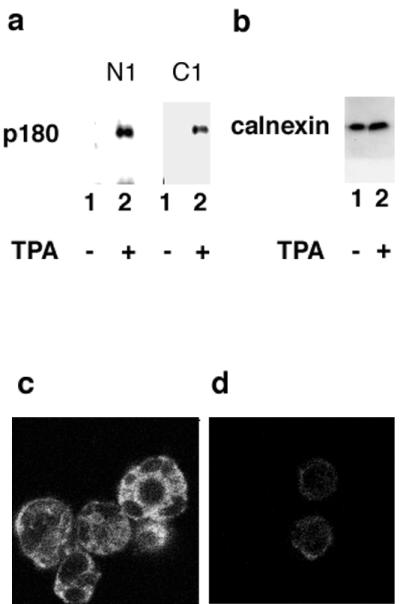
Induction of p180 protein expression in THP-1 cells upon treatment with TPA. (a) Western blotting analysis of THP-1 cell lysates. The p180 protein was detected by affinity-purified antibodies against N1 or C1, as indicated. (b) A MAb against calnexin was used. Lanes (a and b): 1, no TPA treatment; 2, TPA treatment. (c and d) IF analyses on THP-1 cells using the affinity-purified antibody against p180 with TPA treatment (c) and without TPA treatment (d).
Identification of a virion counterpart as the UL48 gene product.
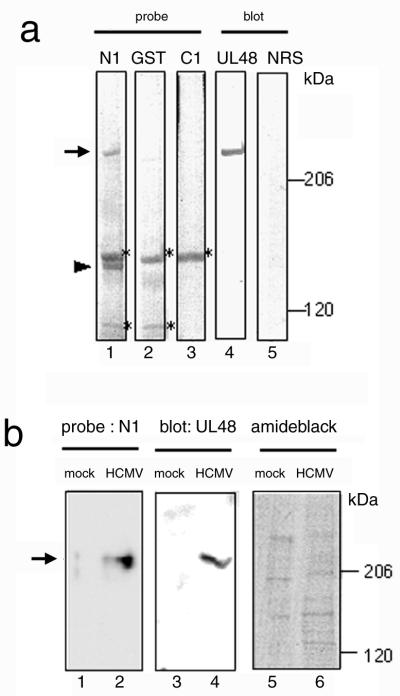
To address which components of HCMV were bound to p180, an in vitro overlay assay using a p180 polypeptide probe was carried out. HCMV proteins were separated by SDS-PAGE, transferred to a membrane, and overlayed with the biotinylated probes. This assay reproducibly detected a 230-kDa positive band using the N1 probe and also, occasionally, a 140-kDa protein (Fig. 6a, lane 1). It appears to be specific because the signal was not observed with the control GST probe (lane 2) or C1 probe (lane 3). This specificity is in agreement with the result from the virus overlay assay of Fig. 2. The binding to the 150-kDa protein was unlikely to be specific because a signal was obtained with the control GST probe (Fig. 6a, lane 2). The binding of the N1 probe to the 230-kDa protein was also detected in HCMV-infected MRC-5 cells at the late phase of infection (Fig. 6b, lane 2) but not in mock-infected cells (lane 1).
FIG. 6.
The recombinant p180 polypeptide specifically bound to the UL48 protein. (a) The purified virus proteins were separated in SDS-5% PAGE gels and subjected to either an in vitro overlay assay (lanes 1 to 3) or Western blotting (lanes 4 and 5). The N1 probe (lane 1), GST probe (lane 2), and C1 probe (lane 3) were used for the in vitro overlay assay, and the anti-UL48 antibody (lane 4) or normal rabbit serum (NRS) (lane 5) were used for Western blotting. Arrow, 230-kDa positive band; arrowhead, 140-kDa band; ∗, protein bands assumed to be nonspecific. (b) Mock-infected (lanes 1, 3, and 5) and HCMV-infected (lanes 2, 4, and 6) fibroblasts at 4 days p.i. were used instead of the virion preparation and analyzed as for panel a. Lanes 1 and 2, N1 probe; lanes 3 and 4, anti-UL48 antibody; lanes 5 and 6, protein staining by amido black.
Among the genes contained in HCMV, the UL48 gene product is supposed to be a candidate for the 230-kDa protein, based on molecular weight. As shown in Fig. 6a, blotting analysis using the anti-UL48 antibody revealed that the antibody bound to a protein showing the same mobility as the 230-kDa protein (Fig. 6a, lane 4). These results suggested that the N1-binding 230-kDa protein was the UL48 gene product. We have not yet identified the other N1-binding protein of 140 kDa, mainly due to existence of multiple virus proteins.
To determine whether HCMV binding to p180 was mediated through interaction with the UL48 protein (pUL48), inhibition experiments using an anti-UL48 antibody were carried out. For this purpose, we generated rabbit polyclonal antibody against the recombinant UL48 polypeptide (UL48/S1) and incubated the HCMV probe with the antibody prior to the virus overlay assay. As shown in Fig. 3b, the binding of the HCMV probe to the endogenous p180 was efficiently reduced by pretreatment with the anti-UL48/S1 antibody in a dose-dependent manner but was not affected by normal rabbit sera. Therefore it was suggested that the HCMV binding to p180 was mediated, at least partially, through interaction with tegument protein pUL48.
Interaction between p180 and UL48 in transfected and infected cells.
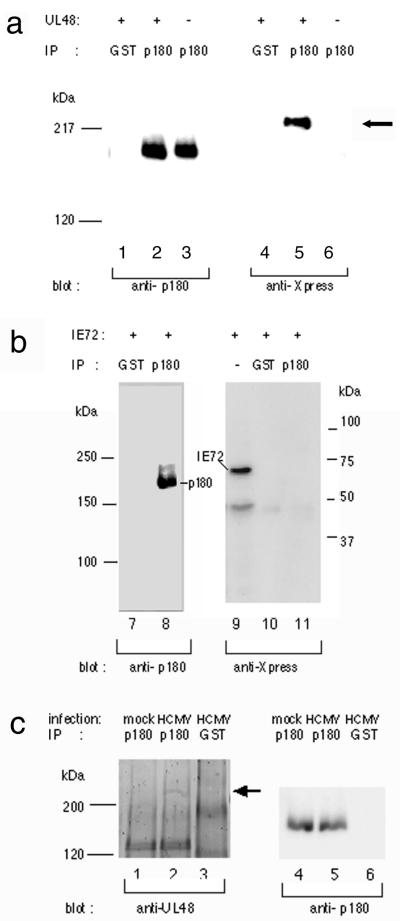
We tested whether endogenous p180 can interact with pUL48 in vivo. pUL48 fused with Xpress epitope was transiently expressed in 293T cells, and cell lysates were immunoprecipitated with either the anti-p180 antibody or the anti-GST antibody. Figure 7a shows that the recombinant pUL48 was coprecipitated with p180. As a control experiment we used recombinant IE1 of HCMV fused with the same tags used in similar coprecipitation assays. IE1 was not detected in the immune complex of the anti-p180 antibody shown in Fig. 7b. Next we carried out similar coprecipitation assays with HCMV-infected cells to determine whether interaction between p180 and UL48 actually occurs during HCMV infection. At 4 days p.i. lysates were prepared from mock-infected and HCMV-infected cells and were sedimented to remove capsids prior to the immunoprecipitation assay to reduce background levels. Although a majority of the UL48 antigen, probably associated with the capsid, was removed from the lysates by the presedimentation procedure (data not shown), a small but distinct amount of the UL48 protein was coprecipitated with p180 (Fig. 7c, lane 2) in HCMV-infected cells but not with GST (lane 3). IE1 was not detected in the immune complex of the anti-p180 antibody (data not shown). Thus these data clearly indicate that nondenatured forms of p180 and pUL48 proteins can interact specifically both in transfected and HCMV-infected cells.
FIG. 7.
The UL48 protein specifically interacted with endogenous p180. (a) 293T cells were transfected with pcUL48 (lanes 1, 2, 4, and 5 [the left lane is considered lane 1]) or vehicle only (lanes 3 and 6). Immunoprecipitation (IP) was carried out using either the anti-p180 antibody (lanes 2, 3, 5, and 6) or the anti-GST antibody (lanes 1 and 4), followed by blotting with anti-Xpress or anti-p180 antibodies. (b) An HCMV IE1-expressing plasmid was used instead of pcUL48 and analyzed as for panel a. (c) At 4 days p.i. mock-infected and HCMV-infected cells were lysed and centrifuged at 50,000 × g for 30 min to remove capsids and analyzed as for panel a. The UL48 protein (arrow) was detected by the anti-UL48 antibody.
Unique relocation of human p180 during the late replication cycle of HCMV infection.
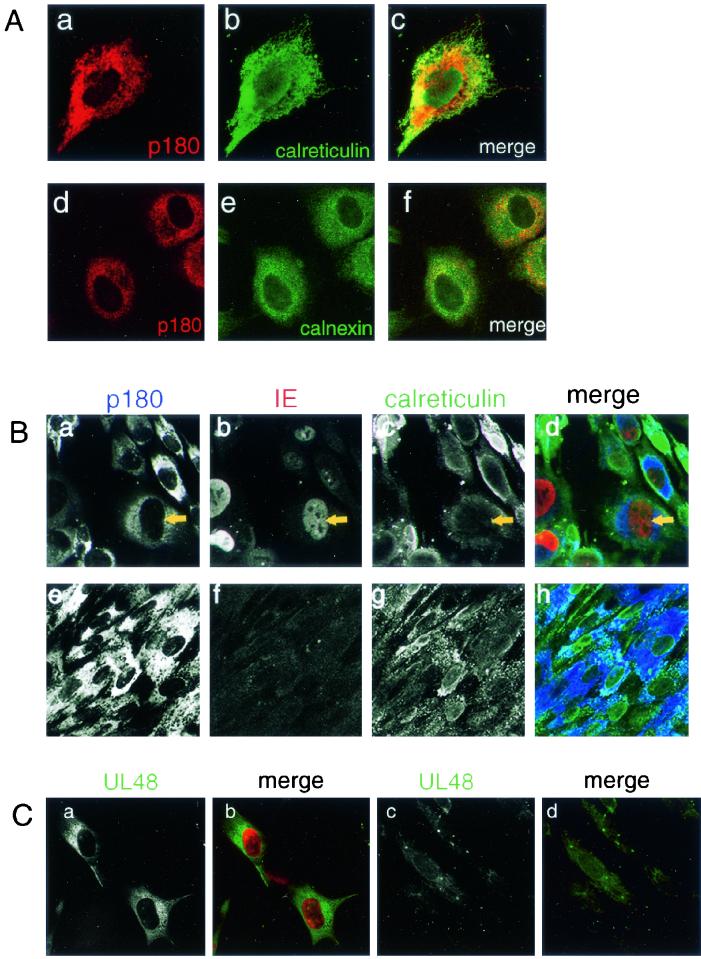
Canine p180 has been originally reported as an endoplasmic reticulum (ER) membrane protein in MDCK cells (28), while chicken ES/130 was possibly associated with the vacuolar system in the embryonic heart (25). To define the subcellular localization of p180 in mock-infected and HCMV-infected cells, affinity-purified antibodies against p180 were used. A perinuclear reticulated staining pattern throughout the cytoplasm was observed in MRC-5 and K-1034 cells (Fig. 8A, a and c, respectively). Double immunostaining analysis revealed the colocalization of human p180 and ER resident proteins calreticulin and calnexin in MRC-5 and K-1034 cells (Fig. 8A, c and f, respectively).
FIG. 8.
Immunofluorescence analyses of the p180 protein in cultured cells. (A) MRC-5 (a to c) or K-1034 (d to f) cells were cultured on a coverslip and processed for IF assay. (a and d) Antibody against p180; (b) anti-calreticulin antibody; (e) MAb against calnexin; (c and f), merged signal. The perinuclear reticular staining patterns for human p180 in MRC-5 (a) and K-1034 cells (d) were observed; the patterns resemble those for ER markers calreticulin and calnexin. (B) HCMV-infected (a to d) or mock-infected (e to h) MRC-5 cells at 3 days p.i. were processed for triple staining. (a and e) Anti-p180 antibody; (b and f) MAb against IE protein; (c and g) anti-calreticulin antibody; (d and h) merged signals. The infected cells exhibiting the enlargement of IE-protein positive nucleus showed a unique relocation of p180 signal to the perinuclear area, which is segregated from that of calreticulin, an ER protein. (C) HCMV-infected (a and b) and mock-infected (c and d) MRC-5 cells at 3 days p.i. were stained with anti-UL48/S1 antibody and a MAb against IE protein. (a and c) Anti-UL48/S1 antibody; (b and d) merged signal. UL48 signal (green) was observed as a juxtanuclear vacuolar pattern in the IE protein (red)-positive cells.
After infection with HCMV, the staining pattern of p180 in MRC-5 cells was not affected unless the replication cycle had progressed into the late phase (data not shown). When the infected cells exhibited CPE such as enlarged nuclei and cytoplasm at 3 days p.i., the p180 signal was relocated to the juxtanuclear area and exhibited a vacuolar pattern (Fig. 8B, a). At this time p.i., ER luminal marker calreticulin showed a pattern distinct from that of p180 and was mainly observed in the periphery of the cells (Fig. 8B, c). A very faint signal throughout the cytoplasm was obtained with the control anti-GST antibody in both HCMV- and mock-infected cells (data not shown). The UL48 signal was also found to show a juxtanuclear vacuolar pattern at this time p.i. (Fig. 8C), which was consistent with the previously reported localization of pUL48 (6).
Ultrastructure of HCMV-infected cells at the late phase of infection.
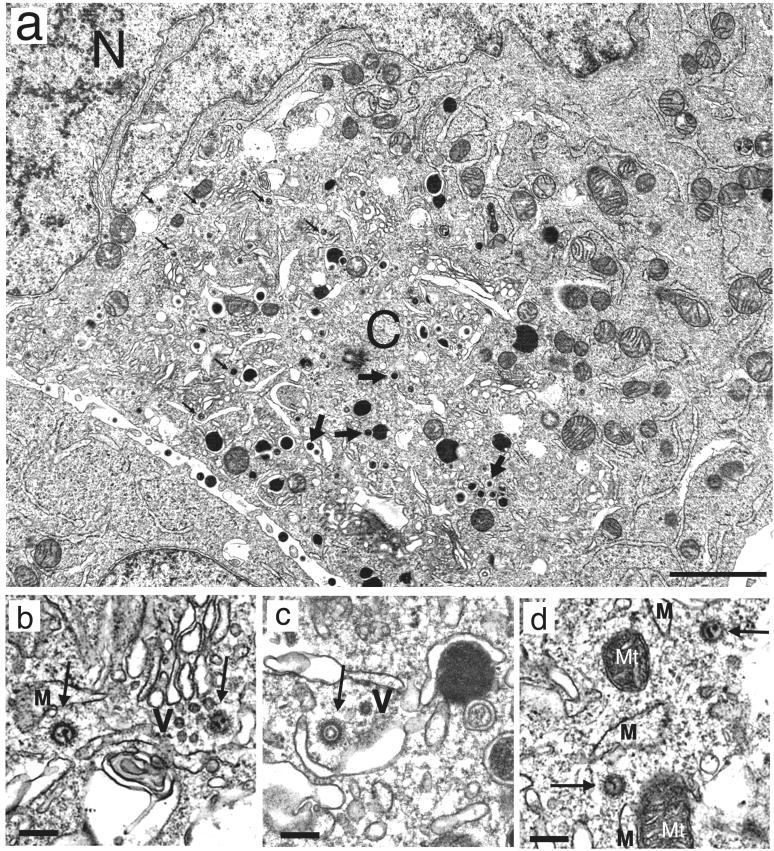
For further characterization of the p180-positive juxtanuclear region in the IF analysis, we analyzed ultrastructurally HCMV-infected cells by transmission electron microscopy (Fig. 9). As previously reported, infected cells exhibited an altered morphology of subcellular membranes, which includes dilation of rough ER, morphologically aberrant Golgi apparatus, and segregation of subcellular organelles (15, 29). These changes first appeared prominent in the area between the nucleus and centriole at 3 days p.i. At this time, naked virions in the cytoplasm were frequently observed (Fig. 9a), as reported frequently (15, 29, 30, 36). The naked virions were observed mainly in the area between the nucleus and centriole, which is assumed to be very close to the p180-positive area defined by the IF analysis. The distribution of naked virions in the juxtanuclear region is in contrast to that of enveloped virions and dense bodies appearing to be mature shapes, which were observed mostly in the distal area. The electron micrographs at a higher magnification revealed that the tegument of naked virions appeared to be decorated with filamentous structures and connected to the cellular membranes or vesicles through a thin filamentous network (Fig. 9b to d), suggesting interaction of the tegument structure with intracellular membranes morphologically at this time p.i. Then, the area exhibiting altered morphology continued to extend gradually (data not shown).
FIG. 9.
Ultrastructural features of HCMV-infected fibroblasts at late phase of infection. HCMV-infected MRC-5 cells (multiplicity of infection, 1 to 3) were harvested 3 or 4 days p.i. and processed for electron microscopy. (a) A number of cytoplasmic naked virions without envelopes were observed (small arrows) in the area between the nucleus (N) and centriole (C). Note that enveloped virions (large arrows) and dense bodies with mature shapes are mostly found in the distal area. The subcellular organelles exhibited altered membrane morphology such as dilation of RER and morphologically aberrant Golgi apparatus. Normal organelles were segregated to the cell periphery. (b to d). Higher-magnification micrographs revealed a thin filamentous network between the fine filamentous tegument of the naked virions (arrows) and the cellular vesicles (V) or membranes (M). Mt, mitochondrion. Bars, 2 μm (a) and 0.2 μm (b to d).
DISCUSSION
Little information on the molecular mechanisms of HCMV transport in the cytoplasm is available to date. To our knowledge, no host factor associated with HCMV particles in the process of intracellular movement has been reported. Since movement of virus is likely to proceed by the cellular transport system (32), identification of host factors which bind to HCMV particles would provide a valuable insight into the mechanism of the virion transport process. In the present paper, we first demonstrated that the human homologue of ES130/p180 binds to HCMV through interaction with the UL48 protein. The interaction between the two proteins was assumed to be specific and was observed both in vitro and in vivo. Moreover, we demonstrated that p180 is highly expressed in the cytoplasm of HCMV-permissive cells. The UL48 gene is conserved among herpesviruses (9) and encodes the largest HCMV polypeptide (3, 14). Because pUL48 is one of the predominant components of the HCMV tegument and is tightly associated with the capsid (14) and because p180 is found to be relocated to the juxtanuclear region at the late phase of infection, the specific interaction of p180 with pUL48 may be involved in the cytoplasmic events of the HCMV replication cycle such as intracellular movement and/or the capsid assembly process. The domain structure of p180 predicted from the primary sequence also leads us to speculate that it is possibly involved in intracellular transport systems as discussed below.
The p180 protein was first identified as a ribosome receptor (28) and is highly conserved among species; however, its physiological role, including a proposed function as a receptor for the ribosome on rough ER, is poorly understood. Based on the deduced primary sequence, it was predicted that p180 was a membrane protein with a short luminal tail. Most of the molecule is projected into the cytoplasmic area (19). The large cytoplasmic domain is divided into two distinct domains structurally, namely, a highly basic N-terminal half including the tandem repeat and a C-terminal acidic coiled-coil domain. It was proposed that the tandem repeat in the N-terminal half was a ribosome-binding domain (28). It is noteworthy that the N-terminal domain was also responsible for HCMV binding, as presented here. It was reported that p180 contains a region homologous to kinectin (25) which was originally isolated as a candidate receptor of the kinesin motor on the ER (37). This strongly suggests that p180 functions in the vesicle transport or early secretory pathway of the host cells. The enhanced cellular secretory pathway and membrane proliferation were demonstrated in Saccharomyces cerevisiae after overexpression of the canine homologue (5). Our preliminary study also showed that an overexpressed human p180 resulted in upregulation of cellular secretory pathways in mammalian cells (K. Ogawa-Goto et al., unpublished results). Because structural features of p180 strongly suggest that it may have a role in the vesicle transport or early secretory pathway in host cells, it is intriguing to speculate that HCMV binds to the host p180 protein through the tegument in order to facilitate cytoplasmic transport of the capsid. The functions of p180 both in the physiological state and in the HCMV replication process remain to be elucidated.
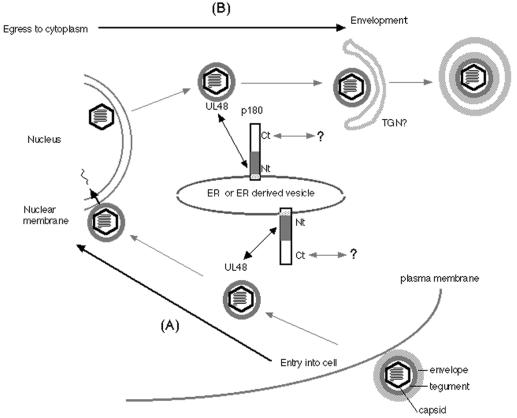
In HSV-1 infection of Vero cells, a possible involvement of a dynein motor and microtubule network has been demonstrated (33), and interaction of the motor with a tegument protein of HSV-1 has also been reported (43). The mechanism of HCMV transport in the cytoplasm is entirely unknown. Although it is supposed that pUL48 binds tightly to the capsid and may have a role in structural integrity (14), no functional analyses have been carried out to date. Recently, Desai proposed a very attractive function for the HSV-1 homologue of UL48 (12). His data elegantly showed that a null mutation of the HSV-1 homologue, UL36, resulted in cytoplasmic accumulation of unenveloped DNA-filled capsids during the late phase, and he proposed that UL36 may function in directing capsids actively to a maturation compartment. It remains unclear, at present, whether the HCMV UL48 protein has the same function as the HSV-1 homologue. Nevertheless, it is intriguing to speculate that the specific binding between p180 and pUL48 may work to facilitate a proper destination of capsid transportation in cytoplasm. Notably, an IF assay of HCMV-infected cells showed that p180 was concentrated at the juxtanuclear region in the late phase of infection. This unique pattern of p180 in HCMV-infected cells seems to be similar to that of UL48, as shown in the present study and as reported previously (6), and also to that of the major tegument protein encoded by the UL32 gene (26). Coincidentally, a number of naked virions were observed in the area between the nucleus and centriole by electron microscopy at this time p.i. This region also appeared to be in the vicinity of a p180-positive area, again implying the possible interaction between p180 and the UL48 protein in the HCMV replication cycle. This observation is probably consistent with recently accumulated findings that the final HCMV envelopment occurs in the cytoplasmic compartments of infected cells (15, 24, 26, 27, 36). The reported calculation that the cytoplasmic domain of p180 has a rod structure 124 nm in length (19) may also account for the structural features of this molecule, which possesses the ability to bind with the tegument. This size is assumed to be within the range of those of the filamentous structures apparently projected from the cellular membranes or vesicles which connect to the tegument layer of naked virions in our electron-microscopic observations, although their identity is still unknown. Figure 10 illustrates our model of the interaction of p180 with HCMV in the replication cycle as predicted from our present findings. The proposed dual functions of tegument proteins before and after nuclear events account for the dual possibilities of p180 interaction with the tegument, namely, (i) transport of HCMV capsids to the nucleus upon entry into the cell and (ii) egress from the nucleus to the final envelopment compartment. Since a coiled-coil structure is a hallmark of protein-protein interactions in various fundamental cellular functions including vesicle transport (8, 39), the carboxyl half of p180, possessing a large coiled-coil domain, might be involved in utilizing the cellular transport system and thereby in facilitating capsid transport in the cytoplasm.
FIG. 10.
Schematic representation of the interaction between p180 and tegument protein during the course of HCMV infection. A model of the interaction between p180 and HCMV through the tegument protein is presented. The cytoplasmic disposition of p180 accounts for a dual possibility of interaction with the tegument before and after nuclear events. First (A), upon entry of HCMV into the host cell, released capsids which are transported to the nucleus may bind to the N-terminal domain of p180 (Nt) located on the ER or ER-derived vesicles. Second (B), after the nuclear assembly process, transported capsids that egressed from the nucleus to the final envelopment compartments may also interact with p180. The coiled-coil structure in the p180 carboxyl domain (Ct) is the most probable candidate for the interaction with other proteins involved in the cellular transport system which results in facilitating capsid transport in the cytoplasm. TGN, trans-Golgi network.
In the present study our virus overlay assay defined a host factor that associates with the HCMV tegument protein. It is conceivable that disruption of the envelope structure during the purification process and exposure of the tegument may have led to the detection of the tegument-binding protein in our virus overlay assay. Since pretreatment of the HCMV probe with anti-UL48 antibody resulted in abolishment of the ability to bind to p180, it is likely that the interaction with the UL48 protein is largely responsible for HCMV binding to p180 under the tested conditions. However, we cannot rule out the possibility that p180 might interact with enveloped virions and UL48 independently. The possibility of p180 binding to tegument proteins is also consistent with our observation that the addition of detergent to the probe solution resulted in increased affinity for the binding of the probe to p180. Unlike what was found for p180, a reduction rather than an increase in the binding affinity was observed for another HCMV-binding 150-kDa protein, implying that the 150-kDa protein may be bound by the envelope. Although its molecular weight is close to that of CD13, it was distinguishable from CD13 based on its distinct behavior in the solubilization step. Further studies of the 150-kDa protein may provide a new information on the HCMV-host interaction.
The cellular secretory pathway was activated by p180 overexpression (5) as mentioned above, and we demonstrated a high level of p180 expression in HCMV-permissive cells. Taken together, these findings suggest that the level of expression of p180 might be an intriguing factor for HCMV replication. Functional analysis, such as analysis of transdominant inhibition of p180 during the infectious and normal conditions, remains to be performed; such an analysis may elucidate the role of the interaction between p180 and pUL48 and also might give us important information about a general role for p180 in cellular secretory pathways. Thus, our virus overlay assay permitted us to explore host factors that associate with HCMV and to provide new insights to the virus-host cell interaction during replication.
Acknowledgments
We thank W. Gibson (Johns Hopkins University) for his generous gift of antibodies and E. Moriishi for technical assistance with electron-microscopic observations.
REFERENCES
- 1.Aebersold, R. H., J. Leavitt, R. A. Saavedra, L. E. Hood, and S. B. Kent. 1987. Internal amino acid sequence analysis of proteins separated by one- or two-dimensional gel electrophoresis after in situ protease digestion. Proc. Natl. Acad. Sci. USA 84:6970-6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ando, Y., T. Iwasaki, T. Sata, S. Soushi, T. Kurata, and Y. Arao. 1997. Enhanced cytopathic effect of human cytomegalovirus on a retinal pigment epithelium cell line, K-1034, by serum-free medium. Arch. Virol. 142:1645-1658. [DOI] [PubMed] [Google Scholar]
- 3.Baldick, C. J., Jr., and T. Shenk. 1996. Proteins associated with purified human cytomegalovirus particles. J. Virol. 70:6097-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batterson, W., D. Furlong, and B. Roizman. 1983. Molecular genetics of herpes simplex virus. VIII. Further characterization of a temperature-sensitive mutant defective in release of viral DNA and in other stages of the viral reproductive cycle. J. Virol. 45:397-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker, F., L. Block-Alper, G. Nakamura, J. Harada, K. D. Wittrup, and D. I. Meyer. 1999. Expression of the 180-kD ribosome receptor induces membrane proliferation and increased secretory activity in yeast. J. Cell Biol. 146:273-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradshaw, P. A., M. R. Duran-Guarino, S. Perkins, J. I. Rowe, J. Fernandez, K. E. Fry, G. R. Reyes, L. Young, and S. K. Foung. 1994. Localization of antigenic sites on human cytomegalovirus virion structural proteins encoded by UL48 and UL56. Virology 205:321-328. [DOI] [PubMed] [Google Scholar]
- 7.Britt, W. J., and C. A. Alford. 1996. Cytomegalovirus, p. 2493-2523. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 8.Burkhard, P., S. V. Strelkov, and J. Stetefeld. 2001. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 11:82-88. [DOI] [PubMed] [Google Scholar]
- 9.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison III, T. Kouzarides, J. A. Martignetti, E. Preddie, S. C. Satchwell, P. Tomlinson, K. M. Weston, and B. G. Barrell. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 10.Compton, T., D. M. Nowlin, and N. R. Cooper. 1993. Initiation of human cytomegalovirus infection requires initial interaction with cell surface heparan sulfate. Virology 193:834-841. [DOI] [PubMed] [Google Scholar]
- 11.Dainese Hatt, P., M. Quadroni, W. Staudenmann, and P. James. 1997. Concentration of, and SDS removal from, proteins isolated from multiple two-dimensional electrophoresis gels. Eur. J. Biochem. 246:336-343. [DOI] [PubMed] [Google Scholar]
- 12.Desai, P. J. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J. Virol. 74:11608-11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forte, J. G., T. M. Forte, and E. Heinz. 1973. Isolation of plasma membranes from Ehrlich ascites tumor cells. Influence of amino acids on (Na++K+)-ATPase and K+-stimulated phosphatase. Biochim. Biophys. Acta 298:827-841. [DOI] [PubMed] [Google Scholar]
- 14.Gibson, W. 1996. Structure and assembly of the virion. Intervirology 39:389-400. [DOI] [PubMed] [Google Scholar]
- 15.Gilloteaux, J., and M. R. Nassiri. 2000. Human bone marrow fibroblasts infected by cytomegalovirus: ultrastructural observations. J. Submicrosc. Cytol. Pathol. 32:17-45. [PubMed] [Google Scholar]
- 16.Jarvis, M. A., C. E. Wang, H. L. Meyers, P. P. Smith, C. L. Corless, G. J. Henderson, J. Vieira, W. J. Britt, and J. A. Nelson. 1999. Human cytomegalovirus infection of Caco-2 cells occurs at the basolateral membrane and is differentiation state dependent. J. Virol. 73:4552-4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kari, B., and R. Gehrz. 1992. A human cytomegalovirus glycoprotein complex designated gC-II is a major heparin-binding component of the envelope. J. Virol. 66:1761-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katano, H., K. Ogawa-Goto, H. Hasegawa, T. Kurata, and T. Sata. 2001. Human-herepsvirus-8-encoded K8 protein colocalizes with the promyelocytic leukemia protein (PML) bodies and recruits p53 to the PML bodies. Virology 286:446-455. [DOI] [PubMed] [Google Scholar]
- 19.Langley, R., E. Leung, C. Morris, R. Berg, M. McDonald, A. Weaver, D. A. Parry, J. Ni, J. Su, R. Gentz, N. Spurr, and G. W. Krissansen. 1998. Identification of multiple forms of 180-kDa ribosome receptor in human cells. DNA Cell Biol. 17:449-460. [DOI] [PubMed] [Google Scholar]
- 20.Mocarski, E. S., Jr. 1996. Cytomegaloviruses and their replication., p. 2447-2492. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 21.Neyts, J., R. Snoeck, D. Schols, J. Balazarini, J. D. Esko, A. V. Schepdael, and E. D. Clercq. 1992. Sulfated polymers inhibit the interaction of human cytomegalovirus with cell surface heparan sulfate. Virology 189:48-58. [DOI] [PubMed] [Google Scholar]
- 22.Nowlin, D. M., N. R. Cooper, and T. Compton. 1991. Expression of a human cytomegalovirus receptor correlates with infectibility of cells. J. Virol. 65:3114-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogawa-Goto, K., Y. Arao, Y. Ito, T. Ogawa, T. Abe, T. Kurata, S. Irie, and H. Akanuma. 1998. Binding of human cytomegalovirus to sulfated glucuronyl glycosphingolipids and their inhibitory effects on the infection. J. Gen. Virol. 79:2533-2541. [DOI] [PubMed] [Google Scholar]
- 24.Radsak, K., M. Eickmann, T. Mockenhaupt, E. Bogner, H. Kern, A. Eis-Hubinger, M. Reschke, A. Martinez, A. Castro, C. Gil, and C. Perez. 1996. Retrieval of human cytomegalovirus glycoprotein B from the infected cell surface for virus envelopment. Arch. Virol. 141:557-572. [DOI] [PubMed] [Google Scholar]
- 25.Rezaee, M., K. Isokawa, N. Halligan, R. R. Markwald, and E. L. Krug. 1993. Identification of an extracellular 130-kDa protein involved in early cardiac morphogenesis. J. Biol. Chem. 268:14404-14411. [PubMed] [Google Scholar]
- 26.Sanchez, V., K. D. Greis, E. Sztul, and W. J. Britt. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J. Virol. 74:975-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanchez, V., E. Sztul, and W. J. Britt. 2000. Human cytomegalovirus pp28 (UL99) localizes to a cytoplasmic compartment which overlaps the endoplasmic reticulum-Golgi-intermediate compartment. J. Virol. 74:3842-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Savitz, A. J., and D. I. Meyer. 1990. Identification of a ribosome receptor in the rough endoplasmic reticulum. Nature 346:540-544. [DOI] [PubMed] [Google Scholar]
- 29.Severi, B., M. P. Landini, and E. Govoni. 1988. Human cytomegalovirus morphogenesis: an ultrastructural study of the late cytoplasmic phases. Arch. Virol. 98:51-64. [DOI] [PubMed] [Google Scholar]
- 30.Severi, B., M. P. Landini, M. Musiani, and M. Zerbini. 1979. A study of the passage of human cytomegalovirus from the nucleus to the cytoplasm. Microbiologica 2:265-273. [Google Scholar]
- 31.Sinzger, C., A. Grefte, B. Plachter, A. S. Gouw, T. H. The, and G. Jahn. 1995. Fibroblasts, epithelial cells, endothelial cells and smooth muscle cells are major targets of human cytomegalovirus infection in lung and gastrointestinal tissues. J. Gen. Virol. 76:741-750. [DOI] [PubMed] [Google Scholar]
- 32.Sodeik, B. 2000. Mechanisms of viral transport in the cytoplasm. Trends Microbiol. 8:465-472. [DOI] [PubMed] [Google Scholar]
- 33.Sodeik, B., M. W. Ebersold, and A. Helenius. 1997. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol. 136:1007-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soderberg, C., T. D. Giugni, J. A. Zaia, S. Larsson, J. M. Wahlberg, and E. Moller. 1993. CD13 (human aminopeptidase N) mediates human cytomegalovirus infection. J. Virol. 67:6576-6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor, H. P., and N. R. Cooper. 1990. The human cytomegalovirus receptor on fibroblasts is a 30-kilodalton membrane protein. J. Virol. 64:2484-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tooze, J., M. Hollinshead, B. Reis, K. Radsak, and H. Kern. 1993. Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur. J. Cell Biol. 60:163-178. [PubMed] [Google Scholar]
- 37.Toyoshima, I., H. Yu, E. R. Steuer, and M. P. Sheetz. 1992. Kinectin, a major kinesin-binding protein on ER. J. Cell Biol. 118:1121-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tugizov, S., E. Maidji, and L. Pereira. 1996. Role of apical and basolateral membranes in replication of human cytomegalovirus in polarized retinal pigment epithelial cells. J. Gen. Virol. 77:61-74. [DOI] [PubMed] [Google Scholar]
- 39.Wade, R. H., and F. Kozielski. 2000. Structural links to kinesin directionality and movement. Nat. Struct. Biol. 7:456-460. [DOI] [PubMed] [Google Scholar]
- 40.Wanker, E. E., Y. Sun, A. J. Savitz, and D. I. Meyer. 1995. Functional characterization of the 180-kD ribosome receptor in vivo. J. Cell Biol. 130:29-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weinshenker, B. G., S. Wilton, and G. A. Rice. 1988. Phorbol ester-induced differentiation permits productive human cytomegalovirus infection in a monocytic cell line. J. Immunol. 140:1625-1631. [PubMed] [Google Scholar]
- 42.Wright, J. F., A. Kurosky, and S. Wasi. 1994. An endothelial cell-surface form of annexin II binds human cytomegalovirus. Biochem. Biophys. Res. Commun. 198:983-989. [DOI] [PubMed] [Google Scholar]
- 43.Ye, G.-J., K. T. Vaughan, R. B. Vallee, and B. Roizman. 2000. The herpes simplex virus 1 UL34 protein interacts with a cytoplasmic dynein intermediate chain and targets nuclear membrane. J. Virol. 74:1355-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]