Abstract
In this study, we have identified a novel mitochondrial ubiquitin ligase, designated MITOL, which is localized in the mitochondrial outer membrane. MITOL possesses a Plant Homeo-Domain (PHD) motif responsible for E3 ubiquitin ligase activity and predicted four-transmembrane domains. MITOL displayed a rapid degradation by autoubiquitination activity in a PHD-dependent manner. HeLa cells stably expressing a MITOL mutant lacking ubiquitin ligase activity or MITOL-deficient cells by small interfering RNA showed an aberrant mitochondrial morphology such as fragmentation, suggesting the enhancement of mitochondrial fission by MITOL dysfunction. Indeed, a dominant-negative expression of Drp1 mutant blocked mitochondrial fragmentation induced by MITOL depletion. We found that MITOL associated with and ubiquitinated mitochondrial fission protein hFis1 and Drp1. Pulse–chase experiment showed that MITOL overexpression increased turnover of these fission proteins. In addition, overexpression phenotype of hFis1 could be reverted by MITOL co-overexpression. Our finding indicates that MITOL plays a critical role in mitochondrial dynamics through the control of mitochondrial fission proteins.
Keywords: E3 ubiquitin ligase, mitochondria, mitochondrial dynamics, mitochondrial fission
Introduction
Ubiquitination is a post-translational modification in which a small conserved peptide, ubiquitin, is appended to target proteins in the cell through a series of complex enzymatic reactions (Hershko and Ciechanover, 1992). The ubiquitin–proteasome system plays an important role in controlling the levels of various cellular proteins and regulates basic cellular processes such as cell cycle progression, signal transduction, cell transformation and immune recognition (Hershko and Ciechanover, 1998). This system is also involved in the protein quality control, which maintains the health of the cell (Sitia and Braakman, 2003).
E3 ubiquitin ligases are a large family of proteins that can be classified into three major structurally distinct types: N-end rule E3, E3 containing the HECT (Homology to E6AP C-Terminus) domain and E3 with the RING (Really Interesting New Gene) finger, including its derivatives, the U-Box and the PHD (Plant Homeo-Domain) (Jackson et al, 2000; Pickart, 2001; Patterson, 2002). PHD constitutes a widely distributed subfamily of zinc fingers and had been initially believed to be involved in chromatin-mediated transcriptional regulation. But recent studies have demonstrated that various proteins containing PHD function as E3 ubiquitin ligases including MEKK1 and Doa10 (Coscoy and Ganem, 2003). Subsequently, several PHD-containing viral proteins have been identified that promote immune evasion by downregulating proteins that govern immune recognition (Ishido et al, 2000a). For example, Kaposi's sarcoma-associated herpesvirus encodes two proteins, MIR (modulator of immune recognition) 1 and 2, which have been shown to contain PHD and function as E3 ubiquitin ligases for immune recognition-related molecules such as MHC class I (Ishido et al, 2000b; Means et al, 2002). Thus, these viral regulators lead to ubiquitination of their targets by functioning as E3 ubiquitin ligases that require the PHD motif. We have recently demonstrated that human genome also encodes a novel PHD-containing protein that functions as an E3 ubiquitin ligase designated as c-MIR (Goto et al, 2003). Furthermore, in a genome screen for related mammalian proteins containing PHD like c-MIR, we discovered a novel membrane-bound E3 ubiquitin ligase designated MITOL, which is specifically localized in mitochondria.
Mitochondria are the center of cellular energy production and essential metabolic reactions, and are involved in the induction of apoptosis by release of cytochrome c. Mitochondria are remarkably dynamic organelles. Time-lapse microscopy of living cells reveals that mitochondria undergo constant migration and morphological changes. The mitochondrial fission and fusion machinery plays an essential role in the dynamics, division, distribution and morphology of the organelle (Yaffe, 1999; Jensen et al, 2000; Griparic and van der Bliek, 2001; Shaw and Nunnari, 2002). Evolutionarily conserved large GTPases Dnm1/Drp1/Dlp1 and Fzo1/mitofusin are involved in mitochondrial fission and fusion, respectively (Hales and Fuller, 1997; Hermann et al, 1998; Labrousse et al, 1999; Sesaki and Jensen, 1999). Mammalian protein hFis1 is the orthologue of the yeast Fis1p known to participate in yeast mitochondrial division (James et al, 2003). However, their regulatory mechanisms are largely unknown.
In this study, we demonstrate that MITOL is involved in the regulation of mitochondrial dynamics through the downregulation of mitochondrial fission proteins. We discuss a crucial role of MITOL in mitochondrial biology.
Results
MITOL is a novel mitochondria-specific E3 ubiquitin ligase
In a human genome screening for MIR family, we identified a novel mitochondrial ubiquitin ligase and named it MITOL. Figure 1A shows the amino-acid sequence of MITOL. MITOL contains a PHD motif at its N-terminus and predicted four-transmembrane domains. We first examined whether PHD in MITOL exhibited E3 ubiquitin ligase activity. For this purpose, two constructs, GST-fused PHD (2–72 amino acids of MITOL) and GST-fused PHD mutant (CS mutant; C65S, C68S), which lacks Zn-binding ability, were generated. In vitro ubiquitination assay using lysate of rabbit reticulocytes containing E1 and E2s revealed a clear polyubiquitin signal in GST-PHD but not in GST or GST-PHD CS mutant (Figure 1B). This indicated that PHD in MITOL has an E3 ubiquitin ligase activity. Northern blot analysis showed the ubiquitous expression of MITOL in various human tissues (Figure 1C). To ascertain expression of MITOL a specific anti-MITOL antibody was raised against a peptide corresponding to amino acids 257–279 of MITOL. Western blot analysis with anti-MITOL antibody demonstrated the expression of MITOL as an ≈30 kDa band in both COS-7 and HeLa cells, but not with control, preimmune serum (Figure 1D). In addition, an enhanced MITOL signal in MITOL expression vector-transfected cells verified the specificity of this antibody and the position of MITOL protein band.
Figure 1.
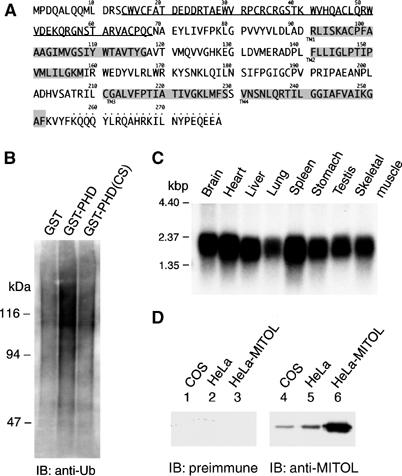
Identification of MITOL, a novel membrane-bound E3 ubiquitin ligase. (A) Amino-acid sequence of human MITOL. PHD motif and predicted four-transmembrane domains are shown by an underline and shadow lines, respectively. Anti-MITOL antibody was raised against the peptide indicated by dots above the amino acids. (B) E3 ubiquitin ligase activity in GST-fused MITOL-PHD, but not CS mutant (C65S, C68S). In vitro ubiquitin ligase assay using purified GST, GST-PHD and GST-PHD CS mutants was performed as described in Materials and methods. (C) Northern blot analysis of MITOL in various human tissues. A full-length human MITOL cDNA was used as a probe. (D) Expression of MITOL protein in COS-7 and HeLa cell lines. Lysates of COS-7, HeLa and MITOL expression vector-transfected HeLa cells were immunoblotted with control preimmune or immune serum containing anti-MITOL antibody.
In order to examine subcellular distribution of MITOL in HeLa cells, immunohistochemical analysis using anti-MITOL antibody was performed and is shown in Figure 2A (upper panels). Endogenous MITOL was colocalized with mitochondrial marker MitoTracker, suggesting that MITOL is a mitochondrial protein. Likewise, HeLa cells expressing Myc-tagged MITOL were immunostained with anti-Myc antibody and MitoTracker. Myc-tagged MITOL was also colocalized with mitochondria (Figure 2A, lower panels). To confirm the mitochondrial localization of MITOL, cytosolic and mitochondrial fractions of normal HeLa or HeLa cells expressing Myc-tagged MITOL were isolated and MITOL localization was determined by immunoblotting using anti-MITOL (Figure 2B, left) or anti-Myc antibodies (Figure 2B, right). As expected, MITOL was localized in the mitochondrial fraction. As the mitochondrial fraction is known to contain a part of endoplasmic reticulum (ER) proteins, mitochondrial and ER-rich fractions were separated and MITOL localization was examined using anti-PDI (ER marker) and anti-Tom20 antibodies (mitochondria marker). As shown in Figure 2C, MITOL was not detected in the ER-rich fraction, indicating the specific localization of MITOL in mitochondria. Carbonate extraction assay showed that MITOL behaved like an integral membrane protein. This result was consistent with predicted four-transmembrane structure (Figure 2D). We next examined whether MITOL localized at the outer or inner membrane of mitochondria. Purified intact mitochondria prepared from HeLa cells were treated with trypsin, which attacked the cytosolic domain of the outer membrane proteins. Treatment with trypsin did not affect the inner membrane protein Tim23, whereas it degraded MITOL as well as the outer membrane protein Tom20 (Figure 2E). Mitochondrial outer membrane proteins were removed from mitochondria by nonionic detergent digitonin treatment. The digitonin solubility of MITOL was consistent with that of Tom20 but not Tim23 (Figure 2E). To determine the topology of the N-terminus, trypsin protection assay on mitochondria isolated from cells expressing an N-terminally HA-tagged MITOL was performed. As shown in Figure 2E (right panel), the N-terminal PHD domain rapidly disappeared from mitochondria by trypsin treatment. These results suggested that MITOL was localized at mitochondrial outer membranes and the N-terminal PHD domain is exposed to the cytoplasm.
Figure 2.
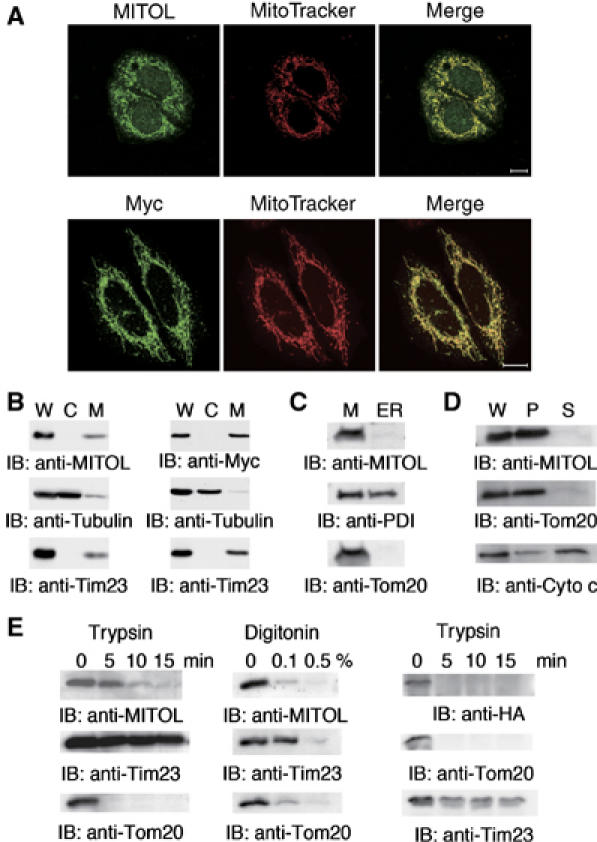
Specific localization of MITOL in the outer mitochondrial membrane. (A) Colocalization of MITOL with mitochondria. (Upper panels) HeLa cells were immunostained with anti-MITOL (green) antibody and MitoTracker (red). (Lower panels) HeLa cells expressing Myc-tagged MITOL were immunostained with anti-Myc antibody (green) and MitoTracker (red). (B) Subcellular fractionation indicates the localization of MITOL in mitochondria. Whole lysates (W) and cytosolic (C) and mitochondrial fractions (M) isolated from HeLa (left) or HeLa cells expressing Myc-MITOL (right) were immunoblotted with anti-MITOL, anti-Myc, anti-tubulin or anti-Tim23 mitochondria marker antibodies. (C) MITOL is not localized in ER. The mitochondrial fraction (M) and the ER-rich fraction (ER) isolated from HeLa cells were immunoblotted with anti-MITOL, anti-PDI (ER marker) or anti-Tom20 (mitochondria marker) antibodies. (D) Carbonate extraction assay indicates that MITOL is an integral membrane protein. Whole lysates (W), supernatant (S; peripheral protein) and pellet (P; integral protein) fractions after carbonate extraction from isolated mitochondria were immunoblotted with anti-MITOL, anti-Tom20 or anti-cytochrome c antibodies. (E) Sensitivity of MITOL to trypsin or digitonin treatment indicates MITOL localization in the mitochondrial outer membrane. Purified mitochondria from HeLa cells (left and middle panels) or cells expressing an N-terminally HA-tagged MITOL (right panel) were treated with trypsin or digitonin, and disappearance of MITOL along with mitochondrial markers was monitored by immunoblotting with anti-MITOL, anti-HA, anti-Tim23 or anti-Tom20 antibodies.
Rapid degradation of MITOL by autoubiquitination activity
To understand the metabolic steady-state level of MITOL expression, the effect of protein synthesis inhibitor cycloheximide (CHX) on MITOL expression was examined. As shown in Figure 3A (left panel), CHX treatment downregulated MITOL expression and this was blocked by addition of proteasome inhibitor MG132. A similar result was obtained in HeLa cells expressing FLAG-MITOL (Figure 3A, right panel). The half-life of FLAG-MITOL was about 60 min and it became difficult to detect the MITOL protein band after 3 h treatment with CHX. On the other hand, treatment with MG132 blocked the CHX-induced downregulation of MITOL (lane 7). Equal amount of tubulin staining demonstrated that the same amount of protein was loaded in each lane. Compared to FLAG-MITOL, endogenous MITOL showed a slow degradation after CHX treatment, suggesting that endogenous MITOL may be regulated by an unknown modification.
Figure 3.
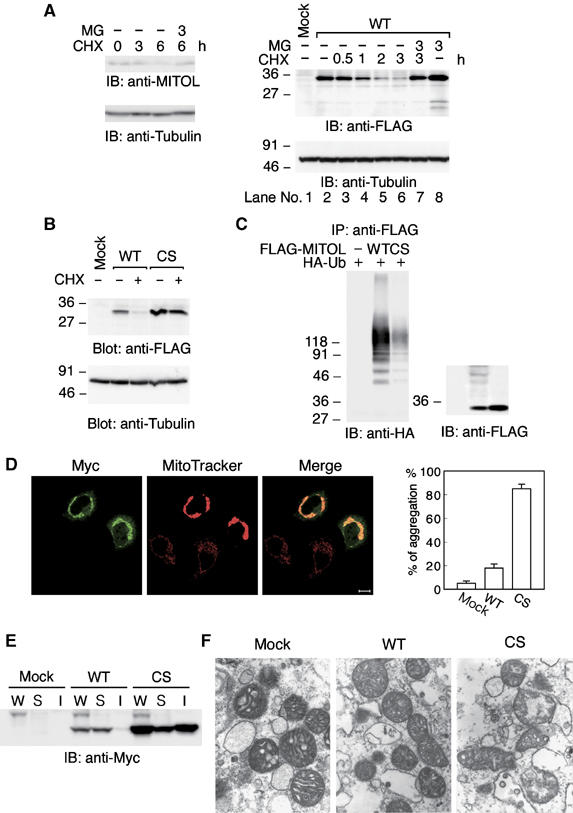
Rapid degradation of MITOL by autoubiquitination activity. (A) Effect of protein synthesis inhibitor CHX on MITOL protein expression. HeLa cells (left panel) or HeLa cells expressing FLAG-MITOL WT (right panel) were treated with or without CHX (10 μg/ml) for indicated times in the presence or absence of MG132 (50 μM) and cell lysates were immunoblotted with anti-MITOL, anti-FLAG or anti-tubulin antibodies. (B) No degradation of MITOL CS mutant. HeLa cells or HeLa cells expressing FLAG-MITOL WT or FLAG-MITOL CS mutant were treated with or without CHX (10 μg/ml) for 3 h and immunoblotted with anti-FLAG or anti-tubulin antibodies. (C) Polyubiquitination of MITOL WT but not MITOL CS mutant. Lysates of HeLa cells cotransfected with control vector, FLAG-MITOL WT or FLAG-MITOL CS mutant with HA-tagged ubiquitin (HA-Ub) were immunoprecipitated with anti-FLAG antibody agarose beads and immunoblotted with anti-HA or anti-FLAG antibodies. (D) Mitochondrial aggregation by MITOL CS mutant. HeLa cells expressing Myc-MITOL WT and CS mutant were stained with anti-Myc antibody and MitoTracker. Bar, 10 μm. Mitochondrial aggregation induced by MITOL WT or CS mutant was measured by counting at least 100 cells (right panel). Error bars represent s.d. n=3. (E) Accumulation of MITOL CS mutant in the insoluble fraction. Whole lysates (W) and soluble (S) and insoluble (I) fractions were isolated from HeLa cells or HeLa cells expressing Myc-MITOL WT or CS mutants and immunoblotted with anti-Myc antibody. Bars, 10 μm. (F) Electron microscopic analysis indicated severe mitochondrial damage in MITOL CS mutant-overexpressing cells. Mitochondrial morphology in control vector, MITOL WT or CS mutant-transfected cells was examined by electron microscopic analysis at 27 600-fold magnification.
To determine whether ubiquitin ligase activity in MITOL-PHD was required for this degradation of MITOL, the effect of CHX treatment on the degradation of CS mutant was similarly studied. Figure 3B shows that CHX treatment failed to degrade the CS mutant. This suggested that the degradation of MITOL was PHD-dependent. We therefore investigated whether MITOL exhibited autoubiquitination activity. Immunoprecipitation of FLAG-MITOL with anti-FLAG antibody from HeLa cells coexpressing FLAG-MITOL and HA-ubiquitin revealed significant polyubiquitination of FLAG-MITOL wild type (WT) but not CS mutant (Figure 3C). This result indicated that MITOL has an autoubiquitination activity. Taken together, these results suggested that MITOL strictly controls its protein expression level by rapid degradation of MITOL through the PHD-dependent autoubiquitination activity.
To investigate the physiological relevance of this rapid turnover of MITOL by autoubiquitination activity, the effects of CS mutant on mitochondrial function and morphology were examined using a transient expression system. We found that compared to MITOL WT, CS mutant significantly induced mitochondrial aggregation (Figure 3D). Statistical analysis showed that over 80% of CS mutant-expressing mitochondria formed aggregates, whereas only 10–20% of WT-expressing mitochondria were aggregated. Western blot analysis indicated an accumulation of CS mutant but not WT in the insoluble fraction of mitochondria (Figure 3E). Electron microscopic analysis revealed the mitochondrial swelling and collapse in CS mutant-expressing cells (Figure 3F). This phenomenon was not observed in WT-expressing mitochondria. This mitochondrial dysfunction may be caused by the accumulation of insoluble or unfolded CS mutant. Taken together, these results suggested that ubiquitin ligase activity of MITOL was necessary for inhibiting accumulation of degenerated MITOL, including other proteins, in mitochondria.
Requirement of MITOL for mitochondrial dynamics through interaction with and ubiquitination of mitochondrial fission factors
To assess the role of MITOL in mitochondria, we established several HeLa cell lines stably expressing FLAG-MITOL WT or FLAG-MITOL CS mutant. We found that the stable expression level of FLAG-MITOL WT or CS mutant was greatly lower than its transient expression (Figure 4A, right panel). This may be owing to the downregulation of FLAG-MITOL through an unknown mechanism to protect cells from toxicity, such as mitochondrial aggregation induced by overexpression of FLAG-MITOL WT or CS mutant. Indeed, no significant mitochondrial aggregation was observed in stable clones of HeLa cell line expressing FLAG-MITOL WT or CS mutant. Instead, HeLa cell lines expressing CS mutant (but not WT) frequently showed an abnormal mitochondrial morphology, such as fragmentation (Figure 4A, left panel). About 30–50% of mitochondria in CS mutant cell line revealed mitochondrial fragmentation. Other two CS mutants also showed a similar mitochondrial fragmentation. To verify this phenomenon, the effects of MITOL knockdown on mitochondrial morphology were examined. HeLa cells transfected with scramble small interfering RNA (siRNA) as control or two siRNAs against MITOL were incubated for 48 h and the expression level of endogenous MITOL was monitored by immunoblot analysis probed with anti-MITOL antibody. As shown in Figure 4B, both the siRNAs against MITOL, but not scramble, reduced MITOL expression by about 70–90%. Using this system, we compared the mitochondrial morphology between control and MITOL-deficient mitochondria and found an increase in mitochondrial fragmentation in MITOL-deficient mitochondria. Statistical analysis showed that over 50% of mitochondria were fragmented in cells transfected with siRNA1 or siRNA2. We generated other three siRNAs against MITOL and found that cells transfected with these three siRNAs showed a similar mitochondrial fragmentation (not shown). This result suggested the enhancement of mitochondrial fission by MITOL depletion. To determine whether mitochondrial fragmentation caused by overexpression of siRNA-mediated depletion of endogenous MITOL is specific to hFis1/Drp1-dependent fission, we examined the effect of dominant-negative Drp1 GTPase-deficient mutant (Drp1K38A) expression on the MITOL-deficient mitochondrial morphology. As shown in Figure 4C, after cotransfection of siRNA and Drp1K38A, the dominant-negative Drp1 mutant blocked mitochondrial fragmentation by siRNA1-mediated MITOL depletion. Statistical analysis demonstrated that Drp1 mutant induced mitochondrial elongation in over 90% of MITOL-depleted cells. These results indicated that MITOL was required for normal mitochondrial morphology and was involved in mitochondrial dynamics probably through the regulation of mitochondrial fission.
Figure 4.
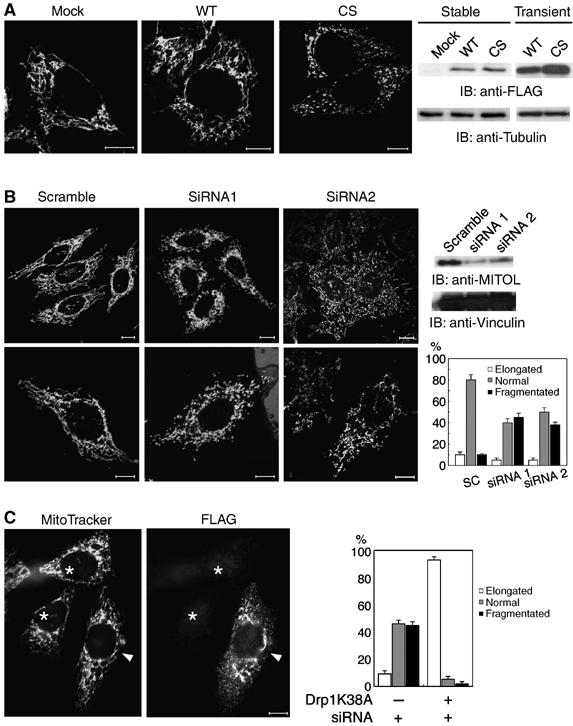
Critical role of MITOL in mitochondrial morphology. (A) Mitochondrial fragmentation in HeLa cell lines stably expressing FLAG-MITOL CS mutant. HeLa cell lines stably expressing control vector, FLAG-MITOL WT or CS mutant were stained with MitoTracker and mitochondrial morphologies were compared. The expression levels of FLAG-MITOL after stable and transient transfection relative to mock-transfected cells were shown by immunoblotting using anti-FLAG antibody (right panel). Bars, 10 μm. (B) Aberrant mitochondrial morphology in MITOL-deficient cells. HeLa cells transfected with scramble, MITOL siRNA1 or siRNA2 were immunoblotted with anti-FLAG and anti-vinculin antibodies (right) or stained with MitoTracker (left). Bars, 10 μm. Percentages of cells showing each mitochondrial morphology were calculated from 100 scramble- or MITOL siRNA-transfected cells. Error bars represent s.d. n=3. (C) Dominant-negative Drp1 mutant blocked mitochondrial fragmentation by MITOL depletion. HeLa cells transfected with FLAG-Drp1 mutant (K38A) and siRNA1 were stained with MitoTracker and anti-FLAG antibody. An arrowhead and asterisks indicate siRNA1-mediated MITOL-depleted cells with or without Drp1 mutant expression, respectively. Bars, 10 μm. Percentages of cells showing each mitochondrial morphology were calculated from 100 MITOL-depleted cells with or without Drp1 mutant expression. Error bars represent s.d. n=3.
Experiments with siRNA-mediated knockdown of MITOL suggested that mitochondrial fission factors such as hFis1 or Drp1 may be substrate for MITOL. To explore this possibility, we investigated whether MITOL could ubiquitinate hFis1. As shown in Figure 5, MITOL WT, but not CS mutant, was found to ubiquitinate hFis1. In addition, co-immunoprecipitation assay revealed that MITOL associated with hFis1 (Figure 5B). To examine the role of MITOL in the control of endogenous hFis1, adenovirus-based constructs expressing MITOL WT and CS mutant were generated. Compared to cells expressing LacZ as control, endogenous hFis1 was decreased in cells expressing MITOL WT. On the other hand, an increase in endogenous hFis1 was detected in cells expressing MITOL CS or MITOL-deficient cells by siRNA1 (Figure 5C). CHX treatment suggested the delayed degradation of endogenous hFis1 in MITOL-deficient cells. To further demonstrate MITOL-dependent turnover of hFis1, pulse–chase experiment was performed. As shown in Figure 5D and E, MITOL overexpression promoted hFis1 turnover and this effect was inhibited by MG132 treatment. Immunoblot analysis probed with anti-FLAG antibody indicated that an equal amount of hFis1 was immunoprecipitated (not shown). Thus, MITOL may control the protein expression level of hFis1 through the ubiquitin–proteasome pathway. Indeed, overexpression of hFis1 causes mitochondrial fragmentation (Figure 6A). Therefore, we next tested whether overexpression phenotype of hFis1 can be reverted by co-overexpression of MITOL. Expectedly, co-overexpression of MITOL WT, but not CS mutant, blocked hFis1-induced mitochondrial fragmentation (Figure 6B and C). Coexpression of MITOL CS mutant induced mitochondrial aggregation consisting of fragmented mitochondria.
Figure 5.
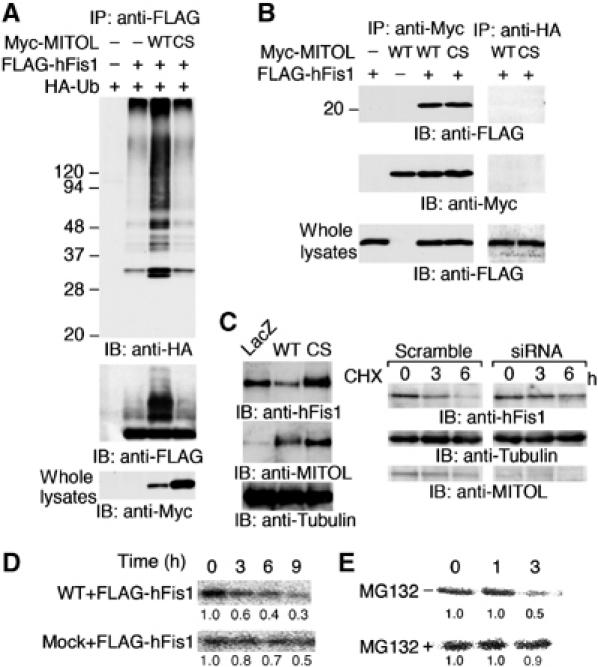
MITOL associates with and ubiquitinates mitochondrial fission protein hFis1. (A) Ubiquitination of hFis1 by MITOL. Lysates of HeLa cells transfected with indicated vectors were immunoprecipitated with anti-FLAG antibody and immunoprecipitates were immunoblotted with anti-HA or anti-FLAG antibodies. Whole lysates were immunoblotted with anti-Myc antibody. (B) Interaction of MITOL with hFis1. Lysates of HeLa cells transfected with indicated vectors were immunoprecipitated with anti-Myc antibody and immunoprecipitates were immunoblotted with anti-FLAG or anti-Myc antibodies. To demonstrate the specificity of the co-immunoprecipitation, anti-HA antibody was used as a negative control. Whole lysates were immunoblotted with anti-FLAG antibody to confirm the expression of FLAG-hFis1. (C) Accumulation of endogenous hFis1 by MITOL dysfunction. Lysates of HeLa cells infected with indicated adenovirus vectors were immunoblotted with anti-hFis1, anti-MITOL or anti-tubulin antibodies (left panel). Lysates of HeLa cells transfected with scramble or MITOL siRNA1 were treated with CHX (10 μg/ml) for indicated times and immunoblotted with anti-hFis1, anti-tubulin or anti-MITOL antibodies (right panel). (D) MITOL expression caused a rapid turnover of hFis1. Pulse–chase experiment was performed. HeLa cells expressing FLAG-hFis1 or FLAG-hFis1/MITOL WT were labeled for 60 min with [35S]Met/Cys labeling mixture and chased in complete DMEM for indicated periods. (E) Effect of MG132 on hFis1 turnover promoted by MITOL overexpression. HeLa cells expressing FLAG-hFis1/MITOL WT in the absence or presence of MG132 were similarly chased for indicated periods. Cells were lysed and immunoprecipitated with anti-FLAG antibody and immunoprecipitated FLAG-hFis1 was visualized by autoradiography. The result (D or E) is representative of three independent experiments. Relative values estimated by NIH image analysis are indicated below.
Figure 6.
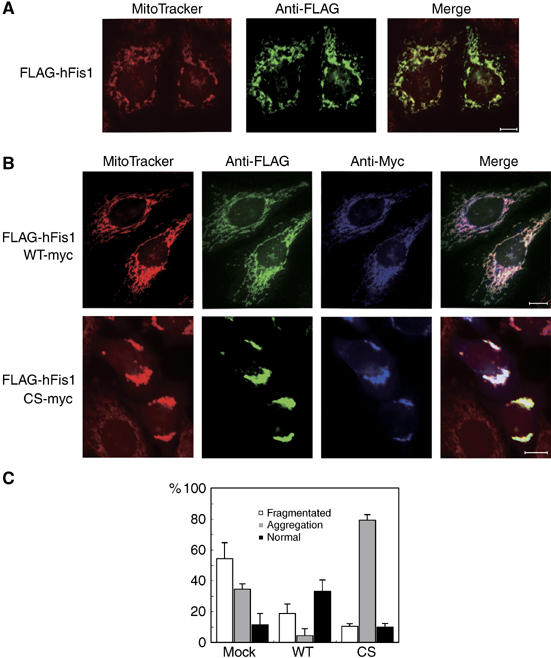
Rescue of hFis1 expression phenotype by MITOL coexpression. (A) Mitochondrial fragmentation by hFis1 expression. HeLa cells expressing FLAG-hFis1 were stained with MitoTracker and anti-FLAG antibody. Bars, 10 μm. (B) Coexpression of MITOL WT, but not CS mutant, blocked hFis1-induced mitochondrial fragmentation. HeLa cells coexpressing Myc-MITOL WT or CS mutant with FLAG-hFis1 were stained with MitoTracker, anti-FLAG and anti-Myc antibodies. Bars, 10 μm. (C) Percentages of cells showing each mitochondrial morphology were calculated from 100 hFis1, hFis1/MITOL WT or hFis1/MITOL CS coexpressing cells. Error bars represent s.d. n=3.
Association of Drp1 with MITOL and MITOL-dependent ubiquitination were similarly observed as shown in Supplementary data 1. Pulse–chase experiment also indicated that MITOL overexpression promoted Drp1 turnover. However, ubiquitination of Drp1 by MITOL was lower than that of hFis1 and expression level of endogenous Drp1 was not affected by adenovirus-mediated expression of MITOL WT (not shown). This may be due to the cytosolic distribution of Drp1 compared to mitochondrial membrane localization of hFis1. Alternatively, hFis1 may be better substrate than Drp1 for MITOL. Taken together, these results suggest that MITOL is involved in mitochondrial dynamics through the control of hFis1 and Drp1 by direct interaction and ubiquitination.
Discussion
In this study, we identified a novel membrane-bound ubiquitin ligase designated MITOL, which is specifically localized in mitochondria. As no mitochondria-specific ubiquitin ligase has been reported to date, the discovery of MITOL is a first indication that mitochondria have a putative ubiquitination system. MITOL appeared to be identical to March-V whose function has not been reported except for a description of its ER localization (Bartee et al, 2004). In their paper, characterization of March-V localization is very poor and our results indicated that their description about March-V is not correct. Here, we have demonstrated that MITOL is involved in mitochondrial dynamics. A new family of proteins containing PHD motif has been shown to constitute a widely distributed subfamily of zinc-finger proteins that function as E3 ubiquitin ligases. Several PHD-containing viral proteins have been identified that promote immune evasion by downregulating proteins that govern immune recognition (Ishido et al, 2000a). Therefore, this family has a common feature to promote or help the survival of certain virus or organelles such as mitochondria in the host cells.
In eukaryotes, mitochondria are dynamic mobile organelles varying in number and shape. The equilibrium between opposing fission and fusion is important for the maintenance of mitochondrial morphologies (Yaffe, 1999; Shaw and Nunnari, 2002). In mammalian cells, the outer membrane protein Fzo1/mitofusin (Chen et al, 2003) and the inner membrane protein OPA1 play a role in mitochondrial fusion (Misaka et al, 2002), whereas the cytosolic protein Drp1 and the outer membrane protein hFis1 are involved in mitochondrial fission (Smirnova et al, 2001; James et al, 2003). However, the regulation of the dynamic balance between fission and fusion is largely unknown so far. In yeast, Mdm30, an E3 ubiquitin ligase containing an F-box motif, has been reported to regulate the protein level of Fzo1 (Fritz et al, 2003). On the other hand, sumoylation of Drp1 elevates its steady-state level at mitochondria and accelerates mitochondrial fragmentation (Harder et al, 2004). Thus, one model to explain how to coordinate the balance between fusion and fission is the control of protein level of these factors involved in fusion and fission by the modification of sumoylation and ubiquitination. Indeed, MITOL-deficient HeLa cells displayed a number of fragmented or very short filamentous mitochondria (Figure 4). Cotransfection and pulse–chase experiments indicated that hFis1 and Drp1 were associated with and ubiquitinated by MITOL. Therefore, MITOL may be closely involved in mitochondrial dynamics via the control of hFis1 and Drp1 protein level through the ubiquitin–proteasome pathway. As mitochondrial fusion factors, as well as fission factors, may also be regulated by MITOL, the relationship between MITOL and fusion factors should be further investigated.
E3 ubiquitin ligases are distributed widely in the cells, including the plasma membrane, cytoplasm, Golgi and ER. In particular, membrane-bound ubiquitin ligases localized at ER have been demonstrated to be involved in the protein quality control through the interaction with chaperone (Sitia and Braakman, 2003). In mitochondria, unfolded proteins have been believed to be degraded by ATP-dependent proteases localized in each compartment in mitochondria (Langer et al, 2001; Arnold and Langer, 2002). However, several mitochondrial proteins such as Tom20 and prohibitin have been reported to be ubiquitinated (Wright et al, 2001; Thompson et al, 2003), suggesting the existence of a specific ubiquitin–proteasome system in mitochondria. As we identified a chaperone as one of MITOL-interacting proteins, it is possible that MITOL interacts with and ubiquitinates unfolded mitochondrial proteins. Mitochondria may possess a putative ubiquitination pathway as a protein quality control system. We are now investigating the role of MITOL in mitochondria quality control.
In our preliminary observation, analysis of time-lapse imaging suggested the severe defect in mitochondrial movement in siRNA-mediated MITOL-depleted cells. In these cells, mitochondria stayed at the same position and often revealed a perinuclear aggregation. Interestingly, one of MITOL-interacting proteins is closely related with motor protein. Thus, MITOL may be involved in the microtubule-based motility of mitochondria. We are currently trying to generate transgenic mice expressing MITOL WT or CS mutant. We have already succeeded in generating transgenic mice overexpressing MITOL WT, but not CS mutant, suggesting that the transgenic mice expressing MITOL CS mutant may be nonviable. Further study would be necessary for understanding the function of MITOL.
Materials and methods
Database searches and cloning of human MITOL cDNA
For the cloning of cDNA coding human MITOL, total RNA isolated from BJAB cells was reverse-transcribed using the Superscript RT kit (Invitrogen) according to the manufacturer's protocol. To determine the 5′ and 3′ ends of the entire coding sequence of MITOL cDNA, 5′ and 3′ rapid amplification of cDNA ends was performed using a GeneRacer Kit (Invitrogen). A full-length cDNA of MITOL was obtained by PCR and subcloned into the pEF-1 (Invitrogen) and pAAV IRES-EGFP vectors and sequenced using a model 310 DNA sequencer (Applied Biosystems). To introduce C-terminal FLAG and Myc epitope tags, MITOL cDNAs were amplified by PCR using adequate primers and subcloned into pApuro vectors. The putative secondary structure and transmembrane topology of MITOL were examined by profile fed neural network system from Heidelberg (PHD) and sosui program. Point mutations of Cys65 and Cys68 of MITOL cDNA to Ser were generated by the site-directed mutagenesis kit (Stratagene).
Detection of MITOL mRNA in human tissues
Poly(A)+RNA blots containing 1 μg of poly(A)+RNA per lane from mouse NorthernLIGHT Blots (Panomics) were hybridized with the 32P-labeled full-length mouse MITOL cDNA. Hybridization was carried out according to the manufacturer's protocol. After washing, the membrane was exposed to X-ray film at −70°C using an intensifying screen (Inatome et al, 2000).
Cell culture and transfection
HeLa and COS-7 cells were grown in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and 100 U/ml penicillin at 5% CO2 at 37°C. For transient assays, expression plasmids were transfected using lipofectamine plus (Invitrogen) according to the manufacturer's instructions.
Small interfering RNA
For RNAi assay, sense and antisense oligonucleotides corresponding to the following target sequences were purchased from Dharmacon: 5′-GCTCTATCTATTGGACAG-3′ (No. 1), 5′-TCTTGGGTGGAATTGCGTT-3′ (No. 2). Scramble oligonucleotide was used as a negative control. The annealed siRNA duplex was transfected into HeLa cells using Lipofectamine 2000 (Invitrogen).
Antibodies
Antibody directed against MITOL was produced by immunizing rabbits with a synthetic peptide, GCKQQQYLRQAHRKILNYPEQEEA, corresponding to amino acids 257–279 of MITOL. Anti-FLAG (M2), anti-α-tubulin and anti-vinculin antibodies were from Sigma. Anti-c-Myc antibody was from Roche. Anti-HA antibody was from Babco. Anti-ubiquitin (P4D1) antibody was from Santa Cruz. The mouse monoclonal antibodies against cytochrome c, Tom20, Tim23 and PDI were purchased from BD Biosciences.
Immunofluorescence microscopy
Cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 1 h at 37°C, then washed twice with 0.2% Tween 20 in PBS, permeabilized with 0.2% Triton X-100 in PBS for 10 min, washed four times with PBS and blocked with 3% bovine serum albumin in PBS, all at room temperature. For double staining, the cells were incubated with appropriate primary antibodies for 1 h at room temperature, washed three times with 0.5% Triton X-100 in PBS and then with appropriate secondary antibodies (Alexa Fluor goat anti-rabbit IgG, Alexa Fluor goat anti-mouse IgG) for 30 min. To visualize the mitochondria, 100 nM MitoTracker (Molecular Probes) was added and incubated for 30 min. The samples were washed as before, mounted using PermaFluor (Immunon) and analyzed using Zeiss LSM510 confocal laser scanning microscope (Hotta et al, 2005).
Electron microscopic analysis
HeLa cells expressing MITOL WT or CS mutant were fixed with 2.5% glutaraldehyde in 0.1 M sodium phosphate buffer (pH 7.4). They were postfixed with 2% OsO4 in the same buffer. After dehydration with a graded series of ethanol, they were substituted by propylene oxide and embedded in Epoxy resin. Silver to gold thin sections were doubly stained with uranyl acetate and lead citrate, and examined with an H-7000 transmission electron microscope (Hitachi, Tokyo, Japan) and photographed at various magnifications.
Immunoprecipitation and immunoblotting
Preparation of cell lysates, immunoprecipitation and immunoblotting were performed as described previously (Kyo et al, 2003). To investigate the ubiquitination of proteins, cells were solubilized in lysis buffer (1% Triton X-100, 50 mM Tris, pH 7.4, 150 mM NaCl, 10 mM EDTA, 1 mM PMSF, 2 μg/ml aprotinin) to dissociate protein complexes. Total cell lysates were prepared by the direct addition of 1 × SDS sample buffer to the monolayers. Immunoprecipitates and total cell lysates were separated by SDS–PAGE and transferred to the PVDF membrane (Millipore) (Mitsui et al, 2002). The blots were probed with the indicated antibodies. In all blots, proteins were visualized by the enhanced chemiluminescence reagent (Western Lightning; PerkinElmer LifeScience).
Plasmid construction
Drp1 cDNA with C-terminal FLAG epitope tag and hFis1 with N-terminus were obtained from total RNA of HeLa cells by RT–PCR and subcloned into pCMV5 expression vector. Point mutations of Lys38 of Drp1 cDNA to Ala were generated by the site-directed mutagenesis. To construct plasmid DNAs for GST fusion proteins, a fragment encoding MITOL PHD domain was amplified by PCR and then subcloned into pGEX 4T-3 vector (Amersham Biosciences).
Subcellular fractionation
Isolation of mitochondria was performed using a mitochondrial fractionation kit (Active Motif). The supernatant and pellets collected were used as ER and mitochondrial fractions, respectively. Protein concentration was measured using a Bio-Rad protein assay (Bio-Rad) with BSA as a standard.
Carbonate extraction assay
To confirm whether MIOL is peripheral protein or integral protein, we performed carbonate extraction. Mitochondrial suspensions in isotonic buffer (250 mM sucrose, 2.5 mM Hepes pH 7.5) were added with an equal volume of 200 mM Na2CO3 (pH 11.5) (final 100 mM) and incubated on ice for 30 min. The suspensions were centrifuged at 144 000 g at 4°C for 1 h. The supernatant (peripheral protein fraction) and the pellets (integral protein fraction) collected were diluted with 100 mM Na2CO3 in isotonic buffer.
Pulse–chase analysis
HeLa cells were incubated for 1 h with Met/Cys-free DMEM followed by addition of 500 μCi 35S-labeled Met/Cys mixture for 1 h. Following labeling, cells were washed in PBS and incubated in complete medium without radioactive amino acids for the indicated times. Cells were washed in PBS, lysed and equal protein amounts subjected to immunoprecipitation with anti-FLAG M2 agarose (Sigma).
Ubiquitination assay
For in vitro autoubiquitination assay, GST fusion proteins were produced as follows. Expression of GST fusion proteins was induced by 0.1 mM isopropyl-1-D-thio-galactopyranoside for 3–6 h. Bacterial pellets were sonicated in 1% Triton X-100 lysis buffer containing protease inhibitors. After being cleared by centrifugation, bacterial lysates were incubated with glutathione-Sepharose beads (Amersham Biosciences). A 10 μg portion of precipitated GST fusion protein was washed three times with lysis buffer, once with ubiquitination buffer (40 mM Tris–HCl, pH 7.5, 5 mM MgCl2, 2 mM ATP, 2 μM dithiothreitol, 25 μM MG132) and incubated in 150 μl of the same buffer supplemented with rabbit reticulocytes lysate and 40 μg of His-tagged ubiquitin (Calbiochem) for 2 h at 30°C. After in vitro ubiquitination, all samples were washed four times with lysis buffer, eluted with SDS-sample buffer and subjected to immunoblot analysis by probing with anti-ubiquitin (P4D1) antibody.
Adenovirus-based vector construction
The adenovirus-based MITOL vectors were generated by the ViraPower Adenoviral Gateway Expression (Invitrogen). 293T cells were transfected with the recombinant adenoviral plasmid using lipofectin (Invitrogen), and adenovirus with the titer of 1010–1012 PFU/ml was collected.
Digitonin/trypsin treatment
Isolated mitochondria were diluted with mitochondrial buffer (70 mM sucrose, 220 mM D-mannitol, 2.5 mM Hepes–KOH pH 7.4) and digitonin was added at a final concentration of 0, 0.1 or 0.5% (w/v). Each sample was incubated on ice for 10 min and added 4 volumes of mitochondrial buffer and centrifuged at 11 000 r.p.m. at 4°C for 10 min. Each pellet was suspended with mitochondrial buffer and analyzed by Western blot. For trypsin treatment, trypsin was added to the mitochondrial fraction at a final concentration of 1% and incubated on ice for 0, 5, 10 or 15 min. After addition of protease inhibitor and centrifugation at 11 000 r.p.m. at 4°C for 10 min, each pellet was suspended with mitochondrial buffer and analyzed by Western blot.
Supplementary Material
Supplementary Figure
Supplementary Figure Legend
Acknowledgments
This study was supported in part by grants-in-aid for scientific research from MEXT and JSPS. We are grateful to Mr T Kozaki for technical assistance. We thank Dr D Bohmann for the HA-ubiquitin expression plasmid and Dr H Hirai for the pAAV IRES-EGFP vector. We also thank Dr S Jahangeer and Dr M Kojima for critical reading of the manuscript.
References
- Arnold I, Langer T (2002) Membrane protein degradation by AAA proteases in mitochondria. Biochim Biophys Acta 1592: 89–96 [DOI] [PubMed] [Google Scholar]
- Bartee E, Mansouri M, Hovey Nerenberg BT, Gouveia K, Fruh K (2004) Downregulation of major histocompatibility complex class I by human ubiquitin ligases related to viral immune evasion proteins. J Virol 78: 1109–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC (2003) Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol 160: 189–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coscoy L, Ganem D (2003) PHD domains and E3 ubiquitin ligases: viruses make the connection. Trends Cell Biol 13: 7–12 [DOI] [PubMed] [Google Scholar]
- Fritz S, Weinbach N, Westermann B (2003) Mdm30 is an F-box protein required for maintenance of fusion-competent mitochondria in yeast. Mol Biol Cell 14: 2303–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto E, Ishido S, Sato Y, Ohgimoto S, Ohgimoto K, Nagano-Fujii M, Hotta H (2003) c-MIR, a human E3 ubiquitin ligase, is a functional homolog of herpesvirus proteins MIR1 and MIR2 and has similar activity. J Biol Chem 278: 14657–14668 [DOI] [PubMed] [Google Scholar]
- Griparic L, van der Bliek AM (2001) The many shapes of mitochondrial membranes. Traffic 2: 235–244 [DOI] [PubMed] [Google Scholar]
- Hales KG, Fuller MT (1997) Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell 90: 121–129 [DOI] [PubMed] [Google Scholar]
- Harder Z, Zunino R, McBride H (2004) Sumo1 conjugates mitochondrial substrates and participates in mitochondrial fission. Curr Biol 14: 340–345 [DOI] [PubMed] [Google Scholar]
- Hermann GJ, Thatcher JW, Mills JP, Hales KG, Fuller MT, Nunnari J, Shaw JM (1998) Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J Cell Biol 143: 359–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A (1992) The ubiquitin system for protein degradation. Annu Rev Biochem 61: 761–807 [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67: 425–479 [DOI] [PubMed] [Google Scholar]
- Hotta A, Inatome R, Yuasa-Kawada J, Qin Q, Yamamura H, Yanagi S (2005) Critical role of CRMP-associated molecule CRAM for Filopodia and growth cone development in neurons. Mol Biol Cell 16: 32–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inatome R, Tsujimura T, Hitomi T, Mitsui N, Hermann P, Kuroda S, Yamamura H, Yanagi S (2000) Identification of CRAM, a novel unc-33 gene family protein that associates with CRMP3 and protein-tyrosine kinase(s) in the developing rat brain. J Biol Chem 275: 27291–27302 [DOI] [PubMed] [Google Scholar]
- Ishido S, Choi JK, Lee BS, Wang C, DeMaria M, Johnson RP, Cohen GB, Jung JU (2000a) Inhibition of natural killer cell-mediated cytotoxicity by Kaposi's sarcoma-associated herpesvirus K5 protein. Immunity 13: 365–374 [DOI] [PubMed] [Google Scholar]
- Ishido S, Wang C, Lee BS, Cohen GB, Jung JU (2000b) Downregulation of major histocompatibility complex class I molecules by Kaposi's sarcoma-associated herpesvirus K3 and K5 proteins. J Virol 74: 5300–5309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson PK, Eldridge AG, Freed E, Furstenthal L, Hsu JY, Kaiser BK, Reimann JD (2000) The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol 10: 429–439 [DOI] [PubMed] [Google Scholar]
- James DI, Parone PA, Mattenberger Y, Martinou JC (2003) hFis1, a novel component of the mammalian mitochondrial fission machinery. J Biol Chem 278: 36373–36379 [DOI] [PubMed] [Google Scholar]
- Jensen RE, Hobbs AE, Cerveny KL, Sesaki H (2000) Yeast mitochondrial dynamics: fusion, division, segregation, and shape. Microsc Res Tech 51: 573–583 [DOI] [PubMed] [Google Scholar]
- Kyo S, Sada K, Qu X, Maeno K, Miah SM, Kawauchi-Kamata K, Yamamura H (2003) Negative regulation of Lyn protein-tyrosine kinase by c-Cbl ubiquitin-protein ligase in Fc varepsilon RI-mediated mast cell activation. Genes Cells 8: 825–836 [DOI] [PubMed] [Google Scholar]
- Labrousse AM, Zappaterra MD, Rube DA, van der Bliek AM (1999) C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol Cell 4: 815–826 [DOI] [PubMed] [Google Scholar]
- Langer T, Kaser M, Klanner C, Leonhard K (2001) AAA proteases of mitochondria: quality control of membrane proteins and regulatory functions during mitochondrial biogenesis. Biochem Soc Trans 29: 431–436 [DOI] [PubMed] [Google Scholar]
- Means RE, Ishido S, Alvarez X, Jung JU (2002) Multiple endocytic trafficking pathways of MHC class I molecules induced by a Herpesvirus protein. EMBO J 21: 1638–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misaka T, Miyashita T, Kubo Y (2002) Primary structure of a dynamin-related mouse mitochondrial GTPase and its distribution in brain, subcellular localization, and effect on mitochondrial morphology. J Biol Chem 277: 15834–15842 [DOI] [PubMed] [Google Scholar]
- Mitsui N, Inatome R, Takahashi S, Goshima Y, Yamamura H, Yanagi S (2002) Involvement of Fes/Fps tyrosine kinase in semaphorin3A signaling. EMBO J 21: 3296–3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson C (2002) A new gun in town: the U box is a ubiquitin ligase domain. Sci STKE 2002: PE4. [DOI] [PubMed] [Google Scholar]
- Pickart CM (2001) Mechanisms underlying ubiquitination. Annu Rev Biochem 70: 503–533 [DOI] [PubMed] [Google Scholar]
- Sesaki H, Jensen RE (1999) Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J Cell Biol 147: 699–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw JM, Nunnari J (2002) Mitochondrial dynamics and division in budding yeast. Trends Cell Biol 12: 178–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitia R, Braakman I (2003) Quality control in the endoplasmic reticulum protein factory. Nature 426: 891–894 [DOI] [PubMed] [Google Scholar]
- Smirnova E, Griparic L, Shurland DL, van der Bliek AM (2001) Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell 12: 2245–2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson WE, Ramalho-Santos J, Sutovsky P (2003) Ubiquitination of prohibitin in mammalian sperm mitochondria: possible roles in the regulation of mitochondrial inheritance and sperm quality control. Biol Reprod 69: 254–260 [DOI] [PubMed] [Google Scholar]
- Wright G, Terada K, Yano M, Sergeev I, Mori M (2001) Oxidative stress inhibits the mitochondrial import of preproteins and leads to their degradation. Exp Cell Res 263: 107–117 [DOI] [PubMed] [Google Scholar]
- Yaffe MP (1999) The machinery of mitochondrial inheritance and behavior. Science 283: 1493–1497 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure
Supplementary Figure Legend


