Abstract
The copA gene product, a putative copper-translocating P-type ATPase, has been shown to be involved in copper resistance in Escherichia coli. The copA gene was disrupted by insertion of a kanamycin gene through homologous recombination. The mutant strain was more sensitive to copper salts but not to salts of other metals, suggesting a role in copper homeostasis. The copper-sensitive phenotype could be rescued by complementation by a plasmid carrying copA from E. coli or copB from Enterococcus hirae. Expression of copA was induced by salts of copper or silver but not zinc or cobalt. Everted membrane vesicles from cells expressing copA exhibited ATP-coupled accumulation of copper, presumably as Cu(I). The results indicate that CopA is a Cu(I)-translocating efflux pump that is similar to the copper pumps related to Menkes and Wilson diseases and provides a useful prokaryotic model for these human diseases.
Keywords: soft metal resistance, Menkes, Wilson disease
Copper is an essential trace element that is required for a number of enzymes, including cytochrome oxidase, superoxide dismutase, and lysyl oxidase (1). While trace amounts of copper are needed to sustain life, excess copper is extremely toxic. It is likely that most, if not all, cells have mechanisms for copper homeostasis. In eukaryotes, copper is taken into the cell by specific uptake systems and transferred to cytosolic chaperone proteins that deliver copper-requiring enzymes, with little free cytosolic copper ion (2). To maintain low intracellular concentrations of copper, specific pumps remove excess copper ion from the cytosol, either by accumulation into organelles or extrusion out of the cell (1).
P-type ATPases form a large family of cation-transporting pumps. These ubiquitous membrane proteins have been classified into five groups according to substrate specificity (3). The best-characterized group transports the ions of hard Lewis acids and can be further subdivided into monovalent ion pumps for ions such as K+, Na+, and H+, and divalent ion pumps for ions such as Ca2+ and Mg2+. A second subfamily of metal-transporting P-type ATPases is involved in homeostasis of the ions of soft Lewis acids, and genes for these soft metal P-type ATPases have been found in archaea, prokaryotes and eukaryotes, including humans (1, 4, 5). The soft metal-transporting P-type ATPases can be further divided into two subgroups. The first has monovalent soft metal ions as substrates and includes Cu(I)- and Ag(I)-transporting pumps, such as the human Menkes (6) and Wilson (7) disease-related proteins and homologues from Saccharomyces cerevisiae (8), Enterococcus hirae (9), Synechococcus (10), and Helicobacter pylori (11, 12). Members of the other subgroup of soft metal P-type ATPases transport divalent soft metal ions, including Zn(II), Cd(II), and Pb(II). These include ZntA from Escherichia coli and CadA from Staphylococcus aureus plasmid pI258 (5). Recently, a related gene conferring Co(II) resistance was identified in Synechocystis PCC 6803 (13).
Only two genes encoding soft metal-translocating P-type ATPases have been identified in the E. coli chromosome (14). One is zntA, which codes for the Zn(II)-, Cd(II)-, and Pb(II)-translocating ATPase (15–17). The other, designated f834, is more closely related to copper pumps such as the Menkes and Wilson disease proteins. In this study, we demonstrate that this gene encodes a copper-translocating P-type ATPase, and we have therefore named the gene copA and the gene product CopA. Disruption of copA resulted in sensitivity to copper salts in E. coli. The copA-disrupted strain was not sensitive to Zn(II), Cd(II), or Pb(II), and a zntA-disrupted strain was not sensitive to Cu(II). A strain with both copA and zntA disrupted was no more sensitive to metal ions than the individual disruptions. Thus, the two soft metal P-type ATPases have nonoverlapping specificities. From its native promoter, expression of copA was induced by addition of copper or silver salts to the medium and was repressed by chelation of metals with 2-hydroxyquinoline. Everted membrane vesicles from cells expressing copA accumulated radioisotopic copper ion. Copper accumulation required ATP and was inhibited by sodium vanadate. No transport was observed without the reductant DTT, consistent with Cu(I) as the probable substrate of the pump.
Materials and Methods
Growth of Cells.
Cells were grown in LB medium (18) at 37°C. Ampicillin (50 μg/ml), kanamycin (50 μg/ml), chloramphenicol (30 μg/ml), isopropyl-β-d-thiogalactopyranoside (0.1 mM), and 5-bromo-4-chloro-3-indolyl-β-d-galactosidase (80 μg/ml) were added as required. To assay inhibition of growth by metal salts, cells were grown overnight in LB, diluted 1:100 in the same medium with the indicated metal salts, and incubated for 6 h with CuSO4 at 37°C with shaking. Growth was monitored from the absorbance at 600 nm.
Strain Construction and Plasmids.
Standard molecular and genetic techniques were used for strain and plasmid construction (18). Plasmid pCopA was constructed by PCR amplification of copA starting 106 base pairs upstream of the putative start codon and ending 369 base pairs downstream. The forward and reverse primers used for PCR were 5′-CTTTACGGACTTTTACCCGCC-3′ and 5′-GCGGCGGCCGCCTTTGGGAAGTTGAAAAC-3′, respectively. The PCR product was cloned into plasmid pGEM-T (Promega), creating plasmid pCopA. Plasmid pCopA was then digested with BstEI, which cuts at a unique site within copA, and the ends were made blunt by using the Klenow fragment of E. coli DNA polymerase I. The kanamycin resistance gene from plasmid pUC4K (Amersham Pharmacia) was excised as an EcoRI fragment, and the ends were also made blunt. This gene was ligated with BstEI-digested pCopA, generating pCopA1. Plasmid pCopA1 was linearized by using SstI and NcoI, and the linear DNA was transformed into the recD strain JCB499 (19). Homologous recombination of the kanamycin resistance gene into the chromosomal copA gene was confirmed by using the above primers. The copA-disrupted gene was transferred to strain W3110 (20) by generalized transduction with P1 bacteriophage, with selection for kanamycin resistance, creating strain DW3110. The same procedure was used to disrupt copA in E. coli LMG194 (Invitrogen), creating LMG194 ΔcopA.
Plasmid pCopA2, in which copA was controlled by the arabinose promoter, was created by PCR amplification of copA by using forward and reverse primers 5′-TGTTCCATGGCACAAACTATCGACCTG-3′ and 5′-TACAAGCTTTTCCTTCGGTTTAAACCGCAG-3′. The PCR product was digested with NcoI and HindIII and cloned into NcoI–HindIII-digested pBAD/Myc-HisA (Invitrogen). In pCopA2, copA is in-frame with the sequence for the Myc epitope and six histidine codons. To construct a copA gene without the Myc–His fusion for use in antibody production, copA was amplified by PCR with forward and reverse primers 5′-TGTTCCATGGCACAAACTATCGACCTG-3′ and 5′-TTGAATTCGCATCCGCAATGATGTACTTATTC-3′. The PCR product was digested with NcoI and EcoRI and cloned into NcoI–EcoRI-digested pBAD/Myc-HisC (Invitrogen), creating plasmid pCopA3.
Preparation of Everted Membrane Vesicles.
Cells were grown overnight at 37°C in 20 ml of LB, diluted 100-fold into prewarmed medium, and allowed to grow to an optical density of 0.8 at 600 nm. For induction studies, cultures of strains W3110 or DW3110 were induced with the indicated concentrations of CuSO4 or other metal salts for 2 h. For transport experiments, cultures of strain LMG194ΔcopA bearing plasmid pCopA2 were induced with 0.0002% arabinose for 2 h at 30°C. Everted membrane vesicles were prepared as described previously (16), except the cells were lysed at 16,000 psi (1 psi = 6.89 kPa). The membrane vesicles were suspended in a buffer consisting of 10 mM Tris⋅HCl (pH 7.5), containing 0.25 M sucrose and 0.2 M KCl and stored at −70°C until use. Protein concentration was determined by using a bicinchoninic acid method (21).
PAGE and Immunoblotting.
Samples were prepared by incubation in SDS sample buffer for 30 min at room temperature and separated by SDS/PAGE (22). To prepare CopA for antibody production, LMG194 pCopA3 was grown to an optical density of 0.8 at 600 nm and induced with 0.02% arabinose for 2 h. Then, everted membrane vesicles were prepared. Membrane protein was separated by SDS/PAGE, the CopA band was excised, and antiserum was produced commercially (Cocalico Biologicals, Reamstown, PA). Immunoblotting was performed by using an enhanced chemiluminescence assay (DuPont/NEN) and exposed on x-ray film at room temperature as described previously (23).
64Cu Transport Assays.
64Cu was obtained from the Mallinckrodt Institute of Radiology, Washington University Medical Center (St. Louis, MO). Transport assays were performed at room temperature by a modification of the method of Camakaris et al. (4). Unless otherwise noted, the reaction mixture (1 ml) contained 40 mM histidine (pH 6.8), 0.2 M KCl, 0.25 M sucrose, 1 mM DTT, 0.5 mg of membrane protein, 10 μM 64CuCl2 (0.5–10 μCi/ml), and, where noted, 5 mM Na2ATP or Na2ADP. The reaction was initiated by addition of 5 mM MgCl2. At intervals, 0.1-ml samples were withdrawn and filtered through nitrocellulose filters (0.22-μm pore size; Whatman). The filters were presoaked in a buffer consisting of 40 mM histidine (pH 6.8), 0.2 M KCl, 0.25 M sucrose, 10 mM MgSO4, and 20 mM CuCl2. Following filtration, the filters were washed with 5 ml of the same buffer and dried, and the radioactivity was quantified in a liquid scintillation counter. The values obtained with the assay mixture without membrane vesicles were subtracted from all points.
Results
Disruption of copA Produces Sensitivity to Copper Ion.
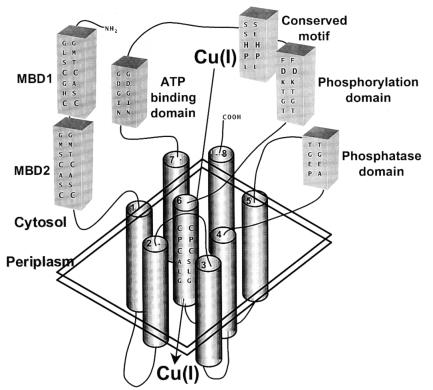
ORF f834 potentially encodes an 834-residue P-type ATPase that is highly similar to copper pumps such as the human Menkes disease proteins (4) (Fig. 1). Since, as shown below, disruption of f834 results in copper ion sensitivity, we have renamed the gene copA and the gene product CopA. In common with other P-type ATPases, CopA has conserved ATP binding (GDGIN), phosphorylation (DKTGT), and phosphatase (TGE) domains. The soft metal P-type ATPases have additional regions that distinguish them from the hard metal pumps. These include N-terminal CXXC motifs that are probable metal binding domains. While the human copper pumps have six N-terminal CXXC sequences, the E. coli CopA has two, G11LSCGHC and G107MSCASC. In contrast, homologues from E. hirae and H. pylorii have only a single N-terminal CXXC sequence. The soft metal pumps have a membrane-embedded CPC(H) sequence. From hydropathy analysis, E. coli CopA can be predicted to have eight transmembrane segments, including C479PC in predicted transmembrane helix 6. There is a conserved HP motif in the soft metal P-type ATPases that corresponds to H562P in CopA.
Figure 1.
Model of the CopA P-type ATPase. CopA is a P-type ATPase that pumps Cu(I) from the cytosol to the periplasm. From the predicted structure of homologous copper pumps (1, 4), CopA can be predicted to have an N-terminal region with two cytosolic CXXC metal binding domains (MBD1 and MBD2) and eight transmembrane segments (TM). Connecting TM4 and TM5 is the conserved phosphatase domain. TM6 is predicted to be part of the translocation domain and has the consensus CPC sequence. Connecting TM6 and TM7 are the phosphorylation and ATP binding domains and a conserved sequence found only in soft metal P-type ATPases. Two sets of conserved primary sequences are shown: on the left is CopA, and on the right is the corresponding sequence from the Menkes protein. (Modified from ref. 4.)
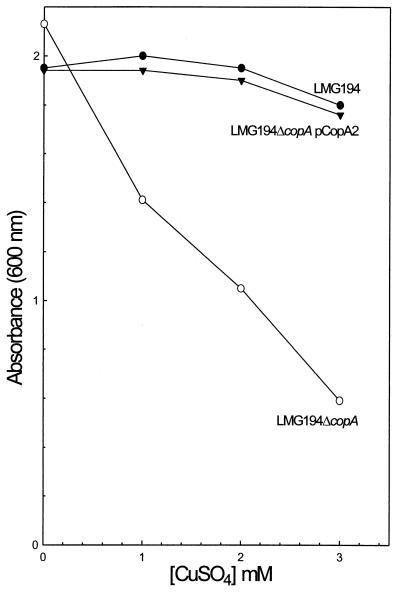
The effect of CuSO4 on the growth in liquid LB medium of the E. coli copA disruptions LMG194ΔcopA was compared with strain E. coli LMG194 (Fig. 2). The disrupted strain exhibited decreased resistance that could be complemented by introduction of plasmid pCopA2. Although copA is controlled by the arabinose promoter in pCopA2, complementation did not require addition of arabinose, suggesting that sufficient CopA was produced even in the absence of inducer. Copper ion sensitivity was observed with strain DW3110, and the sensitive phenotype was complemented by plasmid pY163, which carries the E. hirae copB gene (data not shown). However, the copA-disrupted strain was still relatively resistant to copper salts, suggesting that other genes contribute to intrinsic copper tolerance in E. coli (24). Disruption of copA did not result in a significant increase in sensitivity to Ag(I). A related protein from Salmonella typhimurium, SilP, confers resistance to silver but not copper ions (25).
Figure 2.
CopA confers copper resistance. Copper ion resistance was assayed as described in Materials and Methods for strain LMG194 (wild type) (●), LMG194ΔcopA (copA:Kn) (○), or LMG194ΔcopA pCopA2 (copA) (▾).
CopA and ZntA Do Not Have Overlapping Specificities.
E. coli has only two soft metal P-type ATPases, ZntA for Zn(II), Cd(II), and Pb(II) resistance and CopA for resistance to copper salts. To examine whether the two pumps have overlapping specificity, a double disruption of both genes was constructed. The zntA∷Cm disruption from E. coli SJB101 (15) was transferred into W3110 by generalized transduction by using P1 phage, with selection for resistance to chloramphenicol, producing strain Z3110. Generalized transduction using P1 was also used to introduce the copA∷Kn disruption from strain DW3110 into strain Z3110, generating the double disrupted strain DZ3110 (zntA∷Cm copA∷Kn). The double disrupted strain DZ3110 was no more sensitive to Cu(II) than was DW3110, with only a copA disruption. Similarly, the double disrupted strain was no more sensitive to Zn(II) or Cd(II) than was Z3110, with only a zntA disruption (data not shown). Thus, the observed Zn(II) and Cd(II) sensitivities can be attributed solely to disruption of zntA, and the Cu(II) sensitivity is due solely to disruption of copA. Since expression of both copA and zntA does not produce increased resistance to either monovalent or divalent soft metals than expression of either gene separately, CopA does not catalyze Zn(II) or Cd(II) extrusion, and ZntA does not pump copper ion out of cells.
CopA Is Induced by Copper and Silver Ions.
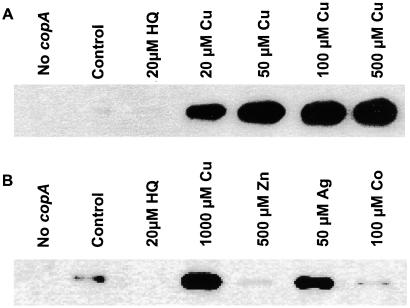
Under control of its endogenous promoter, copA required copper ion for expression. When strains W3110 and E. coli DW3110 (copA∷Kn) were induced by 1 mM CuSO4 for 2 h, no band corresponding to CopA could be observed in Coomassie-stained SDS polyacrylamide gels of the membrane fraction (data not shown), indicating that little CopA is made from the chromosomally encoded gene. However, by immunoblotting, CopA could be observed in membranes from strain W3110 (Fig. 3). Addition of CuSO4 resulted in an approximate 50-fold increase in the amount of CopA compared with the uninduced control, as determined by densitometry. Addition of the chelator 2-hydroxyquinoline to the growth medium almost completely eliminated synthesis of CopA. The effect of 2-hydroxyquinoline could be reversed by addition of CuSO4 to the growth medium. No immunologically crossreacting material was detected in the copA-disrupted strain E. coli DW3110 with or without induction with CuSO4. Ag(I) also induced CopA in W3110, but no induction was observed with Zn(II) or Co(II). These results suggest that expression of copA is transcriptionally regulated by Cu(II) or Ag(I). Although Cu(II) is added to the medium, it is reasonable to assume that it is reduced to Cu(I) intracellularly, and that Cu(I) is the actual inducer. The copA gene is in a monogenic operon, with no genes for potential regulatory proteins immediately upstream or downstream. Within the −10 element of the putative copA promoter, the inverted repeat sequence 5′-AAGGTTTAACCTT-3′ is a potential binding site for a regulatory protein.
Figure 3.
Copper or silver regulates copA expression. Cells of strain W3110 (wild type) and DW3110 (copA:Kn) were grown and induced, and immunoblotting was performed on membranes as described in Materials and Methods. (A) Lane 1, DW3110; lane 2, uninduced W3110; lane 3, W3110 + 20 μM 2-hydroxyquinoline; lanes 4–7, W3110 + 20, 50, 100, or 500 μM CuSO4. (B) Lane 1, DW3110; lane 2, uninduced W3110; lane 3, W3110 + 20 μM 2-hydroxyquinoline; lane 4, W3110 + 1000 μM CuSO4; lane 5, W3110 + 500 μM ZnSO4; lane 6, W3110 + 50 μM AgNO3; lane 7, W3110 + 100 μM CoCl2.
ATP-Dependent 64Cu(I) Transport.
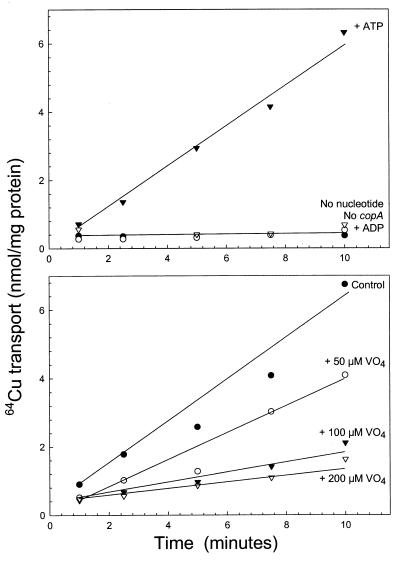
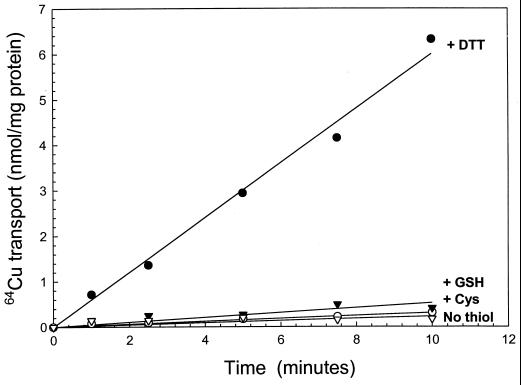
Everted membrane vesicles prepared from strain LMG194ΔcopA expressing copA from plasmid pCopA2 accumulated 64Cu (Fig. 4A). Transport required MgATP. MgADP could not substitute, and cells without copA did not transport 64Cu even in the presence of MgATP. Uptake was inhibited by sodium vanadate (Fig. 4B), an inhibitor of P-type ATPases. No accumulation was observed in the absence of DTT (Fig. 5). Essentially, no transport was found with glutathione or cysteine. The requirement for a strong reductant suggests that the substrate of the pump is the monovalent Cu(I) cation. When Ag(I) was added together with 64Cu, precipitation occurred, so it is not known whether Ag(I) inhibits Cu(I) transport. Because radioisotopic Ag(I) is commercially unavailable, transport of Ag(I) could not be examined.
Figure 4.
Uptake of 64Cu in everted membrane vesicles of E. coli is ATP-dependent. Vesicles were prepared from cells of strain LMG194ΔcopA pCopA2 induced with 0.0002% arabinose as described in Materials and Methods. Transport was assayed with 10 μM 64CuSO4 reduced with 1 mM DTT. (A) ○, No pCopA2. ●, No energy source. ▿, 5 mM MgADP. ▾, 5 mM MgATP. (B) ●, No sodium vanadate. ○, +50 μM sodium vanadate. ▾, +100 μM sodium vanadate. ▿, +200 μM sodium vanadate.
Figure 5.
CopA transport activity requires DTT. Transport activity was assayed as described in the legend to Fig. 4 with 5 mM MgATP. ▿, No thiol. ●, +1 mM DTT. ▾, +1 mM glutathione. ○, +1 mM cysteine.
Discussion
Ions of transition metals such as copper and zinc have essential roles in gene regulation, enzyme structure, and catalysis and are therefore required for growth. In excess, copper and zinc ions are toxic. To satisfy these opposing physiological demands, metal ion homeostasis is maintained by a balance between uptake and efflux (5). In E. coli, Zn(II) is accumulated by the ZnuACB transporter, a member of the ABC transporter superfamily (26). At present no Cu(II) uptake system has been identified in this organism. Efflux systems for these two ions have been identified. ZntA is a P-type ATPase that catalyzes efflux of Zn(II), Cd(II), or Pb(II) (15–17). In this study, we demonstrate that E. coli f834, renamed copA, confers resistance to copper ion but not to other metal ions in E. coli. From sequence analysis, the copA gene product could be predicted to be a copper ion-translocating P-type ATPase. The transport data are consistent with this postulate. Uptake into everted membrane vesicles was observed only when ATP was present as an energy source and was inhibited by vanadate, a classical P-type ATPase inhibitor. The dithiol DTT, a strong reductant, was required for CopA catalyzed 64Cu uptake. A similar requirement for DTT was found for transport of 64Cu by the Menkes protein (27). The prerequisite for a reductant suggests that the substrate of CopA is Cu(I), although DTT may also maintain the cysteine thiolates of CopA in a reduced form.
E. coli CopA exhibits identity with orthologues, for example, 36% identity with CopA from E. hirae: 31% with the human Wilson disease protein and 29% with the Menkes disease protein. However, the similarity is greater within conserved domains (Fig. 1). A crucial question in the biochemical mechanism of the soft metal P-type ATPases is the role of the conserved N-terminal CXXC motifs (4). What function do they serve and why is the motif repeated up to six times in the human proteins? Whereas a number of the bacterial soft metal pumps have a single N-terminal motif (10, 12, 28), E. coli CopA has two. Recent studies suggest the six metal binding motifs of the Menkes protein may not be essential for transport but are required for intracellular trafficking (29). However, since this is not an issue with prokaryotes, the presence of multiple N-terminal motifs in the E. coli orthologue implies that they have a role in transport. Even though the number of N-terminal CXXC motifs differ between the two P-type ATPases, the transport properties of CopA are quite similar to those of the Menkes protein (27). Another possible role for the CXXC motifs may be to sense copper ion, binding it in the cytosol for transfer to the translocation domain. The similarities with the human copper diseases proteins coupled with the ability to analyze copA genetically and to overproduce and apply biochemical assays make CopA an excellent model for the study of human diseases of copper metabolism.
Acknowledgments
This work was supported by United States Public Health Service Grants GM55425 (to B.P.R.) and GM54102 (to B.M.) and National Research Service Award GM18973 (to C.R.). We thank the following people for strains and plasmids: Robert K. Poole for E. coli SBJ101 and Marc Solioz for plasmid pY163.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Pena M M, Lee J, Thiele D J. J Nutr. 1999;129:1251–1260. doi: 10.1093/jn/129.7.1251. [DOI] [PubMed] [Google Scholar]
- 2.Rae T D, Schmidt P J, Pufahl R A, Culotta V C, O'Halloran T V. Science. 1999;284:805–808. doi: 10.1126/science.284.5415.805. [DOI] [PubMed] [Google Scholar]
- 3.Axelsen K B, Palmgren M G. J Mol Evol. 1998;46:84–101. doi: 10.1007/pl00006286. [DOI] [PubMed] [Google Scholar]
- 4.Camakaris J, Voskoboinik I, Mercer J F. Biochem Biophys Res Commun. 1999;261:225–232. doi: 10.1006/bbrc.1999.1073. [DOI] [PubMed] [Google Scholar]
- 5.Rensing C, Ghosh M, Rosen B P. J Bacteriol. 1999;181:5891–5897. doi: 10.1128/jb.181.19.5891-5897.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vulpe C, Levinson B, Whitney S, Packman S, Gitschier J. Nat Genet. 1993;3:7–13. doi: 10.1038/ng0193-7. [DOI] [PubMed] [Google Scholar]
- 7.Bull P C, Thomas G R, Rommens J M, Forbes J R, Cox D W. Nat Genet. 1993;5:327–337. doi: 10.1038/ng1293-327. [DOI] [PubMed] [Google Scholar]
- 8.Fu D, Beeler T J, Dunn T M. Yeast. 1995;11:283–292. doi: 10.1002/yea.320110310. [DOI] [PubMed] [Google Scholar]
- 9.Odermatt A, Suter H, Krapf R, Solioz M. J Biol Chem. 1993;268:12775–12779. [PubMed] [Google Scholar]
- 10.Phung L T, Ajlani G, Haselkorn R. Proc Natl Acad Sci USA. 1994;91:9651–9654. doi: 10.1073/pnas.91.20.9651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ge Z, Taylor D E. FEMS Microbiol Lett. 1996;145:181–188. doi: 10.1111/j.1574-6968.1996.tb08575.x. [DOI] [PubMed] [Google Scholar]
- 12.Melchers K, Herrmann L, Mauch F, Bayle D, Heuermann D, Weitzenegger T, Schuhmacher A, Sachs G, Haas R, Bode G, et al. Acta Physiol Scand Suppl. 1998;643:123–135. [PubMed] [Google Scholar]
- 13.Rutherford J C, Cavet J S, Robinson N J. J Biol Chem. 1999;274:25827–25832. doi: 10.1074/jbc.274.36.25827. [DOI] [PubMed] [Google Scholar]
- 14.Blattner F R, Plunkett G, III, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, et al. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 15.Beard S J, Hashim R, Membrillo-Hernandez J, Hughes M N, Poole R K. Mol Microbiol. 1997;25:883–891. doi: 10.1111/j.1365-2958.1997.mmi518.x. [DOI] [PubMed] [Google Scholar]
- 16.Rensing C, Mitra B, Rosen B P. Proc Natl Acad Sci USA. 1997;94:14326–14331. doi: 10.1073/pnas.94.26.14326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rensing C, Mitra B, Rosen B P. Biochem Cell Biol. 1998;76:787–790. [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 19.Bardwell J C, McGovern K, Beckwith J. Cell. 1991;67:581–589. doi: 10.1016/0092-8674(91)90532-4. [DOI] [PubMed] [Google Scholar]
- 20.Bachmann B J. In: Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Washington, D.C.: American Society for Microbiology; 1987. pp. 1190–1219. [Google Scholar]
- 21.Smith P K, Krohn R I, Hermanson G T, Mallia A K, Gartner F H, Provenzano M D, Fujimoto E K, Goeke N M, Olson B J, Klenk D C. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 22.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Dey S, Dou D, Tisa L S, Rosen B P. Arch Biochem Biophys. 1994;311:418–424. doi: 10.1006/abbi.1994.1256. [DOI] [PubMed] [Google Scholar]
- 24.Silver S, Phung L T. Annu Rev Microbiol. 1996;50:753–789. doi: 10.1146/annurev.micro.50.1.753. [DOI] [PubMed] [Google Scholar]
- 25.Gupta A, Matsui K, Lo J F, Silver S. Nat Med. 1999;5:183–188. doi: 10.1038/5545. [DOI] [PubMed] [Google Scholar]
- 26.Patzer S I, Hantke K. Mol Microbiol. 1998;28:1199–1210. doi: 10.1046/j.1365-2958.1998.00883.x. [DOI] [PubMed] [Google Scholar]
- 27.Voskoboinik I, Brooks H, Smith S, Shen P, Camakaris J. FEBS Lett. 1998;435:178–182. doi: 10.1016/s0014-5793(98)01059-x. [DOI] [PubMed] [Google Scholar]
- 28.Solioz M, Odermatt A. J Biol Chem. 1995;270:9217–9221. doi: 10.1074/jbc.270.16.9217. [DOI] [PubMed] [Google Scholar]
- 29.Voskoboinik I, Strausak D, Greenough M, Brooks H, Petris M, Smith S, Mercer J F, Camakaris J. J Biol Chem. 1999;274:22008–22012. doi: 10.1074/jbc.274.31.22008. [DOI] [PubMed] [Google Scholar]