Abstract
The atpE gene encoding the subunit c of the ATP synthase of Mycobacterium tuberculosis, the target of the new diarylquinoline drug R207910, has been sequenced from in vitro mutants resistant to the drug. The previously reported mutation A63P and a new mutation, I66M, were found. The genetic diversity of atpE in 13 mycobacterial species was also investigated, revealing that the region involved in resistance to R207910 is conserved, except in Mycobacterium xenopi in which the highly conserved residue Ala63 is replaced by Met, a modification that may be associated with the natural resistance of M. xenopi to R207910.
R207910 (also known as TMC207) is the lead compound of a series of recently discovered diarylquinolines (DARQs) (1). This new drug, exquisitely active against a broad range of mycobacteria, may significantly improve the treatment of tuberculosis. As it inhibits a new target, R207910 is active against both drug-sensitive and drug-resistant isolates of Mycobacterium tuberculosis (1).
The initial identification of the target of R207910 relied on sequence analysis of a single mutant of M. tuberculosis and two mutants of the fast-growing organism Mycobacterium smegmatis that were resistant to R207910 and harbored two mutations (D32V and A63P, respectively) in the subunit c of ATP synthase encoded by the atpE gene (1). This enzyme contains two structural domains, F0 and F1 (8). F0 includes 1 subunit a, 2 subunits b, and 9 to 12 subunits c made of two α-helices connected by a short loop and arranged in a symmetrical disk. The two mutations, A63P and D32V, affect the α-helices in subunit c and are located in the vicinity of the key glutamic acid residue (E61) involved in proton transport. Because these previous studies were conducted on a limited number of mutants and mycobacterial species, we have undertaken the investigation of atpE from new M. tuberculosis in vitro mutants and from various mycobacterial species. Here, we report on the isolation of new M. tuberculosis in vitro mutants resistant to R207910 and the characterization of the corresponding subunit c sequences. We also describe the heterogeneity of this protein in the Mycobacterium genus, which was evaluated by sequencing the atpE gene from 13 different mycobacterial species.
The isolation of the mutants resistant to R207910 was carried out by spreading 100 μl of a culture of M. tuberculosis H37Rv (108 to 1010 CFU/ml) onto 7H11 plus OADC (oleic acid, albumin, dextrose, and catalase [Serlabo, Bonneuil sur Marne, France]) agar containing from 0.03 μg/ml (the MIC for H37Rv) to 0.5 μg/ml of R207910. After 3 to 6 weeks of incubation at 37°C, seven mutants of M. tuberculosis H37Rv resistant to R207910 were selected on R207910 at concentrations of 0.12 μg/ml for the mutants BK18, BK19, and BK21; 0.25 μg/ml for BK13, BK14, and BK15; and 0.5 μg/ml for BK11.
The atpE gene, as well as the 78-bp upstream and 121-bp downstream regions, were amplified using the degenerate primers atpBS [5′TGTA(CT)TTCAGCCA(AG)GC(GC)ATGG3′] and atpFAS [5′CCGTT(GC)GG(AGT)A(GCT)GAGGAAGTTG 3′] (Eurogentec, Belgium) (boldface indicates degenerate bases in the primers), designed from the sequences of the atpB and atpF genes located upstream and downstream of atpE in the ATP synthase operon. Using these two primers, we first confirmed the amino acid conservation of subunit c in M. tuberculosis by sequencing atpE in 20 nonrelated susceptible clinical isolates (data not shown). Using the same primers, we determined and compared the sequences obtained from the resistant mutants and found in two of them the presence of the mutation A63P previously described in the strain BK12 resistant to R207910 (1). Strikingly, in the five other mutants, the Ala residue found at position 63 in M. tuberculosis H37Rv was conserved while Ile66 was found to be replaced by a methionine (I66M) (Fig. 1).
FIG. 1.
Multiple sequence alignment for subunit c proteins from 13 mycobacterial species. Species abbreviations: M.abs, M. abscessus (GenBank accession no. DQ306899); M.che, M. chelonae (GenBank accession no. DQ306894); M.mar, M. marinum (GenBank accession no. DQ306898); M.ulc, M. ulcerans (GenBank accession no. DQ306897); M.bov, M. bovis (GenBank accession no. DQ306895); Mtb, M. tuberculosis H37Rv (Swiss-Prot accession no. Q10598); M.avi, M. avium (GenBank accession no. DQ378275); M.int, M. intracellulare (GenBank accession no. DQ378276); M.xen, M. xenopi (GenBank accession no. DQ306893); M.lep, M. leprae (GenBank accession no. DQ306896); M.per, M. peregrinum (GenBank accession no. DQ378277); M.for, M. fortuitum (GenBank accession no. DQ378278); M.sme, M. smegmatis (GenBank accession no. DQ306892). The proton-binding glutamic acid is indicated by an arrow. Gray shading shows conserved amino acids. The mutated positions found in the drug-resistant strains of M. tuberculosis (positions 63 and 66) and M. smegmatis (position 32) are boxed. The mutated residues are indicated below the boxes.
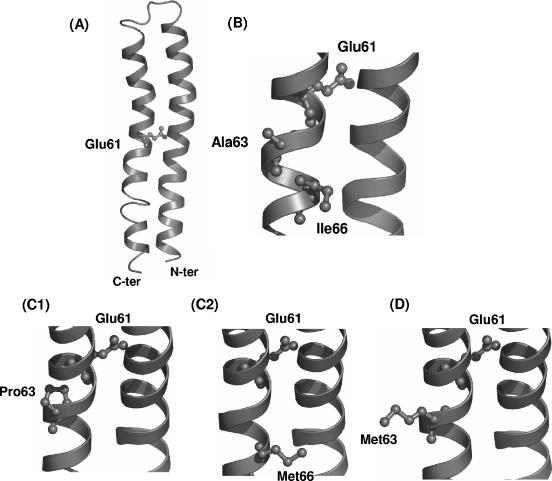
The impact of these mutations was investigated by building a model structure of the monomeric subunit c of M. tuberculosis H37Rv using the homology-modeling server SWISS-MODEL (5-7) with the three-dimensional structure of subunit c from E. coli as a template (Protein Data Bank accession no. 1A91) (4). As shown on Fig. 2, Ala63 and Ile66, which were found to be modified in the two mutants, are positioned in the vicinity (5.7 Å and 9.4 Å, respectively) of the essential residue Glu61, the carboxyl side chain of which permits the proton transfer required for the creation of ATP (2) (Fig. 2A and B). A63P directly affects the α-helix in the Glu61 region (Fig. 2C1), whereas I66M is more distant but introduces significant steric hindrance at the surface of the α-helix in the same area (Fig. 2C2). It can be hypothesized that the two mutations Ala63Pro and Ile66Met, which occur in a region critical for the ATP synthase activity, affect the interactions between R207910 and the c subunits in a region of the ATP synthase where the diarylquinoline could bind.
FIG. 2.
Three-dimensional models of the subunit c from M. tuberculosis H37Rv (A, B, and C) and M. xenopi (D). (A) Ribbon representation of the monomeric subunit c from M. tuberculosis H37Rv (residues 1 to 75). The side chain of residue Glu61 is shown. (B) Closer view of the Glu61 region. The two residues Ala63 and Ile66 found in AtpE of strain H37Rv susceptible to R207910 are shown. (C and D) The same view showing the two mutated residues Pro63 (C1) and Met66 (C2) identified in the H37Rv mutants resistant to R207910 and the Met63 (D) found specifically in the subunit c from M. xenopi, respectively.
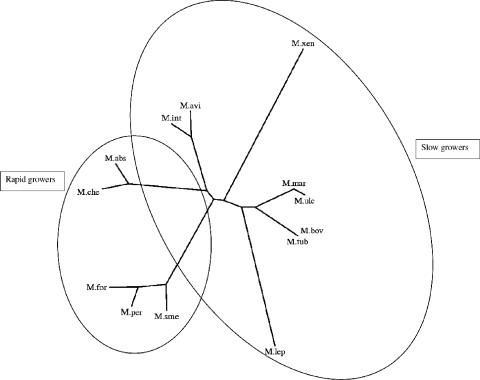
The genetic diversity of atpE was investigated by amplifying with atpBS and atpFAS the atpE genes from 13 mycobacterial species (Table 1). The degree of nucleotide identity found between these genes is high, the lowest identity being 73% (Table 1). The phylogenetic tree represented on Fig. 3 indicates that the atpE sequences can be grouped in seven distinct clusters corresponding well to those established from 16S rRNA sequencing (9) and clearly differentiates the slow growers from the rapid growers. The amino acid sequence alignment shown in Fig. 1 confirms that the degree of identity at the protein level is very high, from 90% for Mycobacterium xenopi to 100% for Mycobacterium bovis compared to M. tuberculosis H37Rv. The amino acid variations are scattered throughout the polypeptide sequence, with a higher level of divergence at the N- and C-terminus extremities. Interestingly, residues D32, A63, and I66, are highly conserved in the species included in this study, except in M. xenopi, for which residue 63 is neither an alanine, as found in the susceptible strain of H37Rv, nor a proline, as found in the mutant strains of H37Rv resistant to R207910, but a methionine (Fig. 1). We confirmed the presence of this methionine at position 63 by sequencing atpE in seven nonrelated strains of M. xenopi. The presence of this specific residue at a position clearly involved in resistance to R207910 in M. tuberculosis may be associated with the high MIC of R207910 (4 μg/ml) observed for M. xenopi, which is a species regarded as naturally resistant to the drug (1). Accordingly, in the model of the subunit c of M. xenopi (Fig. 2D), the bulky side chain of Met63 lies in the vicinity of Glu61, supporting the hypothesis that the role played by Met63 in the natural resistance of M. xenopi to R207910 could be similar to the one played by the two other mutations, A63P and I66M, found in the in vitro M. tuberculosis mutants.
TABLE 1.
Percentages of identity worked out from pairwise alignments of atpE genes from 13 mycobacterial species
| Species | % Identity to:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M. tuberculosis | M. ulcerans | M. abscessus | M. leprae | M. bovis | M. marinum | M. xenopi | M. chelonae | M. smegmatis | M. avium | M. intracellulare | M. fortuitum | M. peregrinum | |
| M. tuberculosis | 100 | 91 | 84 | 84 | 100 | 92 | 78 | 83 | 82 | 86 | 86 | 79 | 81 |
| M. ulcerans | 100 | 83 | 83 | 91 | 99 | 80 | 84 | 84 | 84 | 84 | 77 | 79 | |
| M. abscessus | 100 | 77 | 84 | 85 | 77 | 96 | 82 | 84 | 85 | 77 | 78 | ||
| M. leprae | 100 | 84 | 83 | 74 | 76 | 75 | 80 | 80 | 73 | 75 | |||
| M. bovis | 100 | 92 | 78 | 83 | 82 | 86 | 86 | 79 | 81 | ||||
| M. marinum | 100 | 81 | 84 | 86 | 85 | 85 | 80 | 82 | |||||
| M. xenopi | 100 | 77 | 74 | 76 | 77 | 75 | 76 | ||||||
| M. chelonae | 100 | 80 | 85 | 86 | 77 | 78 | |||||||
| M. smegmatis | 100 | 83 | 83 | 94 | 93 | ||||||||
| M. avium | 100 | 96 | 81 | 82 | |||||||||
| M. intracellulare | 100 | 81 | 82 | ||||||||||
| M. fortuitum | 100 | 95 | |||||||||||
| M. peregrinum | 100 | ||||||||||||
FIG. 3.
Phylogenetic tree of atpE genes from 13 mycobacterial species. The tree was created using PHYLIP (3). Branch lengths correspond to the number of nucleotides exchanges of the atpE genes. Species abbreviations: M.abs, M. abscessus (GenBank accession no. DQ306899); M.che, M. chelonae (GenBank accession no. DQ306894); M.mar, M. marinum (GenBank accession no. DQ306898); M.ulc, M. ulcerans (GenBank accession no. DQ306897); M.bov, M. bovis (GenBank accession no. DQ306895); Mtub, M. tuberculosis H37Rv (Swiss-Prot accession no. Q10598); M.avi, M. avium (GenBank accession no. DQ378275); M.int, M. intracellulare (GenBank accession no. DQ378276); M.xen, M. xenopi (GenBank accession no. DQ306893); M.lep, M. leprae (GenBank accession no. DQ306896); M.per, M. peregrinum (GenBank accession no. DQ378277); M.for, M. fortuitum (GenBank accession no. DQ378278); M.sme, M. smegmatis (GenBank accession no. DQ306892).
Nucleotide sequence accession numbers.
The GenBank accession numbers for the atpE genes from the mycobacteria M. xenopi, M. ulcerans, M. marinum, M. bovis, M. leprae, M. abscessus, M. chelonae, M. smegmatis, M. avium, M. intracellulare, M. fortuitum, and M. peregrinum are DQ306893, DQ306897, DQ306898, DQ306895, DQ306896, DQ306899, DQ306894, DQ306892, DQ378275, DQ378276, DQ378278, and DQ378277, respectively.
Acknowledgments
We thank Anil Koul and Nacer Lounis for helpful discussions. We are indebted to Chantal Truffot-Pernot for her advice and technical assistance.
REFERENCES
- 1.Andries, K., P. Verhasselt, J. Guillemont, H. W. Gohlmann, J. M. Neefs, H. Winkler, J. Van Gestel, P. Timmerman, M. Zhu, E. Lee, P. Williams, D. de Chaffoy, E. Huitric, S. Hoffner, E. Cambau, C. Truffot-Pernot, N. Lounis, and V. Jarlier. 2005. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 307:223-227. [DOI] [PubMed] [Google Scholar]
- 2.Dmitriev, O. Y., F. Abildgaard, J. L. Markley, and R. H. Fillingame. 2002. Structure of Ala24/Asp61→Asp24/Asn61 substituted subunit c of Escherichia coli ATP synthase: implications for the mechanism of proton transport and rotary movement in the F0 complex. Biochemistry 41:5537-5547. [DOI] [PubMed] [Google Scholar]
- 3.Felsenstein, J. 1997. An alternating least squares approach to inferring phylogenies from pairwise distances. Syst. Biol. 46:101-111. [DOI] [PubMed] [Google Scholar]
- 4.Girvin, M. E., V. K. Rastogi, F. Abildgaard, J. L. Markley, and R. H. Fillingame. 1998. Solution structure of the transmembrane H+-transporting subunit c of the F1F0 ATP synthase. Biochemistry 37:8817-8824. [DOI] [PubMed] [Google Scholar]
- 5.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 6.Peitsch, M. C. 1995. Protein modeling by E-mail. Bio/Technology 13:658-660. [Google Scholar]
- 7.Schwede, T., J. Kopp, N. Guex, and M. C. Peitsch. 2003. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 31:3381-3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stock, D., C. Gibbons, I. Arechaga, A. G. Leslie, and J. E. Walker. 2000. The rotary mechanism of ATP synthase. Curr. Opin. Struct. Biol. 10:672-679. [DOI] [PubMed] [Google Scholar]
- 9.Tortoli, E. 2003. Impact of genotypic studies on mycobacterial taxonomy: the new mycobacteria of the 1990s. Clin. Microbiol. Rev. 16:319-354. [DOI] [PMC free article] [PubMed] [Google Scholar]