Abstract
Glutamate receptor interacting protein (GRIP) homologues, initially characterized in synaptic glutamate receptor trafficking, consist of seven PDZ domains (PDZDs), whose conserved arrangement is of unknown significance. The Drosophila GRIP homologue (DGrip) is needed for proper guidance of embryonic somatic muscles towards epidermal attachment sites, with both excessive and reduced DGrip activity producing specific phenotypes in separate muscle groups. These phenotypes were utilized to analyze the molecular architecture underlying DGrip signaling function in vivo. Surprisingly, removing PDZDs 1–3 (DGripΔ1–3) or deleting ligand binding in PDZDs 1 or 2 convert DGrip to excessive in vivo activity mediated by ligand binding to PDZD 7. Yeast two-hybrid screening identifies the cell adhesion protein Echinoid's (Ed) type II PDZD-interaction motif as binding PDZDs 1, 2 and 7 of DGrip. ed loss-of-function alleles exhibit muscle defects, enhance defects caused by reduced DGrip activity and suppress the dominant DGripΔ1–3 effect during embryonic muscle formation. We propose that Ed and DGrip form a signaling complex, where competition between N-terminal and the C-terminal PDZDs of DGrip for Ed binding controls signaling function.
Keywords: DGrip, Echinoid, morphogenesis, muscle guidance
Introduction
Nascent somatic muscles use growth-cone-like projections to navigate towards specialized epidermal cells (tendon cells) during mid-embryonic development of Drosophila. Distinct cell guidance systems have been suggested to control targeting to tendon cells by specific muscle groups. The molecular apparatus sending and interpreting muscle guidance cues are only partially known (Volk and VijayRaghavan, 1994; Frommer et al, 1996; Kramer et al, 2001; Schnorrer and Dickson, 2004; Steigemann et al, 2004; Swan et al, 2004). Some of the best characterized players in this process are the Robo-Slit guidance system proteins (Kramer et al, 2001), which act in specific subsets of somatic muscles to selectively adhere to certain target tendon cells via PS-integrins (Fernandes et al, 1996). Other highly conserved signaling systems such as the EGF receptor pathway (Yarnitzky et al, 1998; Volk, 1999) and Wnt signaling (Volk and VijayRaghavan, 1994; Ghazi et al, 2003) also operate during muscle guidance in both embryos and pupae to select and reinforce developmentally programmed signaling between the muscle cell and its target tendon cell.
We recently found that the glutamate receptor interacting protein (DGrip) is also required for proper targeting of nascent muscles towards an attractive signal expressed at segment borders of Drosophila embryos (Swan et al, 2004), with genetic elimination of dgrip resulting in specific defects in patterning of ventro-lateral muscles (VLM, see also Figure 1A). In contrast, lateral transverse muscles (LTMs), which attach within segments, appeared unaffected in dgrip mutants (Figure 1A). Conversely, strong, pan-muscular overexpression of DGrip causes LTMs to produce projections forming ectopic attachment sites, while VLMs appeared unaffected (Swan et al, 2004).
Figure 1.
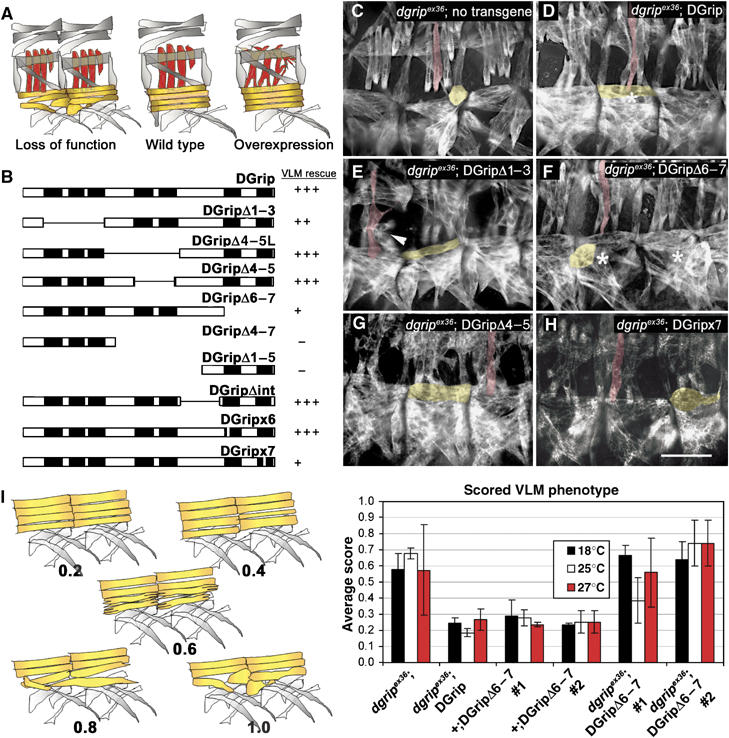
A structure–function map for DGrip in morphogenesis of VLM. (A) Scheme of muscle phenotypes evoked by DGrip loss of function and overexpression. Loss of DGrip function primarily affects VLM (yellow, see (C)) whereas LTM (red) remain unaffected. Strong overexpression of DGrip using 24B-gal4 disturbs LTM morphology (Swan et al, 2004). (B) Scheme of rescue activities of indicated DGrip constructs. Rescue of VLM morphology in dgripex36 mutant background is shown. Pan-muscular expression of wild-type DGrip using twist-gal4 fully rescues the dgripex36 VLM phenotype (+++), while other constructs have a reduced rescue ability (++, +) or exert no effect (−) on muscle rescue. All constructs were characterized in at least two independent lines. (C–H) Muscle myosin stainings in stage 16 embryos. (C) dgripex36, twist-gal4 muscles show typical defects in VLM morphology. One VLM (yellow) and one LTM (red) are labeled for ease of identification. (D) Re-expression of wild-type DGrip using twist-gal4 fully rescues these defects, whereas expression of DGripΔ1–3 in the dgripex36, twist-gal4 background (E) provokes strong dominant LTM (arrowhead) and slight VLM morphology defects. (F, H) Expression of DGrip missing the C-terminal PDZDs (DGripΔ6–7 (F) and DGripx7 (H)) results in only partial rescue of dgripex36 VLMs, with many VLMs appearing atypically round (asterisk in (F) compare to asterisk in (D)). (G) Constructs missing PDZDs 4 and 5 (DGripΔ4–5) behave like wild-type DGrip, fully rescuing the dgripex36 defect, without provoking LTM defects. Scale bar in (H): 30 μm. (I) Quantification of rescue activity for DGripΔ6–7. Left: scheme of VLM defects used as ‘clinical score' (between 0.2 and 1) for quantification. Right: Average scores from over 30 larval hemisegments per condition are plotted, raising temperatures used indicated in colors. While DGripΔ6–7 hardly rescues dgripex36 VLM defects, no dominant effects of DGripΔ6–7 expression are observed in dgripex36/+ heterozygous background.
DGrip consists of seven PSD-95/Discs-large/ZO-1 domains (PDZDs) but no other known protein–protein interaction motifs. The ∼100–150 predicted PDZ proteins in the human genome are thought to direct the polarized localization of many developmentally and physiologically important membrane proteins. Indeed, in recent years, interaction screens have resulted in an explosion in the number of mammalian PDZ proteins identified as binding partners for growth factor receptors, G-protein-coupled receptors, neurotransmitter receptors, ion channels and adhesion molecules (Ranganathan and Ross, 1997; El Far and Betz, 2002).
Mammalian GRIP and GRIP2/ABP were identified by their interaction with AMPA-type glutamate receptors GluRs 2 and 3, and implicated in activity-dependent and subunit-specific GluR trafficking (Dong et al, 1997; O'Brien et al, 1998; Srivastava et al, 1998; Wyszynski et al, 1999, 2002; Liu and Cull-Candy, 2005). While genetic analysis has not yet shown an essential function for GRIP proteins in AMPA receptor clustering, GRIPs are meanwhile thought to participate in numerous cellular functions. GRIP1 mutant mice display kidney agenesis, polydactyly, syndactyly and gross morphological defects of the brain, a phenotype comparable to the human Fraser syndrome (Takamiya et al, 2004). GRIP has also been shown to interact with members of several signaling pathways including ephrins (Bruckner et al, 1999; Lin et al, 1999; Contractor et al, 2002; Hoogenraad et al, 2005) and liprins (Baran and Jin, 2002; Wyszynski et al, 2002; Dunah et al, 2005).
A functional insight into the biological significance of PDZD-ligand interactions has generally been limited by a lack of readily observable phenotypes. We set out to study the functional logic of DGrip, using its penetrant and easily scoreable phenotypes in Drosophila muscle guidance. Mutation and deletion analysis of PDZDs within DGrip strongly suggested that DGrip is indeed an integrative molecule, where PDZD-mediated interactions distributed over DGrip can have positive and negative influence on guidance function. We provide evidence that one particular PDZD ligand—the cell adhesion molecule Echinoid (Ed)—executes both positive and negative interactions on DGrip for muscle guidance. We speculate that a complex interaction between DGrip PDZDs and Ed may spatio-temporally fine tune muscle guidance.
Results
Structure–function analysis for Drosophila Grip using loss- and gain-of-function phenotypes
GRIP proteins are evolutionarily conserved as a string of seven PDZDs, whose functional significance is so far unknown. Genetic elimination of dgrip results in defects of VLM (Swan et al, 2004), which are schematized in yellow in Figure 1A. Instead of forming a single polarized muscle projection, dgrip− VLMs frequently send out two or more projections in essentially randomized directions. When the muscle guidance period ceases, dgrip− VLMs typically appear ‘frozen' in ball-like VLMs extending over only about half of a hemisegment without reaching their target tendon cell. In result, dgrip VLMs form ectopic, integrin-positive adhesion points on the epidermis and other muscles, away from tendon cells (Figure 1A; Swan et al, 2004). In contrast, LTMs (red in Figure 1A), which attach within segments, appear unaffected in dgrip mutants. Complementarily, strong pan-muscular overexpression of DGrip (using the driver 24B-gal4 together with two copies of UAS-dgrip) causes LTMs to produce projections that ectopically attach at segment borders (Swan et al, 2004). In this study, these two phenotypes formed a basis to study the function of individual DGrip PDZDs in vivo.
Muscle-specific re-expression of full-length DGrip cDNA fully rescues the VLM defect (Swan et al, 2004) in embryos hemizygous for dgrip null allele dgripex36 (denoted dgripex36). A structure–function map for DGrip was established by expressing at least two independent transgenic lines of DGrip variants in dgrip background using the muscle-specific twist-gal4 driver (Figure 1B). Muscle defects were directly scored in late stage embryos. In addition, larval muscles, which due to their larger size allow reliable identification of more subtle defects, were analyzed as well. This was possible as the twist-gal4 driver used in this study does not express in larval somatic muscles, restricting effects on muscle morphogenesis to embryonic stages. To test for the influence of expression strength on phenotypes, the Gal4/UAS expression system (Brand and Perrimon, 1993) temperature dependence was utilized by rearing animals at 18, 25 or 29°C to evoke successively higher levels of expression. When necessary, muscle morphogenetic defects were quantified by assigning scores to progressively more severe muscle defects, with more than 30 individual hemisegments being evaluated per genotype (Figures 1I and 2D).
Figure 2.
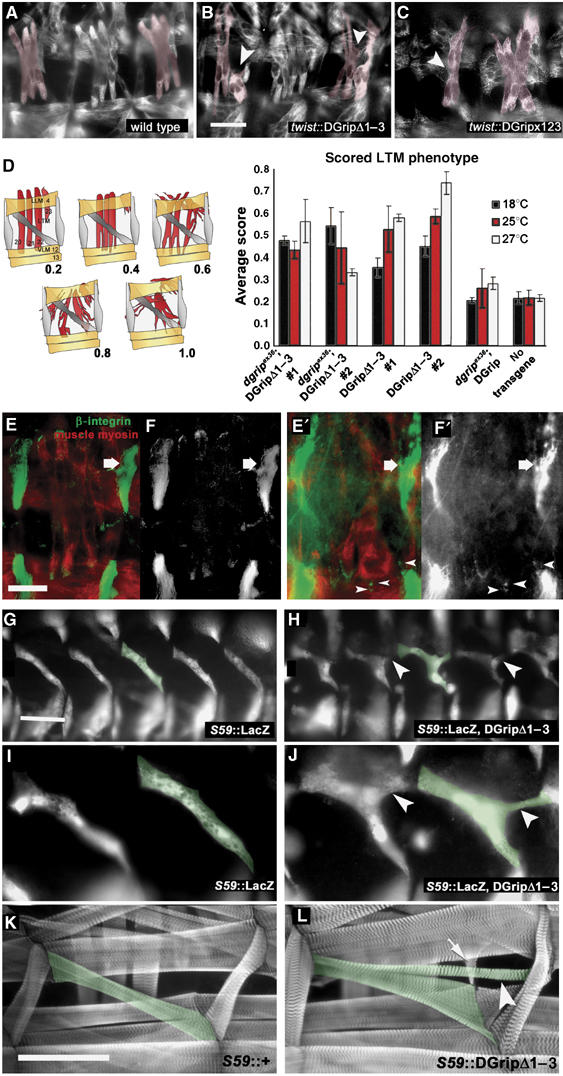
DGripΔ1–3 provokes defects during embryonic muscle guidance. (A–C) Muscle myosin stainings in stage 16 embryos, some LTMs colored in red. (A) wild type; (B) DGripΔ1–3- and (C) DGripx123-expression in wild-type background with the pan-muscular driver twist-gal4 causes LTM defects (arrowheads). LTM defects include splitting into multiple projections and bending away from their target tendon cells. (D) Quantification of LTM morphology defects after DGripΔ1–3 expression scored by average clinical score (n>30, larval hemisegments per condition). At different expression levels (controlled by raising temperature) and with two independent transgenes, DGripΔ1–3 provokes LTM defects neither present in animals comparably expressing wild-type DGrip nor in dgrip mutants. (E, F′) Wild-type embryo (E, F) and twist∷dgripΔ1–3 (E′, F′) embryos costained for integrin (green) and muscle myosin (red) (E, E′), (F and F′) show integrin channel only. In twist-gal4∷dgripΔ1–3 embryos, LTMs form aberrant, integrin positive attachments in ectopic positions (arrowheads). The large integrin-positive attachment sites of segment border attaching muscles are labeled by arrows. (G–I) Genetically labeling embryonic muscle 5 (individual muscle 5 labeled in green) with the S59-gal4 driver reveals guidance defects in DGripΔ1–3 expressing muscles. (H, J) S59-gal4∷GripΔ1–3, lacZ muscles shortly after guidance process showing extra filopodia-like projections (arrowheads) not present in wild-type muscle 5 (G, I). (K, L) Ectopic projections (arrowheads) of muscle 5 (green) established in embryogenesis are still present in larval stages of S59-gal4∷DGripΔ1–3 larvae (L, labeled in green) but not in S59-gal4/+ controls (K). In addition, other muscles can ectopically adhere to DGripΔ1–3 expressing muscles (L, arrow). Scale bar in (B): 20 μm; scale bar in (E): 15 μm; scale bar in (G): 30 μm in (K): 150 μm.
A transgene with PDZDs 4 and 5 deleted (DGripΔ4–5; Figure 1B and G) rescued the dgripex36 VLM misguidance phenotype to wild-type using twist-gal4 even at 18°C (minimal expression conditions), as does the full-length DGrip cDNA (Figure 1D). Thus, PDZDs 4 and 5 appear not to have a determining role in DGrip function. Similarly, DGripΔ4–5L, which additionally removed a region between PDZDs 3 and 4 (Figure 1B) allowed full rescue of dgripex36 muscles.
In contrast, removal of PDZDs 6 and 7 (DGripΔ6–7, Figure 1B and F) produced a protein that could scarcely rescue the dgripex36 muscle phenotype (for detailed analysis see Figure 1I). Thus, PDZDs 6 or 7 of DGrip appear be to involved in DGrip's function in muscle morphogenesis. For this reason, we tested the functional role of ligand binding to these domains. Point mutations disrupting the PDZD 6 or PDZD 7 ligand binding surfaces (see Supplementary data; Daniels et al, 1998; Edwards and Gill, 1999; Lou et al, 2001) were introduced, giving DGripx6 and DGripx7. DGripx6 was able to fully rescue dgripex36 VLMs, suggesting that PDZD 6 is not important for DGrip-dependent muscle function. DGripx7, however, showed impaired ability to rescue dgripex36 VLMs (Figure 1H), very similar to the reduced rescue function of DGripΔ6–7. Thus, ligand binding to PDZD 7, but not to 4, 5 or 6 was found to be important for DGrip function within muscle morphogenesis.
DGripΔ1–3 is an overactive species provoking ectopic projections during muscle guidance
Next, the functional importance of the N-terminal PDZDs was investigated. To this end, the first three PDZDs were deleted and the resulting DGripΔ1–3 expressed in dgripex36 background. DGripΔ1–3 expression clearly restored VLM morphology to a level closer to wild-type when compared with dgripex36 controls, with some minor defects still detectable (Figure 1E, yellow marked VLM). However, DGripΔ1–3 expression also evoked the same slight VLM defects in the dgripex36/+ background, that is, in the presence of one wild-type dgrip gene copy.
VLM defects were never observed when expressing full-length DGrip using twist-gal4, despite the RT–PCR levels of both transgenes being very similar (Supplementary data, for brevity, all subsequent experiments were performed utilizing the twist-gal4 driver at 25°C). This suggested that DGripΔ1–3 was a dominant, overactive DGrip species. In contrast to VLMs, LTMs consistently exhibited very strong defects when expressing DGripΔ1–3 in the dgripex36, dgripex36/+ or wild-type backgrounds: formation and stabilization of multiple projections with splits of embryonic muscles (Figures 1E, arrows, 2B, D and 3B). Defects appeared qualitatively very similar when scored in embryos (Figure 2) or in larvae (Figure 3). When quantified in larval stages, twist-gal4∷DGripΔ1–3 provoked muscle defects were fully evident already at 18°C, expression conditions under which full-length DGrip was unable to provoke any LTM defects (Figure 2D). Thus, DGripΔ1–3 apparently was an overactive DGrip species that could efficiently interfere with muscle morphogenesis.
Figure 3.
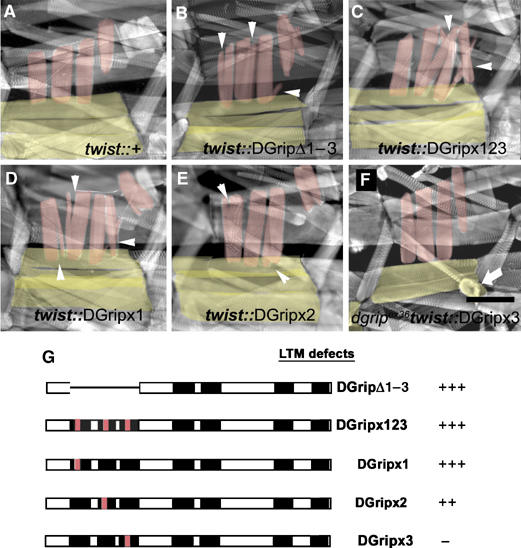
Removal of PDZDs 1–3, or of ligand binding surfaces of PDZD 1 or 2, results in dominantly active Dgrip. The dominant DGripΔ1–3 phenotype is recapitulated by specifically point mutating the ligand binding sites of individual PDZDs. (A–F) Phalloidin labeling of 3rd instar larvae, VLMs in yellow, LTMs in red. (A) Typical bar-shaped LTMs and VLMs in control larvae. (B) Ectopic LTM projections (arrowheads) and mild morphological defects of VLMs in DGripΔ1–3 expressing animal. This phenotype is fully recapitulated (arrowheads) in DGripx123-expressing larvae, where the ligand binding capability of the PDZDs 1–3 is disturbed by point mutations. Mutation of PDZD 1 only (D) results in a similar phenotype, expression of DGrip point-mutated at PDZD 2 only (E) produces slightly weaker dominant defects. (F) Mutation of PDZD 3 does not cause dominant defects, but does not allow proper rescue of dgripex36 VLMs (arrowhead). (G) Summary of LTM defects caused by twist-gal4 driven expression of the indicated dgrip constructs. Scale bar in (F): 150 μm.
The question arose whether DGripΔ1–3 overexpression executed its effects via interfering with proper guidance of nascent LTMs, similar to the manner that VLM guidance suffers from a loss of DGrip function. In fact, when driven with twist-gal4, DGripΔ1–3 provoked the formation of ectopic, integrin-positive attachment sites, obvious in late, muscle myosin-positive embryos (Figure 2E–F). However, the fact that twist-gal4 drives expression in all muscles, interfered with directly scoring guidance behavior in individual muscles. Thus, we decided to use S59-gal4—labeling only muscles 5, 8, 25, 27 and 29 from stage 11 until the end of embryogenesis (Brennan et al, 1999)—to drive lacZ marker protein. When expressing DGripΔ1–3 with this driver, the full set of muscles expressing S59 could be observed (Figure 2G), indicating proper determination of muscle cell fate in the presence of DGripΔ1–3 (Figure 2H). Of the LacZ labeled muscles, nascent muscle 5 (Figure 2G–L, green label) was best suited to score guidance behavior. In fact, muscle 5 overexpressing DGripΔ1–3 nearly always formed a third, ectopic projection attaching at the segment border (Figure 2G–L, arrowheads). These additional projections of S59-gal4∷DGripΔ1–3 expressing muscles were still obvious in larval stages (Figure 2L). We conclude that loss of the first three PDZDs of DGrip renders the protein overactive, ectopically stabilizing projections during embryonic muscle guidance, which were fully propagated into larval muscle morphology.
PDZDs 1 and 2 mediate repression, PDZD 3 de-repression of DGrip activity
As DGripΔ1–3 is obviously an overactive species, we suspected PDZDs 1–3 conferred repression on DGrip activity. Thereby, dominant activity of DGripΔ1–3 may be either mediated by a loss of repressive interactions mediated via PDZDs 1–3 or by a more general structural defect of the protein. To discriminate between these possibilities, we tested if DGripΔ1–3's dominant activity could be mimicked by destroying ligand binding via PDZDs 1, 2 or 3. When the first three PDZD ligand binding surfaces (DGripx1, 2, 3) were mutated together, dominant defects on LTM morphology comparable to DGripΔ1–3 were observed in both embryos (Figure 2C) and larvae (Figure 3C). Dominant LTM defects were also present when only PDZD 1 (DGripx1, Figure 3D) and to a slightly lesser extent when only PDZD 2 (DGripx2, Figure 3E) ligand-binding was abolished. Both DGripx1 and DGripx2 rescued the dgripex36 VLMs, again indicating that these are overactive DGrip species (not shown).
Unlike DGripx1 and DGripx2, DGripx3 did not allow a complete rescue of VLM defects of dgripex36 and showed only negligible dominant defects in LTM guidance (Figure 3F, VLM marked by arrow). Accordingly, DGripx3 appeared to have reduced DGrip activity. However, DGripΔ1–3 and DGripx1, 2, 3 had excessive activity. Thus, ligand binding via PDZD 3 is apparently relevant only in the presence of ligands binding PDZD 1 and 2. PDZDs 1 and 2 in return mediate repression of DGrip function. These findings are easiest explained by postulating that DGrip activity is normally repressed by ligand binding to PDZD 1 and 2, while ligand binding to PDZD 3 is needed to allow efficient de-repression of DGrip activity.
Non-PDZD regions of DGrip are dispensable for muscle guidance function
We had found that PDZDs 4, 5 and 6 when singly mutated (x6) or deleted (Δ4–5) did not affect DGrip guidance function. However, deletion or point mutation of PDZDs 3 and 7 (x3: Figure 3F, x7: Figure 1H, Δ1–3: Figure 1F) only partially compromised DGrip function in vivo. We thus investigated if critical functions of DGrip reside in regions between PDZDs. However, deleting the large regions between PDZD 3 and 4 (in DGripΔ4–5L) and between PDZD 5 and 6 (DGripΔint) did not compromise rescue function (Figure 1B).
Moreover, smaller clusters of PDZDs contributing to DGrip function show no activity in isolation. Neither the first three (DGripΔ4–7) nor the last two (DGripΔ1–5) PDZDs show rescue of VLM misguidance (Figure 1B) or any dominant activity.
DGrip interacts with the cell adhesion molecule Ed via a C-terminal PDZD-binding motif
To identify PDZD-interactors mediating DGrip function in vivo, we performed a yeast-two-hybrid (Y2H) screen using the first three PDZDs of DGrip as baits. Four independent clones encoding fragments of the cell adhesion molecule Ed were retrieved (Figure 4A). All fragments included the C-terminus of the molecule, which contains a type II PDZD-interaction motif (EIIV). Interaction of Ed with DGrip in Y2H was dependent on the EIIV motif (Figure 4C). Moreover, recombinant DGrip expressed in Sf.9 cells (Figure 4B) efficiently interacted with a matrix-bound peptide representing the C-terminal 10 amino acids of Ed (Ed1/2), but not with a scrambled version of it (control).
Figure 4.
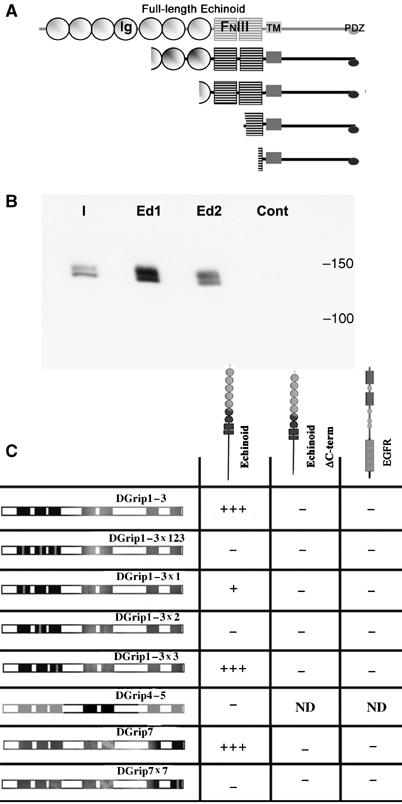
The cell-adhesion molecule Echinoid physically interacts with Dgrip. (A) Y2H screen performed using the first three PDZDs of DGrip as bait returned four independent isolates of the Immunoglobulin (Ig) and Fibronectin type III (FNIII)-domain containing cell adhesion molecule Echinoid (Ed). All isolates contained the transmembrane (TM) region and the entire cytosolic tail, including the EIIV PDZ-ligand motif. (B) Full-length, C-terminally myc-tagged DGrip expressed in Sf.9 cells specifically binds to a 10aa peptide representing the C-terminus of Ed. Shown is the input (I), binding of DGrip-myc to the Ed peptide (Ed1/2, two independent experiments), and binding to a 10aa scrambled control peptide (cont). (C) Y2H experiments reveal a specific pattern of Ed binding to DGrip PDZDs. The Ed cytosolic tail strongly interacts with a construct containing the first three PDZDs of DGrip, or containing PDZD 7 only (+++). This interaction was abolished by point mutation of PDZDs 2 or 7 (−), strongly reduced by point mutation of PDZD 1 (+) and unaffected by point mutation of PDZD 3 (+++). This interaction depended on the EIIV motif at the C-terminal of Ed, as Ed-ΔC-term did not interact with DGrip constructs. DGrip did not interact with the EGFR C-terminus, used here as a control, which has a C-terminal type I PDZ-ligand motif (ETRV).
We mapped Ed binding versus individual PDZDs in the Y2H assay (Figure 4C). Ed binding to DGrip was dependent on an intact PDZD 2 and greatly weakened by point mutation of PDZD 1. Binding was unaffected by point-mutating PDZD 3. No interaction was found between Ed and PDZDs 4 and 5. Thus, Ed binds to PDZD 1 and 2 and might well be involved in the repressive function of these domains. Surprisingly, Ed also interacted with PDZD 7 in a manner that was dependent on the PDZD 7 ligand-binding surface and EIIV motif of Ed (Figure 4C).
Loss of Ed provokes defects in both LTMs and VLMs
Ed is an L1-CAM-like molecule, known as a regulator of both the EGF receptor (Bai et al, 2001; Escudero et al, 2003; Islam et al, 2003; Rawlins et al, 2003a, 2003b; Spencer and Cagan, 2003) and Notch (Ahmed et al, 2003; Escudero et al, 2003) signaling pathways. Ed has not previously been reported as having a phenotype related to muscle development, although both EGFR and Notch signaling are critical for the specification of muscle precursors (Artero et al, 2003). Using the P(lacZ)edk01102 line (lacZ is inserted in the first intron of ed) to mark ed expressing tissues, we found ed expression in nascent muscles (identified by the muscle precursor marker Vg (Ruiz-Gomez, 1998; Figure 5A)) as well as in many other cells. Moreover, antibodies against the intracellular tail of Ed also showed that Ed protein is in fact concentrated at the ends of embryonic VLMs, where these muscles contact the extracellular matrix (Figure 5B), colocalizing here with muscle expressed DGRIP-GFP (Figure 5C). To test for a role of Ed in muscle formation, muscle morphology was examined in several independent ed alleles. The ed1x5 allele (Bai et al, 2001) is predominantly embryonic lethal under our raising conditions, with some animals developing into second instar larvae. Consistent and prominent defects in muscle morphology could be found in both ed1x5 embryos (Figure 5E and E′) and larvae (Figure 5G). The same muscle defects were also observed in strong hypomorph edSlH8 homozygotes (Figure 6I) or ed1x5/edSlH8 animals (Figure 5H). Loss of Ed function resulted in defects of both LTMs and VLMs. VLM defects were reminiscent of partial loss of DGrip function. As such, they showed ‘fasciculated' VLMs also forming ectopic muscle–muscle adhesion within the segment (compare VLMs indicated by arrowheads in echinoid mutants in Figure 5H with partially rescued dgrip mutant in Figure 7G). Muscle-specific overexpression of Ed produced rather mild defects in LTM morphology (Figure 5I).
Figure 5.
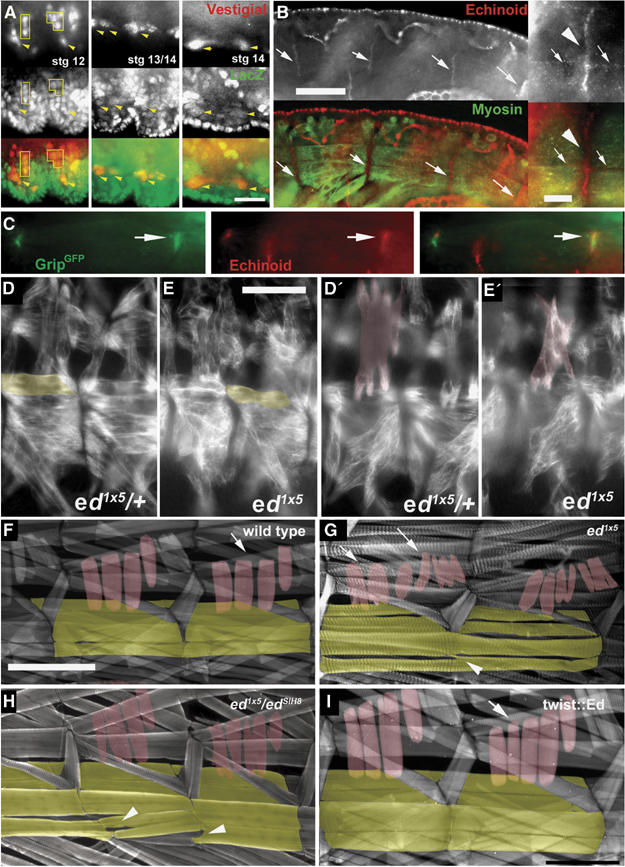
Echinoid in muscle morphogenesis of the Drosophila embryo. (A) Expression of lacZ (green) from the ed locus in P(lacZ)edk01120 combined with labeling of muscle precursors with Vestigial (red). Scale bar in (A): 10 μm. (B) Anti-Ed antibodies (red) stain the ends of morphologically mature VLMs (arrows) visualized by antibodies against muscle myosin (green). Scale bar in (B): 30 μm; Inset: magnified view of Ed accumulation at muscle ends (arrowhead). Some Ed protein is also found at other parts of the muscle membrane (arrows). Scale bar in inset: 5 μm. (C) Ed protein colocalizes with muscle expressed twist-gal4∷DGrip-GFP at muscle ends (arrows). (D–H) ed mutants displaying morphological defects of both VLMs (yellow) and LTMs (red) in embryos (D–E′) and larvae (F–H). (D, D′) ed1x5/+ control embryo showing normal VLMs and LTMs, respectively. (E, E′) ed1x5 embryo shows defects in both VLM and LTM morphology. The same muscle field is shown in two focal planes. (F) Control larva. (G) The strong ed allele ed1x5 produces few homozygous larvae, which survive to 2nd instar. In these larvae, defects in VLMs (arrowhead) and LTMs (arrows) are evident, with both muscle groups forming aberrant projections and ectopic adhesion points. (H) Similar muscle phenotypes are also observed in 3rd instar larvae of the genotype ed1x5 over the strong hypomorphic allele edSlH8 (arrows and arrowheads indicate VLMs and LTMs respectively). (I) Only minor defects of LTMs are evoked by pan-muscular expression of Ed. Scale bar in (E): 35 μm; Scale bar in (F): 200 μm.
Figure 6.
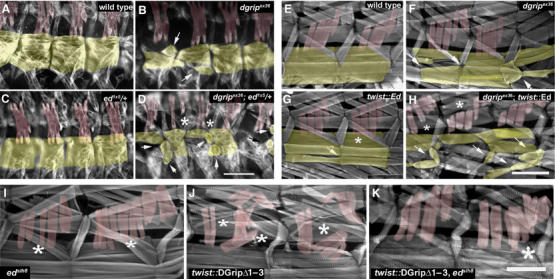
Functional interactions between Echinoid and DGrip in muscle morphogenesis. (A–D) Heterozygosity for ed mutant allele enhances defects in VLM and provokes LTM defects as shown by muscle myosin stainings of stage 16 embryos (A) wild type; (B) dgripex36 mutants with characteristic defects in VLM morphology (arrow); (C) ed1x5/+ embryos show no defects in LTMs or VLMs. (D) dgripex36; ed1x5/+ embryos exhibit more severe VLM defects (arrows) than dgripex36 embryos, where sometimes LTMs might be missing (asterisks). More severe examples of dgripex36; ed1x5/+ embryos (not shown) exhibit completely deranged somatic musculature, where muscle identification is no longer possible. Scale bar in (D): 50 μm. (E–H) Muscle-specific overexpression of Ed enhances defects in dgripex36 mutants. (E) Control larvae, showing bar-like morphology in VLM (yellow) and LTM (red). (F) dgripex36 larvae; (G) twist-gal4∷UAS-ed larvae exhibit few, mild defects in LTM and VLM morphology. Arrow indicates VLM with weakly distorted morphology, asterisks mark slightly split LTMs. (H) twist-mediated expression of Echinoid in the dgripex36 background greatly enhances defects; VLMs are more severely deranged than in dgripex36, and often appear to adhere to other muscles (arrows), whereas LTMs split (asterisks). Scale bar in (H): 300 μm. (I–K) Homozgosity for edSlH8 suppresses DGripΔ1–3 activity. (I) edSlH8 larvae show defects typical for ed zygotic alleles with slight LTM splitting (asterisks) and some malformation of VLMs. (J) twist-gal4∷UAS-dgripΔ1–3 controls with severe malformation of LTMs (asteriks). (K) edSlH8; twist-gal4∷UAS-dgripΔ1–3 larvae consistently showed far milder LTM (asteriks) defects than twist-gal4∷UAS-dgripΔ1–3 processed in parallel (n>30 hemisegments per genotype). Scale bar in (K) 150 μm.
Figure 7.
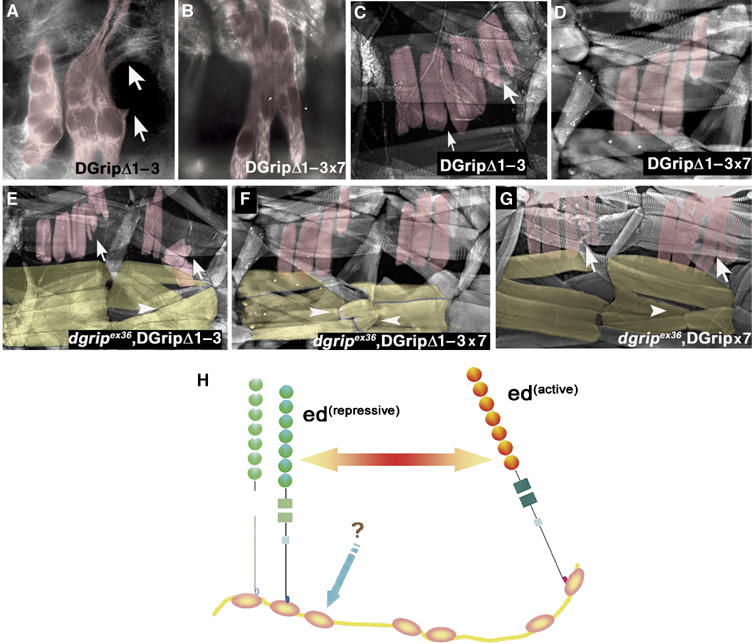
Mutation of the Echinoid-binding DGrip PDZD 7 represses DGripΔ1–3 activity. This figure depicts muscle myosin stainings in embryos (A, B) and phalloidin labeling in larvae (C, D). Point mutation of PDZD 7 reduces DGripΔ1–3 activity. (A–D) The dominant activity of the DGripΔ1–3 protein ((A) embryo; (C) larva), which causes abnormal muscle projections (arrows) is reduced by point mutation of the PDZD 7 ligand binding surface, producing DGripΔ1–3x7, which shows only slight defects in LTM morphology in embryo (B) or larva (D). (E, F) DGripΔ1–3x7 shows only limited rescue ability in dgripex36 VLMs (F, arrowheads) when compared to DGripΔ1–3 (E, arrowheads). (G) DGripx7 produces mild dominant defects of LTMs (arrows) and impaired rescue of VLMs (arrowheads). (H) Model of DGrip–Echinoid functional interaction during muscle morphogenesis. DGrip may act by maintaining the equilibrium between active and repressive Echinoid signaling. Ed binds DGrip at PDZD 2 (and possibly 1), where it is repressed. Interaction of an unknown protein with PDZD 3 relieves this repression, allowing Ed to bind PDZD 7 and activating the complex.
The ed muscle defects could, in principle, reflect a requirement for Ed in forming epidermal tendon cells. However, tendon cells are positioned correctly in ed1x5 zygotic mutants in stainings for tendon cell-specific markers (not shown). It appears very likely that additional removal of maternal Ed could reveal even stronger defects in muscle morphology. However, maternal ed is present in the epidermis in high amounts, making it unlikely that such a function could be demonstrated in the light of epidermal defects to be expected upon removal of maternal Ed.
Genetic interactions between DGrip and Ed signaling in muscles
Given that Ed physically interacts with DGrip, we asked whether DGrip and Ed functionally interacted in vivo. In fact, heterozygosity for ed1x5 strongly enhanced the VLM defects in dgripex36 hemizygous embryos (Figure 6D). In severe cases, a complete disruption of the muscle field, in milder cases a strong enhancement of dgrip muscle defects (compare VLMs labeled by arrows in Figure 6D with B) was observed. LTMs were also defective in dgripex36/Y; ed1x5/+ (Figure 6D, asterisks), while they appeared normal in both ed1x5/+ (Figure 6C) and dgripex36 (Figure 6B). Thus, the reduction of Ed protein levels in embryonic muscles apparently uncovered a subcritical requirement for DGrip in LTM morphogenesis, leading to LTM defects. Therefore, Ed operates in LTM formation even in the absence of DGrip, and its requirement there becomes more obvious in the absence of DGrip. We conclude that DGrip and Ed functionally interact for VLM and, surprisingly, also LTM guidance. dgripex36 muscles, including LTMs, were sensitive to twist-gal4-driven overexpression of Ed (Figure 6H, asterisks LTMs, arrows VLMs), whereas Ed expression in wild-type background produced only very minor defects (Figure 6G, asterisk and arrow). This again suggested that DGrip was functionally linked to Ed. In this context, however, DGrip acted as an inhibitor of excessive Ed-mediated signaling.
Mutations in echinoid suppress dominant activity of DGripΔ1–3
Our data indicated that Ed and DGrip interact physically, and that this complex is involved in controlling muscle morphology. Whether this interaction promoted or inhibited DGrip associated signaling was context-dependent, suggesting that their interaction with one another may be complex. Ed bound PDZDs 1–3 of DGrip as well as PDZD 7. Loss of Ed function should not be able to suppress DGripΔ1–3 activity if Ed binding to DGrip was in fact restricted to PDZDs 1–3. However, the LTM phenotype induced by twist-gal4 mediated expression of DGripΔ1–3 protein was greatly diminished by homozygosity for the edSlH8 chromosome. edSlH8;twist-gal4∷UAS-dgripΔ1–3 animals (Figure 6K) showed LTM phenotypes more closely resembling pure edSlH8 homozygotes rather than identically processed and simultaneously raised twist-gal4∷UAS-dgripΔ1–3 controls (Figure 6J). These interactions suggest that Ed is involved in mediating the unrepressed activity of the DGripΔ1–3 protein.
Ed binds to the PDZDs controlling muscle guidance activity
In our Y2H assay, Ed specifically interacted with PDZD7, as well as with the ‘repressive' PDZDs 1 and 2. As DGripΔ1–3 activity was repressed by reduction in Ed protein, we asked whether the activity of DGripΔ1–3 might be regulated by interactions via PDZDs 6 or 7. To this end, transgenic lines expressing DGripΔ1–3x6 and DGripΔ1–3x7 were constructed and expressed with twist-gal4 in dgripex36 background. The PDZD 6 point mutation did not suppress the dominant action of DGripΔ1–3 but still allowed VLM rescue. DGripΔ1–3x6 behaved identically to DGripΔ1–3, suggesting that PDZD 6 does not mediate DGrip guidance activity after its de-repression in DGripΔ1–3 (not shown). In contrast, DGripΔ1–3x7 showed a severely impaired ability to rescue dgripex36 VLMs in embryos and larvae (Figure 7F, arrowheads) when compared to DGripΔ1–3 (Figure 7E). Consistently, expression of DGripΔ1–3x7 (Figure 7B, embryo, D larva) produced only mild defects in LTMs comparable to the mild defects obtained by expressing DGripx7 protein alone (Figure 7G, arrows), but not to the severe LTM defects observed upon DGripΔ1–3 expression (Figure 7A and E). Thus, ligand binding to PDZD 7 but not to PDZD 6 appeared to be a significant mediator of DGripΔ1–3 activity.
We thus propose that DGrip and Ed functionally interact during muscle guidance. Reduction of Ed protein or defective binding to PDZD 7 of DGrip interfered with the overactivity of DGripΔ1–3, thus suggesting a model (Figure 7H) where DGrip is responsible for the equilibrium between ‘repressive' and ‘active' Ed signaling.
Discussion
We have used genetics to develop a mechanistic model concerning a well-defined function mediated by Drosophila Grip—embryonic muscle guidance. Functional requirements were not transmitted by single domains, but were found to be distributed over the whole length of this 7 PDZD protein in an unexpectedly complex manner. Binding ligands via PDZDs 1 and 2 repressed the activity of the protein, binding to PDZD 3 was involved in de-repression, and PDZ-ligand binding via PDZD 7-mediated DGrip function after its de-repression. Despite the fact that there was no critical dependence on PDZDs 4–5 or interdomains for function, we cannot exclude that interactions over these domains play a subthreshold role. In fact, the DGripΔ1–3x7 construct showed some residual functionality in terms of muscle rescue. Thus, the whole protein might be used as an ‘intelligent frame' designed to execute fine controls such as thresholding functions or coincidence detections. In fact, all attempts to provide DGrip activity or to repress DGrip activities with only partial fragments (DGripΔ4–7, DGripΔ1–5) failed (Figure 1B, our data), leading us to believe that DGrip is responsible for the organization of a macromolecular complex, of which the transmembrane protein Ed is part.
PDZDs are not functionally isolated
Our analysis suggests that a critical number of PDZDs are utilized for DGrip function, with both negative and positive interactions occurring. Such dependence between PDZDs may be due to structural chaperoning (Feng et al, 2003). Alternatively, a fixed orientation might be required for high-affinity binding to its targets as found for tandem PDZDs 1 and 2 in PSD-95 (Long et al, 2003), with a complex of two PDZDs having higher binding affinity than either PDZD alone. Moreover, allosteric changes upon PDZD-ligand binding could change binding affinities of neighboring domains (Fuentes et al, 2004; Peterson et al, 2004) or via bridging interactions where one molecule contacts multiple sites on a PDZ protein to effect conformational change (van Huizen et al, 1998; Schlieker et al, 2004; Wilken et al, 2004). Such mechanisms might be the substrate for integrating ligand binding and functional output over a large ‘multivalent' PDZD protein.
Point mutations of PDZD 1 and PDZD 2 recapitulated the DGripΔ1–3 phenotype in the LTM group of muscles (Figure 3), indicating that the repressive function of the PDZDs 1–3 region is not ‘structural' (i.e. by covering other PDZDs on the protein). Instead, we suggest that ligand interactions are communicated over the whole protein to steer equilibrium between two different functional modes of DGrip signaling.
DGrip interacts with Ed
Ed was identified as a novel DGrip interactor. Ed is cell adhesion protein with 7 Ig and 2 FNIII domains, described to have both adherence and signaling roles in Drosophila tissues (Islam et al, 2003; Rawlins et al, 2003a, 2003b; Wei et al, 2005). It is highly conserved among invertebrates and its closest vertebrate homologues are Nectins, which exhibit 3 Ig domains and end in the PDZ-binding motif E/A-X-Y-V. Functionally, both protein families are similar: although not functionally redundant with Ed (Wei et al, 2005), Nectins are present at mammalian adherens junctions (AJs) along with l-afadin (Tachibana et al, 2000) and, like Ed, regulate Cadherin-based adherence at AJs (Sato et al, 2006). Several lines of evidence link Ed to DGrip:
Ed interacted with DGrip in a yeast two-hybrid screen, dependent on the C-terminal EIIV motif, mediated via PDZDs 1, 2 or 7 (Figure 4). Myc-tagged DGrip specifically interacts with a peptide representing the last 10 amino acids of the Ed protein, including the EIIV PDZ-binding motif.
ed zygotic mutants have defects in the morphogenesis of embryonic muscles qualitatively similar to DGripΔ1–3 overexpression.
the dgripex36 muscle phenotype in embryos is enhanced by heterozygosity for ed1x5. Here, LTMs (unaffected in pure dgripex36) are affected as well (Figure 6D).
dgripex36 mutant muscles (both VLMs and LTMs) are sensitive to Ed overexpression (Figure 6H). These synthetic defects suggest that DGrip, while itself not essential for LTM morphogenesis, regulates Ed in this group of muscles.
homozygosity for hypomorphic edslH8 chromosome strongly reduced the severity of the phenotype evoked by pan-muscular expression of DGripΔ1–3 (Figure 6K), indicating that Ed acts downstream of activated DGrip.
Notably, the pattern of Ed-PDZD binding correlates with the DGrip-dependent LTM phenotype. Expression of DGrip missing PDZDs 1, 2 and 3 together, or ligand binding in PDZD 1 and PDZD 2 only, showed a strong dominant active phenotype (Figures 2 and 3). Mutation of PDZD 2 caused a dominant phenotype in LTMs (Figure 3E). In a yeast-two hybrid test, Ed interacted strongly with PDZD 2 with and PDZD 7 (Figure 4C), and more weakly with PDZD 1.
In imaginal discs, Ed binds two different PDZD proteins via its EIIV motif: Canoe, an F-actin interacting protein and PAR-3/Bazooka. This interaction is mutually exclusive, thereby influencing cell adhesion and the remodeling of subcortical actin at AJs (Wei et al, 2005). Here, we propose a similar mechanism, in that both functional states of Ed are established via binding to the same protein (DGrip) at different sites. In this model, DGrip may assist in maintaining equilibrium between active and inactive signaling states of Ed, which in its inactive state binds to PDZDs 1 and 2, and in its active form to PDZD 7 of DGrip. This interaction appears tissue specific in nature, as DGrip mutants do not display the full spectrum of defects of ed mutants (such as neurogenic phenotypes (Ahmed et al, 2003), our data) and that there are as yet unknown members of the DGrip–Ed complex, such as that which binds to the ‘de-repressing' PDZD 3.
Both Ed loss of function and overexpression can produce similar phenotypes in muscles (Figures 5E, G–I and 6G, I), which are strongly enhanced by the absence of DGrip. Ed is described as a homophilic cell adhesion molecule (Islam et al, 2003; Rawlins et al, 2003a; Spencer and Cagan, 2003), and is maternally expressed in the epidermis, over which nascent muscles ‘crawl' during the muscle guidance process to reach their target apodeme. ed clones in wing discs show cell sorting behavior, causing aggregation and adhesion of only those cells expressing the same complement of cell adhesion molecules (Wei et al, 2005). Thus, both reduction and excess of Ed on the ‘muscle side' of transient muscle–epidermal adhesions could lead to significant changes in the cell adhesion properties of the developing muscle. The experiments shown in this study for DGripΔ1–3 overexpression in muscle 5 (Figure 2G–L) and for VLMs in dgrip mutants (Swan et al, 2004, Figure 5) imply that a tight balance of DGrip activity might particularly be needed to keep navigating muscle projections motile and to avoid their premature stabilization at ectopic epidermis contacts during the ‘steering' process—ultimately instructed by Slit/Robo or other guidance systems. It is likely that Ed and DGrip form complexes enriched at muscle projection membranes to locally control adhesiveness. Ectopic adhesions among muscles cells with aberrant DGrip activity are in fact indicative of changes in muscle adhesiveness (e.g. see arrow in Figure 2L).
Natural variants of mGRIP missing PDZDs 1–3 have been localized to mammalian synapses (Charych et al, 2004), and it has recently been found that the type 5 metalloproteinase MT5-MMP is recruited by GRIP1/2 to growth-cone filopodia and to both mature and developing synapses, where it proteolyses N-cadherins (Monea et al, 2006). GRIP2 was also observed to be a member of a δ-catenin containing complex (Monea et al, 2006). Drosophila Echinoid is known to regulate DE-Cadherin in homeotypic cell–cell junctions (Wei et al, 2005). Given these promising indications, it will prove interesting to see whether in the context Grip proteins became famous for—synapse assembly—similar molecular strategies are used by the GRIP protein as those we describe here in the context of muscle morphogenesis.
Materials and methods
Immunostaining
Staining of embryos and larvae as well as most antibodies used were described recently (Swan et al, 2004). In addition were used: anti-Ed (rabbit, used at 1:250 (Rawlins et al, 2003a)), GFP (mouse, used at 1:200; MolProbes) and β-PS integrin (rabbit, used at 1:50; Nick Brown).
Biochemistry
The detailed procedure is described (Soltau et al, 2004). In brief, a synthetic peptide representing the C-terminus of Ed (NRRVIREIIV) and a scrambled control (RIVRIRIEVN) were generated by peptides&elephants GmbH, Nuthetal, Germany. These were coupled to NHS-activated sepharose at a concentration of 3 mg/ml matrix. Transfected Sf.9 cells were lysed in NTEP-buffer (50 mM Tris/HCl, 150 mM NaCl, 5 mM EDTA, 10 mM iodacetamide, 1 mM PMSF and 0.5% (v/v) Nonidet NP40, pH 7.9) on ice. Sf.9 cell extracts were ‘precleared' 3 h with 400 μl NHS-sepharose-slurry to prevent unspecific binding to the NHS-sepharose. Precleared supernatant was applied to the peptide/NHS-matrix for 1 h at 4°C, the matrix washed five times, eluted by boiling in SDS sample buffer and analyzed by SDS–polyacrylamide gel electrophoresis followed by Western blotting. Anti-Myc-Ab (mouse, 1:500, Santa Cruz) was used for detection.
Supplementary Material
Supplementary data
Supplementary Figure
References
- Ahmed A, Chandra S, Magarinos M, Vaessin H (2003) Echinoid mutants exhibit neurogenic phenotypes and show synergistic interactions with the Notch signaling pathway. Development 130: 6295–6304 [DOI] [PubMed] [Google Scholar]
- Artero R, Furlong EE, Beckett K, Scott MP, Baylies M (2003) Notch and Ras signaling pathway effector genes expressed in fusion competent and founder cells during Drosophila myogenesis. Development 130: 6257–6272 [DOI] [PubMed] [Google Scholar]
- Bai J, Chiu W, Wang J, Tzeng T, Perrimon N, Hsu J (2001) The cell adhesion molecule Echinoid defines a new pathway that antagonizes the Drosophila EGF receptor signaling pathway. Development 128: 591–601 [DOI] [PubMed] [Google Scholar]
- Baran R, Jin Y (2002) Getting a GRIP on liprins. Neuron 34: 1–2 [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415 [DOI] [PubMed] [Google Scholar]
- Brennan K, Baylies M, Arias AM (1999) Repression by Notch is required before Wingless signalling during muscle progenitor cell development in Drosophila. Curr Biol 9: 707–710 [DOI] [PubMed] [Google Scholar]
- Bruckner K, Pablo Labrador J, Scheiffele P, Herb A, Seeburg PH, Klein R (1999) EphrinB ligands recruit GRIP family PDZ adaptor proteins into raft membrane microdomains. Neuron 22: 511–524 [DOI] [PubMed] [Google Scholar]
- Charych EI, Yu W, Li R, Serwanski DR, Miralles CP, Li X, Yang BY, Pinal N, Walikonis R, De Blas AL (2004) A four PDZ domain-containing splice variant form of GRIP1 is localized in GABAergic and glutamatergic synapses in the brain. J Biol Chem 279: 38978–38990 [DOI] [PubMed] [Google Scholar]
- Contractor A, Rogers C, Maron C, Henkemeyer M, Swanson GT, Heinemann SF (2002) Trans-synaptic Eph receptor-ephrin signaling in hippocampal mossy fiber LTP. Science 296: 1864–1869 [DOI] [PubMed] [Google Scholar]
- Daniels DL, Cohen AR, Anderson JM, Brunger AT (1998) Crystal structure of the hCASK PDZ domain reveals the structural basis of class II PDZ domain target recognition. Nat Struct Biol 5: 317–325 [DOI] [PubMed] [Google Scholar]
- Dong H, O'Brien RJ, Fung ET, Lanahan AA, Worley PF, Huganir RL (1997) GRIP: a synaptic PDZ domain-containing protein that interacts with AMPA receptors. Nature 386: 279–284 [DOI] [PubMed] [Google Scholar]
- Dunah AW, Hueske E, Wyszynski M, Hoogenraad CC, Jaworski J, Pak DT, Simonetta A, Liu G, Sheng M (2005) LAR receptor protein tyrosine phosphatases in the development and maintenance of excitatory synapses. Nat Neurosci 8: 458–467 [DOI] [PubMed] [Google Scholar]
- Edwards DC, Gill GN (1999) Structural features of LIM kinase that control effects on the actin cytoskeleton. J Biol Chem 274: 11352–11361 [DOI] [PubMed] [Google Scholar]
- El Far O, Betz H (2002) G-protein-coupled receptors for neurotransmitter amino acids: Cterminal tails, crowded signalosomes. Biochem J 365: 329–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudero LM, Wei SY, Chiu WH, Modolell J, Hsu JC (2003) Echinoid synergizes with the Notch signaling pathway in Drosophila mesothorax bristle patterning. Development 130: 6305–6316 [DOI] [PubMed] [Google Scholar]
- Feng W, Shi Y, Li M, Zhang M (2003) Tandem PDZ repeats in glutamate receptor interacting proteins have a novel mode of PDZ domain-mediated target binding. Nat Struct Biol 10: 972–978 [DOI] [PubMed] [Google Scholar]
- Fernandes JJ, Celniker SE, VijayRaghavan K (1996) Development of the indirect flight muscle attachment sites in Drosophila: role of the PS integrins and the stripe gene. Dev Biol 176: 166–184 [DOI] [PubMed] [Google Scholar]
- Frommer G, Vorbruggen G, Pasca G, Jackle H, Volk T (1996) Epidermal egr-like zinc finger protein of Drosophila participates in myotube guidance. EMBO J 15: 1642–1649 [PMC free article] [PubMed] [Google Scholar]
- Fuentes EJ, Der CJ, Lee AL (2004) Ligand-dependent dynamics and intramolecular signaling in a PDZ domain. J Mol Biol 335: 1105–1115 [DOI] [PubMed] [Google Scholar]
- Ghazi A, Paul L, VijayRaghavan K (2003) Prepattern genes and signaling molecules regulate stripe expression to specify Drosophila flight muscle attachment sites. Mech Dev 120: 519–528 [DOI] [PubMed] [Google Scholar]
- Hoogenraad CC, Milstein AD, Ethell IM, Henkemeyer M, Sheng M (2005) GRIP1 controls dendrite morphogenesis by regulating EphB receptor trafficking. Nat Neurosci 8: 906–915 [DOI] [PubMed] [Google Scholar]
- Islam R, Wei SY, Chiu WH, Hortsch M, Hsu JC (2003) Neuroglian activates Echinoid to antagonize the Drosophila EGF receptor signaling pathway. Development 130: 2051–2059 [DOI] [PubMed] [Google Scholar]
- Kramer SG, Kidd T, Simpson JH, Goodman CS (2001) Switching repulsion to attraction: changing responses to slit during transition in mesoderm migration. Science 292: 737–740 [DOI] [PubMed] [Google Scholar]
- Lin D, Gish GD, Songyang Z, Pawson T (1999) The carboxyl terminus of B class ephrins constitutes a PDZ domain binding motif. J Biol Chem 274: 3726–3733 [DOI] [PubMed] [Google Scholar]
- Liu SJ, Cull-Candy SG (2005) Subunit interaction with PICK and GRIP controls Ca(2+) permeability of AMPARs at cerebellar synapses. Nat Neurosci 8: 768–775 [DOI] [PubMed] [Google Scholar]
- Long JF, Tochio H, Wang P, Fan JS, Sala C, Niethammer M, Sheng M, Zhang M (2003) Supramodular structure and synergistic target binding of the N-terminal tandem PDZ domains of PSD-95. J Mol Biol 327: 203–214 [DOI] [PubMed] [Google Scholar]
- Lou X, Yano H, Lee F, Chao MV, Farquhar MG (2001) GIPC and GAIP form a complex with TrkA: a putative link between G protein and receptor tyrosine kinase pathways. Mol Biol Cell 12: 615–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monea S, Jordan BA, Srivastava S, DeSouza S, Ziff EB (2006) Membrane localization of membrane type 5 matrix metalloproteinase by AMPA receptor binding protein and cleavage of cadherins. J Neurosci 26: 2300–2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien RJ, Lau LF, Huganir RL (1998) Molecular mechanisms of glutamate receptor clustering at excitatory synapses. Curr Opin Neurobiol 8: 364–369 [DOI] [PubMed] [Google Scholar]
- Peterson FC, Penkert RR, Volkman BF, Prehoda KE (2004) Cdc42 regulates the Par-6 PDZ domain through an allosteric CRIB-PDZ transition. Mol Cell 13: 665–676 [DOI] [PubMed] [Google Scholar]
- Ranganathan R, Ross EM (1997) PDZ domain proteins: scaffolds for signaling complexes. Curr Biol 7: R770–R773 [DOI] [PubMed] [Google Scholar]
- Rawlins EL, Lovegrove B, Jarman AP (2003a) Echinoid facilitates Notch pathway signaling during Drosophila neurogenesis through functional interaction with Delta. Development 130: 6475–6484 [DOI] [PubMed] [Google Scholar]
- Rawlins EL, White NM, Jarman AP (2003b) Echinoid limits R8 photoreceptor specification by inhibiting inappropriate EGF receptor signalling within R8 equivalence groups. Development 130: 3715–3724 [DOI] [PubMed] [Google Scholar]
- Ruiz-Gomez M (1998) Muscle patterning and specification in Drosophila. Int J Dev Biol 42: 283–290 [PubMed] [Google Scholar]
- Sato T, Fujita N, Yamada A, Ooshio T, Okamoto R, Irie K, Takai Y (2006) Regulation of the assembly and adhesion activity of E-cadherin by nectin and afadin for the formation of adherens junctions in Madin-Darby canine kidney cells. J Biol Chem 281: 5288–5299 [DOI] [PubMed] [Google Scholar]
- Schlieker C, Mogk A, Bukau B (2004) A PDZ switch for a cellular stress response. Cell 117: 417–419 [DOI] [PubMed] [Google Scholar]
- Schnorrer F, Dickson BJ (2004) Muscle building; mechanisms of myotube guidance and attachment site selection. Dev Cell 7: 9–20 [DOI] [PubMed] [Google Scholar]
- Soltau M, Berhorster K, Kindler S, Buck F, Richter D, Kreienkamp HJ (2004) Insulin receptor substrate of 53 kDa links postsynaptic shank to PSD-95. J Neurochem 90: 659–665 [DOI] [PubMed] [Google Scholar]
- Spencer SA, Cagan RL (2003) Echinoid is essential for regulation of Egfr signaling and R8 formation during Drosophila eye development. Development 130: 3725–3733 [DOI] [PubMed] [Google Scholar]
- Srivastava S, Osten P, Vilim FS, Khatri L, Inman G, States B, Daly C, DeSouza S, Abagyan R, Valtschanoff JG, Weinberg RJ, Ziff EB (1998) Novel anchorage of GluR2/3 to the postsynaptic density by the AMPA receptor-binding protein ABP. Neuron 21: 581–591 [DOI] [PubMed] [Google Scholar]
- Steigemann P, Molitor A, Fellert S, Jackle H, Vorbruggen G (2004) Heparan sulfate proteoglycan syndecan promotes axonal and myotube guidance by slit/robo signaling. Curr Biol 14: 225–230 [DOI] [PubMed] [Google Scholar]
- Swan LE, Wichmann C, Prange U, Schmid A, Schmidt M, Schwarz T, Ponimaskin E, Madeo F, Vorbruggen G, Sigrist SJ (2004) A glutamate receptor-interacting protein homolog organizes muscle guidance in Drosophila. Genes Dev 18: 223–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana K, Nakanishi H, Mandai K, Ozaki K, Ikeda W, Yamamoto Y, Nagafuchi A, Tsukita S, Takai Y (2000) Two cell adhesion molecules, nectin and cadherin, interact through their cytoplasmic domain-associated proteins. J Cell Biol 150: 1161–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamiya K, Kostourou V, Adams S, Jadeja S, Chalepakis G, Scambler PJ, Huganir RL, Adams RH (2004) A direct functional link between the multi-PDZ domain protein GRIP1 and the Fraser syndrome protein Fras1. Nat Genet 36: 172–177 [DOI] [PubMed] [Google Scholar]
- van Huizen R, Miller K, Chen DM, Li Y, Lai ZC, Raab RW, Stark WS, Shortridge RD, Li M (1998) Two distantly positioned PDZ domains mediate multivalent INADphospholipase C interactions essential for G protein-coupled signaling. EMBO J 17: 2285–2297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk T (1999) Singling out Drosophila tendon cells: a dialogue between two distinct cell types. Trends Genet 15: 448–453 [DOI] [PubMed] [Google Scholar]
- Volk T, VijayRaghavan K (1994) A central role for epidermal segment border cells in the induction of muscle patterning in the Drosophila embryo. Development 120: 59–70 [DOI] [PubMed] [Google Scholar]
- Wei SY, Escudero LM, Yu F, Chang LH, Chen LY, Ho YH, Lin CM, Chou CS, Chia W, Modolell J, Hsu JC (2005) Echinoid is a component of adherens junctions that cooperates with DE-Cadherin to mediate cell adhesion. Dev Cell 8: 493–504 [DOI] [PubMed] [Google Scholar]
- Wilken C, Kitzing K, Kurzbauer R, Ehrmann M, Clausen T (2004) Crystal structure of the DegS stress sensor: how a PDZ domain recognizes misfolded protein and activates a protease. Cell 117: 483–494 [DOI] [PubMed] [Google Scholar]
- Wyszynski M, Kim E, Dunah AW, Passafaro M, Valtschanoff JG, Serra-Pages C, Streuli M, Weinberg RJ, Sheng M (2002) Interaction between GRIP and liprin alpha/SYD2 is required for AMPA receptor targeting. Neuron 34: 39–52 [DOI] [PubMed] [Google Scholar]
- Wyszynski M, Valtschanoff JG, Naisbitt S, Dunah AW, Kim E, Standaert DG, Weinberg R, Sheng M (1999) Association of AMPA receptors with a subset of glutamate receptor-interacting protein in vivo. J Neurosci 19: 6528–6537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarnitzky T, Min L, Volk T (1998) An interplay between two EGF-receptor ligands, Vein and Spitz, is required for the formation of a subset of muscle precursors in Drosophila. Mech Dev 79: 73–82 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary Figure


