Abstract
Cell membranes are fascinating supramolecular aggregates that not only form a barrier between compartments but also harbor many chemical reactions essential to the existence and functioning of a cell. Here, it is proposed to review the molecular dynamics and mosaic organization of the plasma membrane, which are thought to have important functional implications. We will first summarize the basic concepts of Brownian diffusion and lipid domain formation in model membranes and then track the development of ideas and tools in this field, outlining key results obtained on the dynamic processes at work in membrane structure and assembly. We will focus in particular on findings made using fluorescent labeling and imaging procedures to record these dynamic processes. We will also discuss a few examples showing the impact of lateral diffusion on cell signal transduction, and outline some future methodological challenges which must be met before we can answer some of the questions arising in this field of research.
Keywords: actin meshwork, cell membrane, fluorescence microscopy, lateral diffusion, membrane microdomain
Introduction
The nature of the physical constraints preventing the free molecular diffusion, which enabled the first self-replicating forms of life on Earth to develop, is still a matter of debate. However, the creation of a lipid bilayer envelope forming the basic structural unit common to all organisms was a crucial step in evolution. Cell membranes are fascinating two-dimensional supramolecular assemblies resulting from low-energy interactions occurring among a wide range of components. The fluid mosaic model (Singer and Nicolson, 1972) was of great heuristic value, and contributed to our thinking about membrane organization by providing timely insights into the random pattern of cell membrane organization resulting from free diffusion processes. However, experimental evidence subsequently indicated that the lateral motion of membrane components was not free after all, but constrained by various mechanisms such as direct and indirect interactions with cytoskeleton elements. The role of lipids was actively investigated with a view to understanding how lateral phase separation occurs in simple model membranes. The existence of lipid domains in cell membranes was suggested quite early by the results of several original studies (Karnovsky et al, 1982). However, many questions remain unanswered, as processes such as transversal asymmetry and lateral segregation are known to occur in membranes on various spatiotemporal scales (Sprong et al, 2001).
The idea that membranes act as barriers dates back to the time when cell observations were carried out using the first optical microscopes. Later, the use of fluorescence labeling and more advanced imaging techniques made it possible to refine this barrier concept, but the fundamental limit is still there: the Rayleigh limit which makes it impossible to obtain images showing details smaller than the wavelength of light. It is precisely at the spatial and temporal limits where instruments fail that the membrane concept becomes fuzzy and unclear. Fortunately, modern physics, chemistry and biology have been able to draw a fairly detailed picture of membrane organization although the various techniques used for this purpose are far from being ideal, as each of them has its own assets and drawbacks.
Among these tools, optical techniques based on fluorescent contrast principles are of great value because they can be used to observe living cells under physiological conditions. Fluorescence gives an excellent signal to noise ratio which compensates for the Rayleigh limit. Of special interest is the case where only a few fluorescent molecules are present in the field of view without overlapping. When dealing with this configuration, which is called the single molecule regime, the good signal to noise ratio associated with fluorescence labeling can be combined with a high degree of localization accuracy. With this regime, the trajectories of individual molecules can be traced and measured with the precision required to examine membrane organization. Studies on individual molecules' trajectories and properties are more interesting than ensemble measurements, as they provide information about single events. This makes it possible to draw up frequency histograms of the actual distribution of the values of experimental parameters, rather than just obtaining mean distribution values. Distributions undeniably contain more useful information than average values alone. The details of the underlying distributions become crucially important when the system under study is inhomogeneous, as in the case of the cell membrane.
One of the reasons for the controversy about lipid rafts is the difficulty in observing objects that are both so small and so dynamic. Although seeing is believing, one has to acknowledge that most of the tools available for studying the spatio-temporal architecture of live cell membranes either do not involve images at all, or have extended the concept of images far beyond what can actually be seen by the eyes.
The aim of this paper is to review the dynamics of membrane organization in live cell membranes, while at the same time describing the development and the application of the advanced biophotonic technologies associated with these studies. First, we propose to recall the Brownian motion that occurs in all biological systems and what contributes to generating heterogeneity in cell membranes. Next, we will illustrate how biophotonic methods have contributed to determining the time scales on which membrane organization occurs and it will be attempted to define what must be done to be able to observe these complex molecular dynamics in intact membranes. Lastly, we will suggest some of the key points on which future research is likely to focus and some of the challenges that lie ahead.
The fundamentals of plasma membrane dynamics
Brownian motion, diffusion and membrane organization
Brownian motion is a principle that applies to all biological systems (Berg, 1983): as the result of thermal agitation processes, molecules are constantly on the move, colliding with each other and bouncing back and forth (Figure 1). Diffusion is simply the macroscopic effect of Brownian motion. This means that diffusion processes have the following main features: (1) the diffusion rates are temperature-dependent, (2) as collisions with other molecules slow down diffusion processes, the higher the molecular density of a medium is, the lower the diffusion rate will be and most importantly, (3) as the random forces generated by collisions have no preferred direction, diffusion will cause a tendency towards homogeneity.
Figure 1.
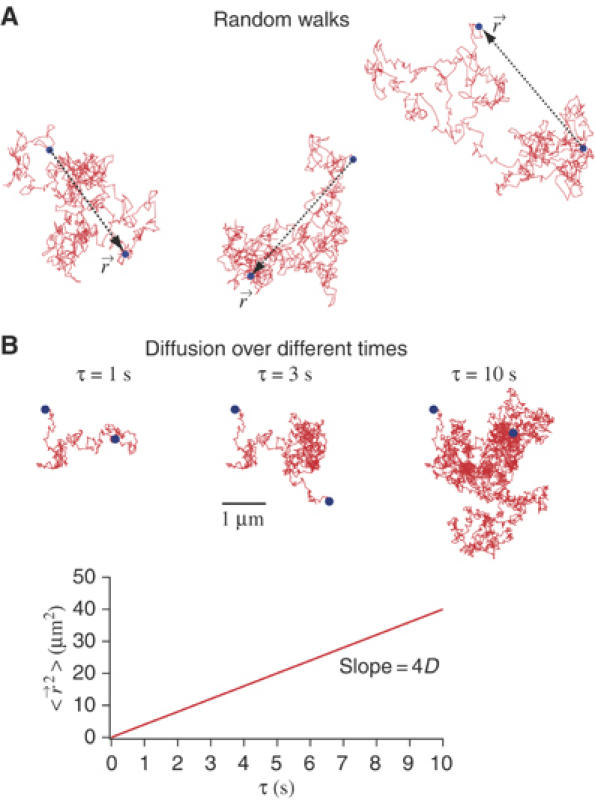
Brownian motion and diffusion. (A) Molecules undergo random-walk motion owing to collisions with solvent molecules. If we observe a molecule during a time interval τ, we will see that it is displaced by a position vector 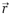 . If we repeat this observation over and over again during τ, the average displacement
. If we repeat this observation over and over again during τ, the average displacement 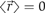 (〈 〉 denotes the average over a large ensemble of trajectories), as the random forces exerted by the solvent molecules on the molecule have no preferred direction. To characterize the Brownian motion, we should use, instead of the average displacement, the average displacement squared (also referred as to the mean squared displacement, MSD),
(〈 〉 denotes the average over a large ensemble of trajectories), as the random forces exerted by the solvent molecules on the molecule have no preferred direction. To characterize the Brownian motion, we should use, instead of the average displacement, the average displacement squared (also referred as to the mean squared displacement, MSD), 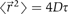 , where D denotes the diffusion coefficient of the molecule (4 is the factor in the case of a two-dimensional random walk). (B) The three pictures show the trajectory of a molecule having a diffusion coefficient D=1 μm2 s−1 during various time intervals.
, where D denotes the diffusion coefficient of the molecule (4 is the factor in the case of a two-dimensional random walk). (B) The three pictures show the trajectory of a molecule having a diffusion coefficient D=1 μm2 s−1 during various time intervals.
In fact, Brownian diffusion contributes to the continuous spreading of molecules from regions with high molecular concentrations to those with lower concentrations. This process tends to smooth out any heterogeneity (Figure 2A). The seminal fluid mosaic model developed on these lines (Singer and Nicolson, 1972), which focuses on the diffusion mobility of membrane lipids and proteins, suggests that membranes show a lack of lateral organization. On the basis of this model, it was subsequently suggested that no long-range interactions occur within the lipid bilayer matrix. Although the authors clairvoyantly noted that ‘absence of long-range order should not be taken to imply an absence of short-range order in the membrane', it seemed unlikely at this stage that anything other than homogeneity could result from Brownian diffusion processes (Shinbrot and Muzzio, 2001).
Figure 2.
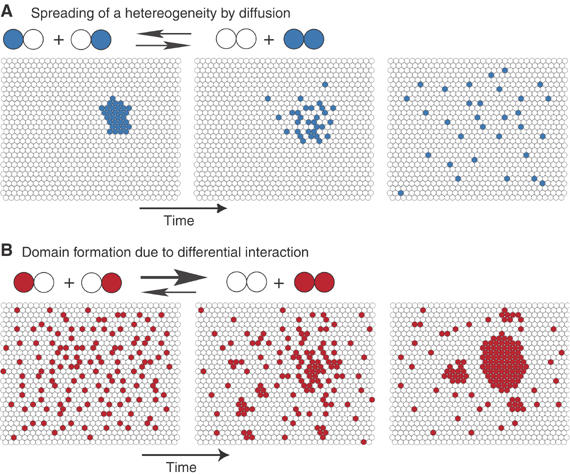
Diffusion and domain formation. (A) In a fluid membrane where the constituents have no differential interactions, that is, where two blue–white pairs are exchanged, giving two pairs of the same color (blue–blue and white–white), the process is energetically costless, and domains will tend to vanish owing to the diffusion processes at work. (B) If the exchange resulting in two like pairs is energetically favorable, however, domains can form. At thermodynamic equilibrium, molecules still diffuse inside and outside the domains, and some of them are transiently trapped in domains.
This raises questions as to how diffusion can be quantified in a two-dimensional fluid medium such as a lipid bilayer. The diffusion coefficient D is usually introduced to characterize the rate at which a molecule spreads over a surface (Figure 1B). A smaller molecular size, a less viscous fluid, or a higher temperature would each increase the amount of motion likely to occur and increase the diffusion coefficient D. When Brownian diffusion occurs in a homogeneous fluid membrane, the diffusion coefficient of a given molecular species is a constant, which is invariant with time and space. Neither the sampling time nor the length scale of the observations performed will therefore affect the results obtained. The situation is different in the case of hindered diffusion in a heterogeneous membrane, as we will see below.
Lipid phase formation in model membranes
Studies on model membranes composed of specific lipid mixtures have brought to light some important features of domain formation and their implications at cell membrane level (London, 2005). Differential interactions between components are the main driving force responsible for the segregation of components into domains. Mismatches between the hydrophobic thicknesses of molecular components, cohesive forces (van der Waals interactions) and the lipid chain entropy are thought to result in a wide range of differential interactions among membrane components. In the simplest mixture of two chemically or structurally distinct lipids (Figure 2B), domain formation will result from the favored exchanges occurring at the microscopic level between similar components.
Alhough the entropy argues in favor of a random pattern of molecular distribution, the results of total free energy minimization studies have suggested that molecular segregation processes are at work (Almeida et al, 2005). At equilibrium, larger domains are favored rather than smaller ones because the edge energy, which is unfavorable to domain formation, is negligible in comparison with the bulk energy of the domains. This results in a phase separation, where immiscible components form two separate regions, each of which has homogeneous thermodynamic parameters. Studies of cholesterol-based mixtures have shown the existence of a state where lipids are tightly packed and ordered, but where the lateral diffusion process is almost as fast as in the classical liquid-disordered (Ld) state (London, 2005). The coexistence of this cholesterol-containing state known as the liquid-ordered (Lo) phase and the Ld phase has strongly stimulated current thinking about the physical characteristics of cell membrane domains.
Phase separation has significant effects on the lateral diffusion of a molecule. If a lipid mixture has nonideal mixing properties, the entropy constantly balances any minimization of the unfavorable interactions. Nevertheless, the process of molecular segregation will still be randomized in the bilayer owing to the persistence of Brownian motion. Single molecules will explore the different phases (Korlach et al, 1999; Dietrich et al, 2001), but will be transiently trapped in the domains when favored interactions occur with the nearest neighbors and when there exists a low probability of exchange at the boundaries between phases. All these events result in a complex pattern of diffusion that is somewhat different from that described in strictly Brownian diffusion terms.
Cell membranes: a mosaic rather than a fluid-phase structure
Despite the great value of the principles of domain formation obtained by studying model membranes at equilibrium, one should be cautious about applying them directly to cell membranes, as the latter are far more complex systems than model membranes. In addition, domains in cell membranes are not necessarily large enough to be called phases, but can be viewed as condensed complexes (McConnell and Vrljic, 2003). However, substantial evidence is now available supporting previous assumptions about heterogeneity and local order in the overall pattern of membrane organization in both model and cell membranes.
At first, the great diversity of lipid species suggested that a wide range of combinatorial interactions must occur among them (van Meer, 2005). Live cell membrane domains cannot be homogeneous in terms of their length, for instance. There are many local variations in composition, asymmetric organization of the leaflets and difference in the membrane thickness. In addition, the complexity of the interactions between lipids and proteins is not restricted to hydrophobic transmembrane regions: crystallographic resolution of membrane protein structures has shown that both the extra- and intracellular domains of transmembrane proteins can establish direct interactions with the respective membrane surfaces (Engelman, 2005).
Domain interfaces in live cell membranes might be stabilized by proteins that associate with their lipid components, as in the case of the N-Ras protein observed at the Ld/Lo phase boundary in an artificial model (Nicolini et al, 2006). This suggests that interfacial adsorption processes may lead to a favorable decrease in line tension between domains.
Enzyme activities, membrane recycling, and signaling events constantly disturb the equilibrium of cell membranes. These events might increase the local concentration of a subset of molecules, and thus create heterogeneity and generate small domains. The results of in silico studies have suggested for example that a combination between the vesicular traffic to and from the plasma membrane and the presence of dynamic barriers impeding lateral mobility might generate membrane patchiness (Gheber and Edidin, 1999). Studies in which the apparent sizes and intensities of class I HLA protein patches were quantified in plasma membranes before and after inhibiting the membrane trafficking processes strongly support this idea (Tang and Edidin, 2001). On similar lines, a recent theoretical study has also shown that small domains can be stabilized by continuous recycling (Turner et al, 2005).
Therefore, starting with the fluid mosaic model where membrane proteins were taken to be dispersed monomers existing in low concentrations in a homogeneous lipid bilayer, we have ended up with a more mosaic and highly dynamic picture of membrane organization, where proteins form preferential associations in a crowded environment and the membrane thickness is variable. Although it is now generally agreed that lipid and protein diversity and local nonequilibrium effects generate a high level of heterogeneity in cell membranes, understanding the size- and time scales on which these heterogeneities occur still remains a challenging issue (Edidin, 2003; Simons and Vaz, 2004; Kusumi et al, 2005; van Meer, 2005).
A comparative study on methods of investigating membrane dynamics
After the seminal study establishing the fluid nature of the cell membrane (Frye and Edidin, 1970), the need arose for suitable tools to quantify the spatio-temporal parameters characterizing the membrane organization. The most useful tool for studying dynamic processes in live cells is still light microscopy (Lagerholm et al, 2005). These methods used tags (conjugated dyes, beads, etc.) to follow the diffusion behavior of a given molecule. As such, these approaches require careful interpretation. We show in Table I a comparison of different methods used to investigate membrane dynamics.
Table 1.
Current methods for investigating membrane dynamics
| Method | Principle and observable(s) | Comments |
|---|---|---|
| FRAP | Level and rate of fluorescence recovery with time following the photobleaching of a limited area of observation | • Well established for FRAP in spot mode |
| • Ensemble average method | ||
| • Translational diffusion | • Weakly sensitive to separate subpopulations with different diffusion characteristics | |
| • Mobile fraction | • Observation area usually larger than the diffraction limit | |
| FCS and ICS | Fluorescence fluctuations induced by low numbers of diffusing fluorescent molecules into a limited area of observation | • Averaging of thousands of single-molecule diffusion events within short acquisition times |
| • Low probe concentration (nM range) | ||
| • Molecular concentration | • Highly sensitive to clustering (photon counting histogram analysis) | |
| • Translational diffusion of multiple subpopulations | • Single & multiple colors | |
| • Molecular clustering | • In standard approach, it is difficult to determine the diffusion of slow-diffusing molecules | |
| • Count rate per molecule | • Observation area usually close to the diffraction limit | |
| SPT and SDT | Scattered light from a single bead particle | • Excellent spatial precision (dependent on the signal/noise ratio) |
| Or fluorescence signal from a single emitter | • Temporal resolution limited by the frequency of image acquisition | |
| • Spatio-temporal identification of the diffusion heterogeneities | ||
| • Experimental bias favoring slow-diffusing particles/molecules | ||
| • Trajectories of individual particles or fluorescent molecules | • Statistics quality strongly related to the length of the trajectories | |
| • Translational diffusion in different cell area | • Valence of the tagged molecules often weakly determined | |
| • Access to different mode of motion | ||
| FCS, fluorescence correlation spectroscopy; FRAP, fluorescence recovery after photobleaching; ICS, image correlation spectroscopy; SDT, single-dye tracing; SPT, single-particle tracking. | ||
Fluorescence recovery after photobleaching
Among these methods, fluorescence recovery after photobleaching (FRAP) makes use of the fact that fluorescent molecules lose their ability to emit photons when exposed to repetitive cycles of excitation and emission. In the FRAP method, a brief intense excitation light irreversibly photobleaches fluorophores located in a small area of the membrane (Figure 3A). Subsequent movement of surrounding nonbleached fluorescent molecules into the photobleached area is recorded at low laser power. By monitoring the levels and rates of fluorescence recovery with time, one can determine kinetic parameters such as the mobile fraction Mf and the diffusion coefficient D. However, various processes such as membrane flow, molecular interactions and trafficking may simultaneously contribute to the overall recovery kinetics, which makes it difficult to interpret the data obtained. Another drawback of FRAP is its lack of spatial resolution although the focal spot can be diffraction limited. The molecules that refill the photobleached spot may also originate from a membrane region located quite far from the active spot. Although FRAP is a simple and powerful method, it needs to be combined with appropriate mathematical models in order to be able to interpret the data properly and analyze the complex dynamic processes involved. Recent advances in fluorescent protein technology and confocal microscopy have made it possible to extend these approaches to intracellular dynamic studies such as those on trafficking processes (Lippincott-Schwartz et al, 2000). An alternative to the FRAP method, the fluorescence loss in photobleaching (FLIP) method, is based on repetitively photobleaching one part of the cell while images of the entire cell are being collected.
Figure 3.
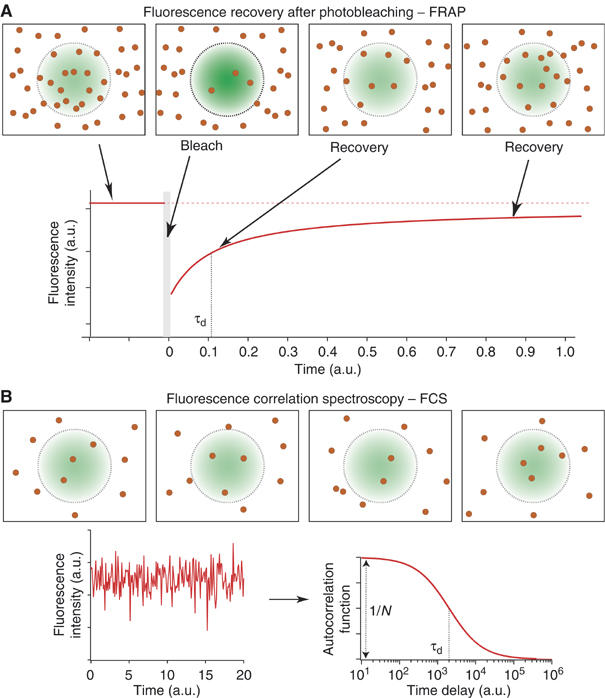
Schematic diagram of the FRAP and FCS techniques. Both methods involve the use of a laser to illuminate a small region of the membrane and a confocal set-up to detect the fluorescence emitted by fluorophores. (A) In FRAP, the amount of fluorescence is recorded at low laser intensity in the region of interest (defined here by the laser spot area). A high-intensity pulse is used to bleach most of the molecules within the spot, and the fluorescence is then measured at low laser intensity. Diffusing molecules then replenish the bleached area, leading to an increase in the detected fluorescence intensity. The characteristic fluorescence recovery time reflects the diffusion time. If some molecules were immobile in the bleached region, the level of intensity after recovery will be lower than before the bleaching. (B) In FCS, fluorescence fluctuations occur as the result of a small number of molecules diffusing in and out of the observation area defined by the laser spot. To investigate the diffusion processes, the autocorrelation function of the fluorescence intensity (ACF) is calculated. The ACF has two main features: its width at half maximum gives the diffusion time and its amplitude is inversely proportional to the average number of molecules present in the spot area.
Single-particle tracking and single-dye tracing
Methods for direct observation of diffusion of individual molecules, such as single-particle tracking (SPT) and single-dye tracing (SDT) methods (Figure 4), have proved to be powerful alternatives to FRAP. In these single-molecule techniques, molecular motion is not masked by the ensemble averaging (Saxton and Jacobson, 1997) and can be studied in all its diversity. In SPT, a submicrometer-sized particle (latex beads ∅ 200–500 nm or colloidal gold ∅ 40–100 nm) that binds to a molecule via a specific interaction is imaged at a high video rate. Tracking individual particles requires computer-assisted analytical methods for accurate location (to within a few tens of nm) of the relevant particles in each image, before linking them up with the subsequent images (Sheetz et al, 1980). A variety of dynamic parameters can then be computed from the individual reconstituted trajectories. The main advantage of SPT is the good signal to noise ratio consistently obtained at high acquisition rates with this method because the signal originates from the incident light scattered by the particle. This process is linear with the excitation power and not subject to photobleaching. One has to be careful about interpreting the data, however, because of the following drawbacks. The bead surface is several orders of magnitude larger than the tracked molecules, and nonspecific interactions in the dense extracellular matrix may substantially slow down its motion. It is also difficult to control the bead/receptor interactions occurring because of noncovalency (the beads may escape and re-engage with another receptor, or multivalent detection can trigger unwanted biological responses and/or membrane domain reorganization resulting from receptor crosslinking). New detector developments combined with the use of organic dyes or fluorescent proteins have helped to solve these problems (Schutz et al, 2000; Lommerse et al, 2004). Although these materials are less invasive than latex or gold beads, they photobleach quite rapidly; the unique optical properties of quantum dots (QD) (high quantum yields, large molar extinction coefficients, size-dependent tunable emission and high photostability) make them appealing candidate tags for use with SDT (Dahan et al, 2003; Michalet et al, 2005).
Figure 4.
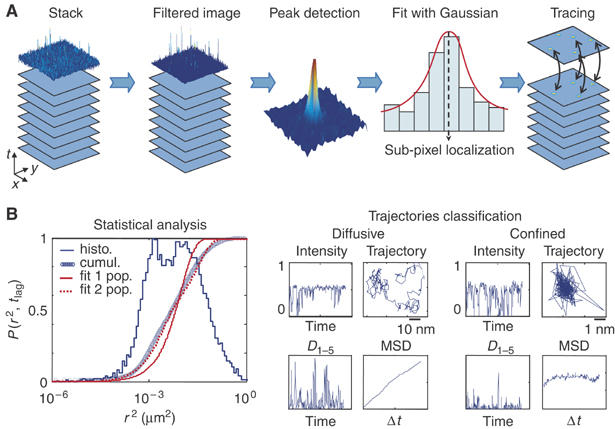
Schematic diagram of the steps involved in SPT/SDT data analysis. (A) From the stack of images acquired to the particle track file. Particles are first detected in each image. The raw images are preprocessed (by performing deconvolution, noise reduction, spatial alignment) to enable accurate determination of each peak position (Gauss-Newton fit). Particle tracing is then performed by connecting all the peak positions from frame to frame to draw up space-time trajectories. (B) Statistical and individual trajectory analysis. Data can be analyzed in various ways. One approach consists of building back histograms of relevant parameters, such as the intensity or MSD, to obtain statistics, and assigning subpopulations for instance, by pooling all the elementary information obtained on each of the particles in the population under investigation. A complementary approach consists of investigating trajectories individually, in order to classify different types of dynamic behavior either between or within traces, accounting for putative transitions between different dynamic states, including the reversible confinement state, for instance. The two examples of 10-s trajectories acquired at a rate of 30 ms/frame mostly depict free diffusion and confinement.
Total-internal-reflection fluorescence (TIRF) microscopy also offers to directly monitor the behaviors of molecules in living cells (Axelrod, 2001). Because illumination is restricted to a thin region of the specimen, background fluorescence can be drastically reduced, allowing single molecule detection in the membrane.
Fluorescence correlation spectroscopy
Advances in fluorescence signal detection methods focusing on single molecules have led to the resurgence of fluorescence correlation spectroscopy (FCS) methods for studying molecular diffusion processes in live cells (Bacia et al, 2006). FCS is based on the fluorescence intensity fluctuations associated with molecules passing through a diffraction-limited observation volume (Figure 3B). Each burst of photons originates from the passage of single fluorescent molecules. The autocorrelation function (ACF) describes the fluorescence fluctuations of the signal. This function, which is determined in real time, gives the average diffusion time taken by the molecule to cross the observation volume, along with the average number of molecules present within this observation volume. FCS therefore provides information about the density of the fluorescent probes and contrary to FRAP, deals solely with the diffusion occurring within the observation volume and requires very low probe concentration levels. However, FCS is only sensitive to fluorescence fluctuations and ignores immobile molecules, owing to the possible occurrence of photobleaching during the residence time of a fluorophore in the confocal volume. It measures only relatively fast-moving molecules (D>0.1 μm2 s−1), although recent advances have extended its range of application to include slow-diffusion processes (D>0.001 μm2 s−1) (Digman et al, 2005). Like the other techniques described above, FCS has to be combined with modeling and statistical tools to be able to accurately interpret complex types of diffusion behavior (Wawrezinieck et al, 2005) or to quantify molecular multimerization (Sanchez-Andres et al, 2005). One particularly promising approach to live cell analysis consists in performing temporal cross correlations between two different fluorescent species to determine the molecular interactions taking place within the observation volume (Larson et al, 2005).
Moreover, image correlation spectroscopy (ICS), which is an extension of FCS, involves spatial correlation analysis of fluorescence fluctuations within an image sample using a laser-scanning microscope (Petersen et al, 1993). Applied on living cells, ICS methods provide quantitative measures of the cluster density as well as the degree of aggregation of fluorescent molecules at the cell surface. Image cross-correlation spectroscopy (ICCS) and now spatiotemporal image correlation spectroscopy (STICS) are powerful derivative approaches to measure quantitatively the interactions of molecules and the dynamics of the resulting clusters in plasma membrane (Costantino et al, 2005).
Origin of constrained lateral diffusion in cell membranes
Early diffusion measurements immediately showed that different environments and/or patterns of membrane organization would have differential effects on the lateral diffusion behavior of membrane components. The first evidence on these lines was provided by data obtained using an elegant FRAP approach performed at variable observation spots, which suggested that corrals are formed in live fibroblasts on various submicrometric scales (Yechiel and Edidin, 1987). We will now describe how various constraining mechanisms have been identified in studies on lateral diffusion.
The cytoskeleton meshwork
The role of the cytoskeleton meshwork (Figure 5A) as a barrier to the lateral mobility of transmembrane proteins was first established based on the finding that faster lateral diffusion occurred in the presence of a disorganized cytoskeleton meshwork (Sheetz et al, 1980). These barriers were found to be located 2–3 nm below the membrane bilayer based on barrier-free path measurements performed with laser optical traps on MHC Class I variants with different truncated cytoplasmic domains (Edidin et al, 1994). In addition, Monte–Carlo simulations on diffusion in the presence of a network confirmed that diffusion was hindered by the network but not on the short-range scale (Saxton, 1989).
Figure 5.
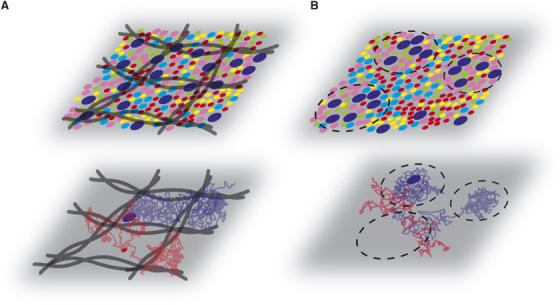
Cytoskeleton meshwork and dynamic partition models. (A) In the cytoskeleton meshwork model, the fences consist of actin filaments (black filaments) which compartmentalize the molecules into domains with comparable compositions. During the diffusion process, the blue molecule is confined between fences before crossing the physical barrier, whereas the red molecule crosses the barriers as if they were nonexistent. (B) In the dynamic partition model, the membrane consists of domains with different compositions. A snapshot of the membrane shows local heterogeneities. Here the molecules partition dynamically, and are sometimes trapped in domains. During their diffusion, blue molecules are confined only transiently in domains in which they partition preferentially, whereas red ones avoid the same domains. Some domains disappear with time, and new ones can also form.
Advanced SPT/SDT technologies have greatly helped to understand further how a cytoskeleton meshwork contributes to subdivision of the plasma membrane into domains (Kusumi et al, 2005). It was suggested that like the barriers surrounding fields, this meshwork consists of fences and pickets that hinder lateral diffusion: the actin-based cytoskeleton meshwork forms the fences and transmembrane proteins anchored to the cytoskeleton form the pickets. In the fence model, the imperfect fluctuating lattice of membrane-associated proteins to actin-based cytoskeleton creates barriers to lateral diffusion (Sako and Kusumi, 1994). In the picket model, the cytoskeleton-anchored transmembrane proteins act as pickets along the cytoskeleton fences and restrict the lateral diffusion of membrane molecules in the absence of interaction with this cytoskeleton. It is worth noting that the molecules (GPI-anchored proteins or lipids) present in the outer leaflet of the plasma membrane are also sensitive to pickets (Fujiwara et al, 2002). Based on individual trajectory analysis, the compartment size was found to range between 30 and 230 nm. Using an alternative approach involving FCS measurements performed at variable illumination spots, a similar cytoskeleton meshwork was found to confine a GFP-tagged transferrin receptor but not lipid analogs present in the outer leaflet (Wawrezinieck et al, 2005; Lenne et al, 2006).
In the fence and picket models, molecules diffuse rapidly within each domain delineated by the cytoskeleton meshwork and escape from one domain to the next by hopping or passing through transient gaps in the barrier. All in all, these data provide a plausible explanation both for the fact that the D values recorded in plasma membrane are smaller by factors of 5–50 than those observed in an artificial membrane, and for the decrease in D observed upon molecular complex formation (oligomerization-induced trapping) (Kusumi et al, 2005).
Dynamic partitioning into membrane microdomains
By definition, the characteristics of a microdomain differ from those of the whole membrane (Figure 5B). Among microdomains, the lipid raft, which has attracted considerable attention, is thought to show greater molecular order and to be involved in diverse cellular functions (Brown and London, 2000; Simons and Vaz, 2004). However, attempts at directly seeing raft structures in live cells have failed, and therefore no clearcut conclusions have been reached on this point. In fact, microdomains are probably very small and extremely diverse in terms of their composition, and may result from a set of weak molecular interactions. This makes them difficult to detect by simply looking for higher local concentrations of the putative markers used.
However, the presence of microdomains would certainly affect the diffusion behavior of the molecules of which membranes are composed. Cholesterol-dependent microdomains 26±13 nm in size were observed over long periods of time by combining SPT with local viscous drag measurements (Pralle et al, 2000). Other studies have established that microdomains show dynamic behavior and are subject to disruption and coalescence. Interestingly, convergent findings have been obtained suggesting that, when detected by SPT, aggregated raft-forming molecules induce the formation of transient confinement zones 200–300 nm in size at the surface of murine fibroblasts (Saxton and Jacobson, 1997). In addition, FRAP measurements on influenza hemagglutinin mutants differing in the stability of their interactions with lipid rafts have indicated that resident raft proteins have a lower diffusion rate than non-raft proteins (Shvartsman et al, 2003).
However, authors performing large-scale confocal FRAP assays failed to distinguish between the diffusion characteristics of putative raft and non-raft proteins (Kenworthy et al, 2004). These results suggest that long-range diffusion measurements would be more informative if combined with other experimental methods. For instance, by performing FRAP at different temperatures, it was possible to subdivide clearly the proteins of the apical membrane of epithelial cells into raft and non-raft groups, depending on their diffusion characteristics (Meder et al, 2006).
FCS, as already mentioned, is a powerful technique particularly well suited to quantifying diffusion parameters with a high temporal resolution. FCS analysis has made it possible to distinguish clearly between the cholera toxin B subunit bound to ganglioside GM1 and the dialkylcarbocyanine dye diI in native cell membranes (Bacia et al, 2004). An original FCS approach was recently developed to measure the diffusion times of a species with high temporal resolution through different observation areas (Wawrezinieck et al, 2005). The FCS diffusion laws give not only the diffusion coefficient but also information about the type of membrane organization. This approach was subsequently used to directly identify and characterize both the lipid- (raft microdomain) and cytoskeleton-dependent (actin meshwork) membrane organization in vivo. Putative raft markers were found to be dynamically compartmented within tens of milliseconds into small microdomains (∅<120 nm) that were sensitive to cholesterol and sphingomyelin levels; whereas actin-based cytoskeleton barriers were mainly responsible for the confinement of the transferrin receptor. Importantly, free Brownian diffusion was observed in all the molecules investigated when both the raft microdomains and actin cytoskeleton meshwork were disrupted, which suggests that these processes are the two main compartmentalizing forces at work in the plasma membrane (Lenne et al, 2006).
Membrane environment, complex formation and lateral mobility
FRAP studies on GFP-tagged membrane proteins have provided new insights into the mechanisms possibly impeding the diffusion rates of proteins in cell membranes. Local synthesis and transport specificities contribute to differences in the composition of organelles from that of the plasma membrane (van Meer, 2005).
Contrary to what occurs in the plasma membrane, most of the ER- and Golgi-resident GFP-tagged proteins studied have been found to be highly mobile and to contain only a small immobile fraction, which suggests that their lateral diffusion is not constrained (Cole et al, 1996). However, relatively low D values were observed with the GFP-tagged H2Ld MHC class I and TAP (transporter associated with antigen processing) molecules present in the ER. This finding suggested that during assembly and peptide loading, the MHC class I molecules were retained in the ER because they were stably associated with TAP molecules in large complexes until efficient peptide loading occurred (Marguet et al, 1999).
It is worth noting here that contrary to the initial hydrodynamic model (Saffman and Delbruck, 1975), which predicted a weak, logarithmic dependence of the diffusion coefficient D on molecular size, the results of a recent study have suggested that the D value may be strongly influenced by protein dimensions (Gambin et al, 2006). It would now be useful to reassess the local changes in D resulting not only from physical barriers but also from molecular complex formation. This opens new perspectives for investigating the dynamics of molecular complexes using appropriate FCS analytical methods to quantify multimerization distribution (Sanchez-Andres et al, 2005) and heteromultimeric complex equilibrium, based on fluorescence cross correlations (Larson et al, 2005).
Membrane dynamics during signaling
The binding of an extracellular ligand to a receptor present in the plasma membrane triggers an appropriate cellular response through a cascade of reactions. The first steps in this cascade often involve direct interactions between the activated receptor and the nearest effectors. The possible involvement of lateral diffusion in the signal transduction process has been formulated in the mobile receptor hypothesis (Cuatrecasas, 1974).
Lateral diffusion in the plasma membrane plane could be involved in propagation of the initial signals. It has been established that stimulating the epidermal growth factor (EGF) receptor (EGFR) locally with EGF could lead to the phosphorylation of all the EGFRs in the whole plasma membrane within less than 5 min (Verveer et al, 2000; Sawano et al, 2002). This lateral propagation of phosphorylation may occur via a diffusion reaction rather than a long-range diffusion mechanism. By tracking EGFR with the EGF conjugated to fluorescent quantum dots, it was established that the EGFR located on cellular filopodia undergo systematic retrograde transport after being activated upon EGF binding. This may constitute the mechanism whereby filopodia detect the presence and concentration of EGF molecules located far from the cell body (Lidke et al, 2005).
The idea that membrane diffusion is involved in receptor signaling processes has also been suggested by studies on the G-protein-coupled receptor (GPCR) family. FRAP experiments have shown that the binding of agonist but not antagonist ligands to 5-hydroxytryptamine 1A receptor (5-HT1AR) increased the lateral diffusion of the receptor (Pucadyil et al, 2004), which suggests that a pre-existing receptor/G-protein complex may be dissociated as the result of ligand binding. On the other hand, cell signaling has also frequently been reported to induce the lateral confinement of specific molecules. One consequence of this confinement is that it promotes specific molecular interactions. Restricted diffusion of phosphatidylinositol 4,5-bisphosphate (PIP2) was reported to contribute to local depletion of the PIP2 by phospholipase C upon activation by a Gq protein-coupled receptor (Cho et al, 2005).
Confinement of receptor and signaling partners in the plasma membrane, resulting in the formation of specific microdomains, has recently been observed in the case of activated T cells (Bunnell et al, 2002; Douglass and Vale, 2005; Gaus et al, 2005). These microdomains excluded and limited the free diffusion of molecules in the membrane but also could trap and immobilize specific proteins (Douglass and Vale, 2005). These membrane arrangements were found to depend on receptor-mediated signaling, cholesterol level and actin cytoskeleton organization. Using LAURDAN as a probe, these microdomains were characterized by two-photon microscopy as condensed membrane phases with a higher molecular order than the rest of the plasma membrane (Figure 6) (Gaus et al, 2005). It would be interesting to compare the local lateral diffusion process occurring in these membrane structures that are located in the immunological synapse with those described in detail for neurotransmitter receptors at the neurological synapse (Dahan et al, 2003; Tardin et al, 2003).
Figure 6.
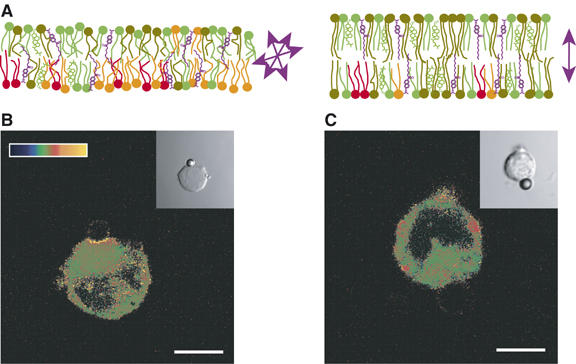
Membrane condensation at the T-cell receptor (TCR) activation site in Jurkat T cells. (A) The Laurdan probe (violet) detects the polarity of its environment and thus water penetration into the hydrophobic part of a lipid bilayer. Laurdan orientation varies in a disordered (left) and ordered (right) lipid bilayer. Arrows indicate the Laurdan transition dipole. (B and C) Generalized polarization (GP) images of Laurdan-labeled Jurkat cells conjugated with α-TCR/CD3-coated (B) or α-TfR-coated (C) beads. GP images were pseudocolored (see scale of GP in B, ranging from 0.5 to 1), and the insets show differential interference contrast (DIC) images. Only the cells activated with a bead coated with anti-CD3 antibodies show membrane condensation at the activation site in the plasma membrane, which display high GP values. Scale bar, 5 μm. These images are provided courtesy of K Gaus and T Harder.
Future challenges for studying membranes in unperturbed live cells
Biologists still dream about specifically imaging or detecting the molecule under investigation with minimum perturbations. In order to characterize events precisely, new approaches and tools are still required. Interactions between light and matter (Figure 7) could in fact be of great interest. The photothermal interference contrast method, for example, has made it possible to examine individual receptors labeled with 10 nm gold nanoparticles on cell surfaces (Cognet et al, 2003). In addition, although few Raman techniques are sensitive enough to provide means of studying cell membranes, stimulated Raman microscopy (Zumbusch et al, 1999), known as the coherent anti-Stokes Raman scattering (CARS) procedure, was recently successfully applied to the study of biomembranes (Li et al, 2005; Potma and Xie, 2005; Wurpel et al, 2005). In this case, the CARS contrast reflects the density of the aliphatic C–H molecular bonds, but the CARS method still requires around 106 molecular bonds per confocal volume (millimolar concentrations). Although the CARS and other nonlinear optical techniques (Pons et al, 2003; Roke et al, 2003) are highly promising methods, they have not yet reached the level of maturity of fluorescence microscopy.
Figure 7.
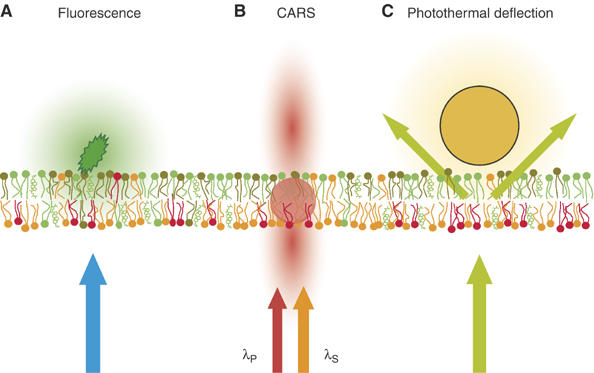
Possible interactions between light and matter in cell membranes. (A) A fluorescent molecule inserted into the plasma membrane emits a specific wavelength (green in the figure) when illuminated by an excitation beam (blue in the figure). (B) CARS requires no labeling, as the vibration levels present in any molecular bond are used. The CARS radiation from the membrane is emitted mostly in the forward and backward directions (red in the figure) when the molecular sample is targeted by two laser pulses (λP and λS) with a frequency difference equal to the vibration frequency of the bond in question (usually C–H to generate a membrane contrast). (C) In photothermal deflection experiments, a tiny (∅ 10 nm) gold bead attached to the membrane absorbs the incoming light (green in the figure) and creates a thermal gradient which scatters the light.
This predominance of the fluorescence approaches will stay popular for studying live cells. Unfortunately, most molecules of interest are not intrinsically fluorescent and they must be tagged. As the cDNA encoding the GFP was characterized, a wide variety of monomeric fluorescent proteins have provided attractive potential candidates for monitoring dynamic processes in which different molecular species are simultaneously involved (for a review, see Shaner et al, 2005). The use of fluorescent quantum dots is emerging as a promising alternative to classical fluorescent tags (GFPs and organic fluorophores) (Michalet et al, 2005). However, there is still a need to improve the functionalization of QD surfaces, the flexibility for bioconjugations and single irreversible molecular associations between individually tracked molecules (Medintz et al, 2005).
Achieving fast single-molecule imaging is probably the most difficult issue facing us at present. Better detectors are needed and hopes are being placed on the possibility of combining back-illuminated CDD with electron illumination CCD techniques to provide the highest possible quantum efficiency (>90%), high frame rates and means of detecting single photons. In conventional optical far field techniques, the spatial resolution is ultimately Rayleigh limited. To overcome this limitation, two strategies have been applied: creating a sub-wavelength excitation volume or collecting the high spatial frequencies (angle of emission) emitted by sub-wavelength scatterers. Strategies of the first type have involved the use of either interference in 4PI microscopy (Hell and Stelzer, 1992), stimulated emission depletion (Klar et al, 2000) or reversible saturable optical fluorescence transitions (Hofmann et al, 2005). With the second strategy, a diffraction pattern or nanostructure is required to be able to convert the high spatial frequency (evanescent waves) into running waves that can be ultimately collected by conventional objective lenses. On these lines, some promising observations have been carried out on plasma membrane domains in fixed, dried cells using scanning near-field optical microscopic methods (SNOM) (Hwang et al, 1998; Ianoul et al, 2005).
Conclusion
Although the first studies on cell membrane dynamics were published years ago, our knowledge of this topic is still incomplete. This field of research has benefited greatly from membrane modeling studies, but we sometimes forget that there is a large intellectual and experimental gap between model and cell membranes. As we have pointed out, one of the main differences between model and cell membranes is the stationary state of the former. In this context, we would like to quote Edidin: ‘Whatever we learn from models should be tempered by the fact that the cells are always right.' (Edidin, 2003). At the technical level, it is critical when studying cell membranes to know how experimentation perturbs the system under investigation, and to take this factor into account when interpreting experimental findings.
There is no doubt that membrane microdomains will continue to be a key issue in studies on cell membrane organization and dynamics. Remaining questions include those relating to the heterogeneity, the distribution size, the half-lives and the dynamics of microdomains. Do cell membranes in the ordered state show the very same properties as those in the Lo phase, and are they created by the coalescence of ‘elementary' rafts? There are many other questions, related to the physical parameters causing a protein to prefer a certain lipid phase or environment, whether and how cytoskeleton barriers and membrane microdomains may influence each other, how membrane microdomains are regulated both quantitatively and qualitatively during cell development and differentiation, and how these regulatory processes may influence such cell functions as signal transduction.
Finding answers to many of these questions will depend on the development of new technologies, as well as the use of multidisciplinary approaches involving teams of mathematicians, physicists, chemists and biologists. Studies on the dynamics of cell membranes and their contribution to cell function will predictably constitute a particularly active field of research in the postgenomic age.
Acknowledgments
We apologize to those of our colleagues whose pioneering work could not be cited owing to space limitations. This reference list contains mainly recent studies and readers are referred to these studies and the references therein for further information. We thank all the members of our laboratories involved in this research and J Blanc for editing the English. This work was supported by institutional grants from INSERM and CNRS, and by specific grants from ARC, FRM, MENRT, EU FEDER and CNRS.
References
- Almeida PF, Pokorny A, Hinderliter A (2005) Thermodynamics of membrane domains. Biochim Biophys Acta 1720: 1–13 [DOI] [PubMed] [Google Scholar]
- Axelrod D (2001) Total internal reflection fluorescence microscopy in cell biology. Traffic 2: 764–774 [DOI] [PubMed] [Google Scholar]
- Bacia K, Kim SA, Schwille P (2006) Fluorescence cross-correlation spectroscopy in living cells. Nat Methods 3: 83–89 [DOI] [PubMed] [Google Scholar]
- Bacia K, Scherfeld D, Kahya N, Schwille P (2004) Fluorescence correlation spectroscopy relates rafts in model and native membranes. Biophys J 87: 1034–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg HC (1983) Random Walks in Biology. Princeton: Princeton University Press [Google Scholar]
- Brown DA, London E (2000) Structure and function of sphingolipid- and cholesterol-rich membrane rafts. J Biol Chem 275: 17221–17224 [DOI] [PubMed] [Google Scholar]
- Bunnell SC, Hong DI, Kardon JR, Yamazaki T, McGlade CJ, Barr VA, Samelson LE (2002) T cell receptor ligation induces the formation of dynamically regulated signaling assemblies. J Cell Biol 158: 1263–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Kim YA, Yoon JY, Lee D, Kim JH, Lee SH, Ho WK (2005) Low mobility of phosphatidylinositol 4,5-bisphosphate underlies receptor specificity of Gq-mediated ion channel regulation in atrial myocytes. Proc Natl Acad Sci USA 102: 15241–15246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cognet L, Tardin C, Boyer D, Choquet D, Tamarat P, Lounis B (2003) Single metallic nanoparticle imaging for protein detection in cells. Proc Natl Acad Sci USA 100: 11350–11355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole NB, Smith CL, Sciaky N, Terasaki M, Edidin M, Lippincott-Schwartz J (1996) Diffusional mobility of Golgi proteins in membranes of living cells. Science 273: 797–801 [DOI] [PubMed] [Google Scholar]
- Costantino S, Comeau JW, Kolin DL, Wiseman PW (2005) Accuracy and dynamic range of spatial image correlation and cross-correlation spectroscopy. Biophys J 89: 1251–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuatrecasas P (1974) Membrane receptors. Annu Rev Biochem 43: 169–214 [DOI] [PubMed] [Google Scholar]
- Dahan M, Levi S, Luccardini C, Rostaing P, Riveau B, Triller A (2003) Diffusion dynamics of glycine receptors revealed by single-quantum dot tracking. Science 302: 442–445 [DOI] [PubMed] [Google Scholar]
- Dietrich C, Bagatolli LA, Volovyk ZN, Thompson NL, Levi M, Jacobson K, Gratton E (2001) Lipid rafts reconstituted in model membranes. Biophys J 80: 1417–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digman MA, Brown CM, Sengupta P, Wiseman PW, Horwitz AR, Gratton E (2005) Measuring fast dynamics in solutions and cells with a laser scanning microscope. Biophys J 89: 1317–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglass AD, Vale RD (2005) Single-molecule microscopy reveals plasma membrane microdomains created by protein–protein networks that exclude or trap signaling molecules in T cells. Cell 121: 937–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edidin M (2003) The State of lipid rafts: from model membranes to cells. Annu Rev Biophys Biomol Struct 32: 257–283 [DOI] [PubMed] [Google Scholar]
- Edidin M, Zuniga MC, Sheetz MP (1994) Truncation mutants define and locate cytoplasmic barriers to lateral mobility of membrane glycoproteins. Proc Natl Acad Sci USA 91: 3378–3382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman DM (2005) Membranes are more mosaic than fluid. Nature 438: 578–580 [DOI] [PubMed] [Google Scholar]
- Frye LD, Edidin M (1970) The rapid intermixing of cell surface antigens after formation of mouse–human heterokaryons. J Cell Sci 7: 319–335 [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Ritchie K, Murakoshi H, Jacobson K, Kusumi A (2002) Phospholipids undergo hop diffusion in compartmentalized cell membrane. J Cell Biol 157: 1071–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambin Y, Lopez-Esparza R, Reffay M, Sierecki E, Gov NS, Genest M, Hodges RS, Urbach W (2006) Lateral mobility of proteins in liquid membranes revisited. Proc Natl Acad Sci USA 103: 2098–2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaus K, Chklovskaia E, Fazekas de St Groth B, Jessup W, Harder T (2005) Condensation of the plasma membrane at the site of T lymphocyte activation. J Cell Biol 171: 121–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheber LA, Edidin M (1999) A model for membrane patchiness: lateral diffusion in the presence of barriers and vesicle traffic. Biophys J 77: 3163–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell S, Stelzer EHK (1992) Properties of a 4Pi confocal fluorescence microscope. J Opt Soc Am A 9: 2159–2166 [Google Scholar]
- Hofmann M, Eggeling C, Jakobs S, Hell SW (2005) Breaking the diffraction barrier in fluorescence microscopy at low light intensities by using reversibly photoswitchable proteins. Proc Natl Acad Sci USA 102: 17565–17569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J, Gheber LA, Margolis L, Edidin M (1998) Domains in cell plasma membranes investigated by near-field scanning optical microscopy. Biophys J 74: 2184–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ianoul A, Grant DD, Rouleau Y, Bani-Yaghoub M, Johnston LJ, Pezacki JP (2005) Imaging nanometer domains of beta-adrenergic receptor complexes on the surface of cardiac myocytes. Nat Chem Biol 1: 196–202 [DOI] [PubMed] [Google Scholar]
- Karnovsky MJ, Kleinfeld AM, Hoover RL, Klausner RD (1982) The concept of lipid domains in membranes. J Cell Biol 94: 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenworthy AK, Nichols BJ, Remmert CL, Hendrix GM, Kumar M, Zimmerberg J, Lippincott-Schwartz J (2004) Dynamics of putative raft-associated proteins at the cell surface. J Cell Biol 165: 735–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar TA, Jakobs S, Dyba M, Egner A, Hell SW (2000) Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc Natl Acad Sci USA 97: 8206–8210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korlach J, Schwille P, Webb WW, Feigenson GW (1999) Characterization of lipid bilayer phases by confocal microscopy and fluorescence correlation spectroscopy. Proc Natl Acad Sci USA 96: 8461–8466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumi A, Nakada C, Ritchie K, Murase K, Suzuki K, Murakoshi H, Kasai RS, Kondo J, Fujiwara T (2005) Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: high-speed single-molecule tracking of membrane molecules. Annu Rev Biophys Biomol Struct 34: 351–378 [DOI] [PubMed] [Google Scholar]
- Lagerholm BC, Weinreb GE, Jacobson K, Thompson NL (2005) Detecting microdomains in intact cell membranes. Annu Rev Phys Chem 56: 309–336 [DOI] [PubMed] [Google Scholar]
- Larson DR, Gosse JA, Holowka DA, Baird BA, Webb WW (2005) Temporally resolved interactions between antigen-stimulated IgE receptors and Lyn kinase on living cells. J Cell Biol 171: 527–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenne PF, Wawrezinieck L, Conchonaud F, Wurtz O, Boned A, Guo XJ, Rigneault H, He HT, Marguet D (2006) Dynamic molecular confinement in the plasma membrane by microdomains and the cytoskeleton meshwork. EMBO J (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Wang H, Cheng JX (2005) Quantitative coherent anti-stokes Raman scattering imaging of lipid distribution in coexisting domains. Biophys J 89: 3480–3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidke DS, Lidke KA, Rieger B, Jovin TM, Arndt-Jovin DJ (2005) Reaching out for signals: filopodia sense EGF and respond by directed retrograde transport of activated receptors. J Cell Biol 170: 619–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Roberts TH, Hirschberg K (2000) Secretory protein trafficking and organelle dynamics in living cells. Annu Rev Cell Dev Biol 16: 557–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommerse PH, Blab GA, Cognet L, Harms GS, Snaar-Jagalska BE, Spaink HP, Schmidt T (2004) Single-molecule imaging of the H-ras membrane-anchor reveals domains in the cytoplasmic leaflet of the cell membrane. Biophys J 86: 609–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- London E (2005) How principles of domain formation in model membranes may explain ambiguities concerning lipid raft formation in cells. Biochim Biophys Acta 1746: 203–220 [DOI] [PubMed] [Google Scholar]
- Marguet D, Spiliotis ET, Pentcheva T, Lebowitz M, Schneck J, Edidin M (1999) Lateral diffusion of GFP-tagged H2Ld molecules and of GFP-TAP1 reports on the assembly and retention of these molecules in the endoplasmic reticulum. Immunity 11: 231–240 [DOI] [PubMed] [Google Scholar]
- McConnell HM, Vrljic M (2003) Liquid–liquid immiscibility in membranes. Annu Rev Biophys Biomol Struct 32: 469–492 [DOI] [PubMed] [Google Scholar]
- Meder D, Moreno MJ, Verkade P, Vaz WL, Simons K (2006) Phase coexistence and connectivity in the apical membrane of polarized epithelial cells. Proc Natl Acad Sci USA 103: 329–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medintz IL, Uyeda HT, Goldman ER, Mattoussi H (2005) Quantum dot bioconjugates for imaging, labelling and sensing. Nat Mater 4: 435–446 [DOI] [PubMed] [Google Scholar]
- Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S (2005) Quantum dots for live cells, in vivo imaging, and diagnostics. Science 307: 538–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolini C, Baranski J, Schlummer S, Palomo J, Lumbierres-Burgues M, Kahms M, Kuhlmann J, Sanchez S, Gratton E, Waldmann H, Winter R (2006) Visualizing association of N-Ras in lipid microdomains: influence of domain structure and interfacial adsorption. J Am Chem Soc 128: 192–201 [DOI] [PubMed] [Google Scholar]
- Petersen NO, Hoddelius PL, Wiseman PW, Seger O, Magnusson KE (1993) Quantitation of membrane receptor distributions by image correlation spectroscopy: concept and application. Biophys J 65: 1135–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons T, Moreaux L, Mongin O, Blanchard-Desce M, Mertz J (2003) Mechanisms of membrane potential sensing with second-harmonic generation microscopy. J Biomed Opt 8: 428–431 [DOI] [PubMed] [Google Scholar]
- Potma EO, Xie XS (2005) Direct visualization of lipid phase segregation in single lipid bilayers with coherent anti-stokes Raman scattering microscopy. Chem Phys Chem 6: 77–79 [DOI] [PubMed] [Google Scholar]
- Pralle A, Keller P, Florin EL, Simons K, Horber JK (2000) Sphingolipid–cholesterol rafts diffuse as small entities in the plasma membrane of mammalian cells. J Cell Biol 148: 997–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucadyil TJ, Kalipatnapu S, Harikumar KG, Rangaraj N, Karnik SS, Chattopadhyay A (2004) G-protein-dependent cell surface dynamics of the human serotonin1A receptor tagged to yellow fluorescent protein. Biochemistry 43: 15852–15862 [DOI] [PubMed] [Google Scholar]
- Roke S, Schins J, Muller M, Bonn M (2003) Vibrational spectroscopic investigation of the phase diagram of a biomimetic lipid monolayer. Phys Rev Lett 90: 128101. [DOI] [PubMed] [Google Scholar]
- Saffman PG, Delbruck M (1975) Brownian motion in biological membranes. Proc Natl Acad Sci USA 72: 3111–3113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sako Y, Kusumi A (1994) Compartmentalized structure of the plasma membrane for receptor movements as revealed by a nanometer-level motion analysis. J Cell Biol 125: 1251–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Andres A, Chen Y, Muller JD (2005) Molecular brightness determined from a generalized form of Mandel's Q-parameter. Biophys J 89: 3531–3547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawano A, Takayama S, Matsuda M, Miyawaki A (2002) Lateral propagation of EGF signaling after local stimulation is dependent on receptor density. Dev Cell 3: 245–257 [DOI] [PubMed] [Google Scholar]
- Saxton MJ (1989) The spectrin network as a barrier to lateral diffusion in erythrocytes. A percolation analysis. Biophys J 55: 21–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton MJ, Jacobson K (1997) Single-particle tracking: applications to membrane dynamics. Annu Rev Biophys Biomol Struct 26: 373–399 [DOI] [PubMed] [Google Scholar]
- Schutz GJ, Kada G, Pastushenko VP, Schindler H (2000) Properties of lipid microdomains in a muscle cell membrane visualized by single molecule microscopy. EMBO J 19: 892–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Steinbach PA, Tsien RY (2005) A guide to choosing fluorescent proteins. Nat Methods 2: 905–909 [DOI] [PubMed] [Google Scholar]
- Sheetz MP, Schindler M, Koppel DE (1980) Lateral mobility of integral membrane proteins is increased in spherocytic erythrocytes. Nature 285: 510–511 [DOI] [PubMed] [Google Scholar]
- Shinbrot T, Muzzio FJ (2001) Noise to order. Nature 410: 251–258 [DOI] [PubMed] [Google Scholar]
- Shvartsman DE, Kotler M, Tall RD, Roth MG, Henis YI (2003) Differently anchored influenza hemagglutinin mutants display distinct interaction dynamics with mutual rafts. J Cell Biol 163: 879–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K, Vaz WL (2004) Model systems, lipid rafts, and cell membranes. Annu Rev Biophys Biomol Struct 33: 269–295 [DOI] [PubMed] [Google Scholar]
- Singer SJ, Nicolson GL (1972) The fluid mosaic model of the structure of cell membranes. Science 175: 720–731 [DOI] [PubMed] [Google Scholar]
- Sprong H, van Der Sluijs P, van Meer G (2001) How proteins move lipids and lipids move proteins. Nat Rev Mol Cell Biol 2: 504–513 [DOI] [PubMed] [Google Scholar]
- Tang Q, Edidin M (2001) Vesicle trafficking and cell surface membrane patchiness. Biophys J 81: 196–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardin C, Cognet L, Bats C, Lounis B, Choquet D (2003) Direct imaging of lateral movements of AMPA receptors inside synapses. EMBO J 22: 4656–4665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner MS, Sens P, Socci ND (2005) Nonequilibrium raftlike membrane domains under continuous recycling. Phys Rev Lett 95: 168301. [DOI] [PubMed] [Google Scholar]
- van Meer G (2005) Cellular lipidomics. EMBO J 24: 3159–3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verveer PJ, Wouters FS, Reynolds AR, Bastiaens PI (2000) Quantitative imaging of lateral ErbB1 receptor signal propagation in the plasma membrane. Science 290: 1567–1570 [DOI] [PubMed] [Google Scholar]
- Wawrezinieck L, Rigneault H, Marguet D, Lenne P-F (2005) Fluorescence correlation spectroscopy diffusion laws to probe the submicron cell membrane organization. Biophys J 89: 4029–4042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurpel GW, Rinia HA, Muller M (2005) Imaging orientational order and lipid density in multilamellar vesicles with multiplex CARS microscopy. J Microsc 218: 37–45 [DOI] [PubMed] [Google Scholar]
- Yechiel E, Edidin M (1987) Micrometer-scale domains in fibroblast plasma membranes. J Cell Biol 105: 755–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumbusch A, Holtom GR, Xie XS (1999) Vibrationnal microscopy using coherent anti-stokes Raman scattering. Phys Rev Lett 82: 4014–4017 [Google Scholar]


