Abstract
The molecular basis for isoniazid resistance in Mycobacterium tuberculosis is complex. Putative isoniazid resistance mutations have been identified in katG, ahpC, inhA, kasA, and ndh. However, small sample sizes and related potential biases in sample selection have precluded the development of statistically valid and significant population genetic analyses of clinical isoniazid resistance. We present the first large-scale analysis of 240 alleles previously associated with isoniazid resistance in a diverse set of 608 isoniazid-susceptible and 403 isoniazid-resistant clinical M. tuberculosis isolates. We detected 12 mutant alleles in isoniazid-susceptible isolates, suggesting that these alleles are not involved in isoniazid resistance. However, mutations in katG, ahpC, and inhA were strongly associated with isoniazid resistance, while kasA mutations were associated with isoniazid susceptibility. Remarkably, the distribution of isoniazid resistance-associated mutations was different in isoniazid-monoresistant isolates from that in multidrug-resistant isolates, with significantly fewer isoniazid resistance mutations in the isoniazid-monoresistant group. Mutations in katG315 were significantly more common in the multidrug-resistant isolates. Conversely, mutations in the inhA promoter were significantly more common in isoniazid-monoresistant isolates. We tested for interactions among mutations and resistance to different drugs. Mutations in katG, ahpC, and inhA were associated with rifampin resistance, but only katG315 mutations were associated with ethambutol resistance. There was also a significant inverse association between katG315 mutations and mutations in ahpC or inhA and between mutations in kasA and mutations in ahpC. Our results suggest that isoniazid resistance and the evolution of multidrug-resistant strains are complex dynamic processes that may be influenced by interactions between genes and drug-resistant phenotypes.
Isoniazid (INH) is one of the most effective and specific agents for the treatment of infections with Mycobacterium tuberculosis. INH is the cornerstone of treatment for drug-susceptible tuberculosis, and it is also widely used to treat latent M. tuberculosis infections. Recent increases in INH-resistant (INHr) and multidrug-resistant (MDR) tuberculosis are jeopardizing the continued utility of this drug (13, 61). Furthermore, the development of INH resistance is a common first step in the evolution to MDR (11). Thus, there has been considerable interest in identifying the molecular basis of INH resistance in clinical M. tuberculosis isolates.
INH is a prodrug that requires activation by the catalase-peroxidase enzyme encoded by the katG gene (65). Activated INH appears to disrupt the synthesis of essential mycolic acids by inhibiting the NADH-dependent enoyl-ACP reductase enzyme encoded by inhA (45). INH resistance is likely to arise through multiple molecular mechanisms, only a subset of which have been fully characterized. Between 40 and 95% of INHr clinical M. tuberculosis isolates have mutations in katG, 75 to 90% of which are located in codon 315, with 10 to 25% of mutations located in other katG loci (12, 35, 37, 42-44, 57, 64). Mutations in katG315 may be favored because mutations at this location appear to decrease INH activation without abolishing catalase-peroxidase activity, a potential virulence factor (22, 41, 49, 59). M. tuberculosis may compensate for katG mutations by overexpressing the ahpC gene (52, 54). INH resistance can also develop through alterations or overexpression of the INH drug target InhA, and 0 to 5% of INHr M. tuberculosis isolates have mutations in the inhA open reading frame (ORF), while 8 to 20% have mutations in the inhA promoter (38, 42, 43, 64). Mutations in ndh, a gene encoding an NADH dehydrogenase, were recently found to confer resistance to INH and ethionamide in M. bovis (58). The ndh mutants had altered NADH/NAD ratios, which appeared to protect them from INH-mediated toxicity. Mutations in at least 16 other genes have been reported to be associated with INH resistance in clinical isolates (42-44). However, the roles of these genes in INH resistance (if any) remain unclear. Furthermore, approximately 10% to 25% of INH-resistant strains do not contain mutations in any known gene targets for INH resistance (42-44, 64). These results demonstrate the need for further investigations in this field.
The sequencing studies that form the basis of our understanding of INH resistance-associated mutations in clinical M. tuberculosis isolates have had a number of important limitations. Most studies have examined relatively small numbers of isolates or have failed to include sufficient numbers of drug-susceptible controls (10, 12, 25, 27-29, 31, 38-40, 43, 44, 47, 55, 64). Even studies that were somewhat larger did not usually include enough INHr and INH-susceptible (INHs) isolates to demonstrate statistically significant associations between specific mutations and INH resistance (10, 12, 25, 27-30, 33, 38, 43, 44, 47), and none of these have been large enough to perform more detailed analyses. Furthermore, much of what is known about INH resistance-associated mutations was discovered by testing collections of predominantly MDR isolates (14, 31, 47). However, as demonstrated in a large study of ethambutol (EMB) resistance (18), MDR isolates may not have the same distribution of mutations as monoresistant M. tuberculosis isolates (4, 19, 25).
Here we present an in-depth study of INH resistance-associated mutations in a large and geographically diverse set of INHs and INHr clinical isolates of M. tuberculosis. Our goals were to determine if any point mutation previously classified as an INH resistance-associated mutation was detectable in INHs isolates and to test for statistically valid associations between mutations and drug resistance. Interactions between different mutations were also studied as a mean to better understand the biology of INH resistance. Finally, we aimed to test the hypotheses that certain mutations were concentrated in MDR isolates compared to monoresistant isolates and that MDR isolates had an excess accumulation of multiple INH resistance-associated mutations. Our results have important implications in understanding how INH resistance arises and continues to evolve in MDR isolates.
MATERIALS AND METHODS
M. tuberculosis clinical isolates.
A total of 1,011 clinical M. tuberculosis isolates were obtained from reference laboratories in Australia, Colombia, India, Mexico, New York City, Spain, and Texas (Table 1). The 56 samples from Australia consisted of all isolates resistant to at least INH collected in the state of Victoria at the Victorian Mycobacterium Reference Laboratory in Melbourne during 2001 and 2002 and 26 randomly selected pan-susceptible control isolates from the same period (27). The samples from Colombia consisted of 133 isolates resistant to at least INH, 159 pan-susceptible isolates, and 7 rifampin (RIF)-monoresistant isolates that were selected from the strain bank of the Mycobacterial Group, Instituto Nacional de Salud, composed of clinical M. tuberculosis isolates collected from all regions of Colombia between 1992 and 2000. The samples from India consisted of all 35 drug-resistant isolates of any type identified in the Vallabhbhai Patel Chest Institute in New Delhi between January 2001 and January 2002. These included 10 streptomycin (STR)-monoresistant, 22 INHr (2 of which were mono-INHr), and 10 randomly selected pan-susceptible control isolates from the same period. The samples from Mexico consisted of 121 INHr (42 of which were mono-INHr) and 76 INHs clinical isolates randomly selected from genotyped isolates collected between 1991 and 1996 in Mexico City, Huauchinango, Puebla, Orizaba, and Veracruz, 3 of which were monoresistant to STR and 73 of which were pan-susceptible controls (7, 16, 23), obtained from the Laboratory of Clinical Microbiology, Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, Mexico City. The samples from Spain consisted of all 109 isolates collected at the General Penitentiary Hospital, Microbiology Laboratory, from a longitudinal study of tuberculosis performed between January 1993 and June 1994 (8). These samples comprised 11 INHr isolates (1 of which was mono-INHr), 1 STR-monoresistant isolate, and 97 pan-susceptible controls. The 65 samples from Texas consisted of all the INHr isolates (30 mono-INHr) collected by the Texas Department of Health and the University of Texas Health Center at Tyler between May 1992 and August 1994 and 91 INHs isolates collected in a population-based study of Tarrant County between May 1992 and December 1996 (60, 62) which were submitted to the mycobacteriology research laboratory of the Central Arkansas Veterans Healthcare System for genotype analysis. Samples from New York City consisted of all INHr isolates (22 of which were monoresistant) for which DNA was available among isolates collected at the Montefiore Medical Center, Bronx, N.Y., between 1989 and 1996 and 124 INHs randomly selected controls (1).
TABLE 1.
Clinical M. tuberculosis isolates included in the study
| Geographic origin | No. (%) of isolates
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Total | Pan-susceptible | INHr | Mono-INHr | MDR | Resistant to indicated no. of drugs
|
||||
| 1 | 2 | 3 | 4 | ||||||
| Australia | 56 | 26 (46) | 29 (52) | 20 (36) | 4 (7) | 21 (38) | 6 (11) | 3 (5) | 0 |
| Colombia | 302 | 159 (53) | 133 (44) | 3 (1) | 128 (46) | 10 (3) | 32 (11) | 68 (23) | 33 (11) |
| India | 45 | 10 (21) | 22 (49) | 2 (4) | 16 (36) | 12 (27) | 12 (27) | 8 (17) | 3 (7) |
| Mexico | 197 | 73 (37) | 121 (61) | 43 (22) | 65 (33) | 46 (23) | 54 (26) | 15 (8) | 9 (5) |
| New York | 146 | 119 (82) | 22 (15) | 3 (2) | 18 (12) | 7 (5) | 8 (5) | 2 (1) | 10 (7) |
| Spain | 109 | 97 (89) | 11 (10) | 1 (1) | 9 (8) | 2 (2) | 6 (5) | 4 (4) | 0 |
| Texas | 156 | 91 (58) | 65 (13) | 30 (19) | 32 (21) | 30 (19) | 10 (7) | 8 (5) | 17 (11) |
| Total | 1011 | 575 (57) | 403 (40) | 102 (10) | 272 (27) | 128 (13) | 128 (13) | 108 (11) | 72 (7) |
Drug susceptibility testing and strain typing.
All isolates included in this study were subjected to susceptibility testing and DNA fingerprinting as described below. Each center or laboratory performed susceptibility testing with at least INH, RIF, STR, and EMB by the agar proportions method (24) (Colombia, India, New York, and Spain), the BACTEC MGIT 960 method (20) (Australia), or the radiometric BACTEC 460 method (30) (Mexico and Texas). The breakpoint concentrations used for INH were 0.2 μg/ml for the agar proportions method and 0.1 μg/ml for the BACTEC MGIT 960 system and BACTEC 460 methods. DNA fingerprinting (either IS6110 or variable-number tandem repeat fingerprinting and spoligotyping [1, 6, 15, 21]) demonstrated that this study population included 343 unique and 265 clustered INHs isolates and 196 unique and 207 clustered INHr isolates.
Detection of INH resistance-associated mutations in clinical isolates of M. tuberculosis.
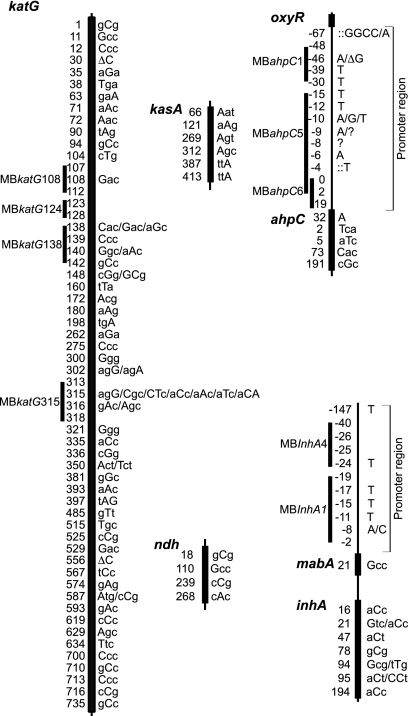
We tested each isolate for virtually all single nucleotide polymorphism (SNP) mutations in the M. tuberculosis katG, kasA, mabA, inhA, oxyR, ahpC, and ndh genes (or their upstream promoter regions) that have been found in association with INH resistance in previously published studies (Fig. 1) (3, 10, 12, 14, 25, 28, 29, 31, 34, 35, 37, 38, 40, 42-44, 46, 48, 51, 52, 54, 56, 58, 59, 65). A total of 204 INH resistance-associated alleles were detected by using 106 high-throughput hairpin-shaped-primer (HP) assays (see Table S1 in the supplemental material) as described previously (17). Regions with high concentrations of potential SNPs (e.g., the promoter regions of inhA and ahpC) and positions 103 to 113, 123 to 128, and 137 to 143 of katG (which account for 36 alleles) were screened using nine molecular beacon assays that were designed to detect only wild-type DNA sequences in those regions (see Table S2 in the supplemental material). Resistance-associated SNPs in negative molecular beacon assays (which indicated that the sequence diverged from the wild-type sequence) were confirmed by DNA sequencing. Additional screening for mutations in katG315 was confirmed by using a previously described molecular beacon assay (40). Finally, the katG gene was tested for the presence of large deletions that could also be associated with INH resistance by scanning for regions where adjacent HP assays suggested the absence of DNA target sequences. Each deletion in katG was confirmed by Southern blot analysis.
FIG. 1.
Alleles screened in this study. The ORF of each gene is shown as a thick line. Intergenic regions are indicated by thin lines. The position of each allele examined in this study is indicated to the left of the lines. Alleles tested inside an ORF are indicated by their codon positions; alleles tested within an intergenic region are indicated by their nucleotide positions relative to the start of the following ORF, except for ahpC, which is shown relative to the transcription start. The specific mutations examined in the HP assays are indicated in capital letters to the right of each gene. Several mutations are indicated when more than one mutant allele was tested at the same codon or nucleotide position. Deletions are indicated with the symbol “Δ,” and insertions are indicated with colons. Question marks (?) indicate the presence of a mutation detected and confirmed by a molecular beacon assay, but the exact nucleotide change was not identified because the sequencing results are inconclusive at that position. The total sequence examined by each molecular beacon is indicated by a line labeled with the name of that molecular beacon, starting with an MB prefix. Sequence accession numbers for katG, ahpC, kasA, ndh, and inhA are Z97193, Z79701, Z70692, BX842577, and Z79701, respectively. Genes and upstream regions are not drawn to scale. The sequences of the primers, molecular beacons, and sequencing primers used to detect or confirm these alleles are shown in Tables S1 to S3 in the supplemental material.
DNA sequencing and other confirmatory testing.
During the course of the study, we performed 220 different sequencing reactions (using BigDye Terminator kits from ABI) to confirm the alleles assigned by the HP assays and to identify the specific mutations indicated by the molecular beacon assay results. Sequencing primer sequences and conditions are indicated in Table S3 in the supplemental material. We also retested the susceptibility patterns to INH, RIF, STR, and EMB of 103 isolates, including all INHs isolates with mutations in katG, inhA, ahpC, or ndh. Totals of seven, one, four, and three discrepancies in the repeat susceptibility results were detected for INH, RIF, EMB, and STR susceptibility, respectively. The drug resistance of each isolate was reconfirmed after performing a third drug susceptibility test in all cases. These final drug susceptibility results were used in all analyses.
Statistical analysis.
Basic descriptive statistics and graphics were employed. Chi-square tests were used to test for individual associations and differences between proportions. Logistic regression was used to build models, with the area under the receiver operating curve used as a general measure of goodness of fit. Chi-square tests for trend were also employed. SAS, version 8.02, was used for all statistical analyses.
RESULTS
Distribution of mutations in INHs and INHr isolates.
In this study, we tested 609 INHs and 402 INHr M. tuberculosis isolates for 240 alleles in five genes that were previously reported to be associated with INH resistance. The most common mutations were found at katG315, as reported previously (3, 40, 42, 44, 56, 57). Mutations in katG315 were identified in 184/403 (46%) INHr isolates (Table 2). The katG S315T mutation was detected in one INHs isolate in this study; however, this isolate was not viable for repeat testing. We suspect that this isolate was truly INHr, and we reclassified it as INHr for subsequent analyses. Five additional INHr isolates (1%) were found by HP PCR scanning and Southern blot analysis to contain deletions of various sizes in the katG gene (data not shown). These five isolates were all catalase negative (32), as would be predicted by the gene deletions. No katG deletions were detected in any of the INHs isolates. Mutations at other katG locations were uncommon, comprising only 17/238 (7%) katG mutants (4% of the total INHr isolates). katG G316S and L587P mutations were found in INHs isolates, while all other katG mutations were found only in INHr isolates.
TABLE 2.
Mutations detected among INHr and INHs isolates
| Mutationa | No. of isolates
|
Mutationa | No. of isolates
|
|||||
|---|---|---|---|---|---|---|---|---|
| INHr | INHs | Total | INHr | INHs | Total | |||
| katG mutations | −46 g→A | 14 | 11 | 25 | ||||
| 123-128-?b | 2 | 2 | −46 ΔG | 1 | 1 | |||
| R128P (cgg→cCg) | 1 | 1 | −39 c→T | 4 | 4 | |||
| N138H (aac→Cac) | 1 | 1 | −30 c→T | 1 | 1 | |||
| N138D (aac→Gac) | 2 | 2 | −15 c→T | 1 | 1 | |||
| N138S (aac→aGc) | 1 | 1 | −12 c→T | 2 | 2 | |||
| A172T (gcg→Acg) | 2 | 2 | −10 c→A | 2 | 2 | |||
| S315R (agc→agG) | 1 | 1 | −10 c→G | 2 | 2 | |||
| S315T (agc→aCc) | 184 | 184 | −10 c→T | 7 | 7 | |||
| S315N (agc→aAc) | 9 | 9 | −9 g→A | 2 | 1 | 3 | ||
| S315I (agc→aTc) | 6 | 6 | −9 g→?b | 2 | 2 | |||
| S315T (agc→aCA) | 21 | 21 | −8 a→?b | 1 | 1 | |||
| G316D (ggc→gAc) | 1 | 1 | −6 g→A | 4 | 4 | |||
| G316S (ggc→Agc) | 2 | 2 | −4::T | 3 | 3 | |||
| W321G (tgg→Ggg) | 1 | 1 | ORF mutations | |||||
| L336P (ctg→cGg) | 1 | 1 | T5I (acc→aTc) | 1 | 1 | |||
| R515C (cgc→Tgc) | 1 | 1 | D73H (gac→Cac) | 7 | 6 | 13 | ||
| L587P (ctg→cCg) | 1 | 1 | Total | 54 | 19 | 73 | ||
| D735A (gac→gCc) | 1 | 1 | ||||||
| Total | 235 | 3 | 238 | inhA mutations | ||||
| Promoter mutationsd | ||||||||
| kasA mutations | −17 g→T | 3 | 1 | 4 | ||||
| D66N (gat→Aat) | 1 | 1 | 2 | −15 c→T | 31 | 31 | ||
| G269S (ggt→Agt) | 36 | 87 | 123 | −8 t→A | 3 | 3 | ||
| G312S (ggc→Agc) | 7 | 12 | 19 | −8 t→C | 1 | 1 | ||
| Total | 44 | 100 | 144 | ORF mutations | ||||
| I21V (atc→Gtc) | 1 | 1 | ||||||
| ndh mutations | I21T (atc→aCc) | 2 | 2 | |||||
| V18A (gtg→gCg) | 12 | 16 | 28 | I47T (att→aCt) | 1 | 1 | ||
| R268H (cgc→cAc) | 2 | 2 | S94A (tcg→Gcg) | 5 | 5 | |||
| Total | 14 | 16 | 30 | I194T (atc→aCc) | 2 | 1 | 3 | |
| Total | 48 | 3 | 51 | |||||
| oxyR-ahpC mutations | ||||||||
| Promoter mutationsc | All mutations | 395 | 141 | 536 | ||||
| −67 g→A | 1 | 1 | ||||||
The mutant codon is indicated, followed by the nucleotide change, shown in capital letters. When the mutation occurs in an intergenic region, the nucleotide change is indicated relative to the start site of the following ORF.
Sequences that were confirmed to be mutants by a molecular beacon assay but whose exact mutation is not known because sequencing results were inconclusive or DNA was unavailable for sequencing.
Positions in the ahpC promoter are situated relative to the transcriptional start residue.
Positions in the inhA promoter are situated relative to the start of the mabA ORF (mabA is the first gene in the mabA-inhA operon).
A total of 144 mutations were detected in kasA, at positions 66, 269, and 312, in 100/608 (16%) INHs isolates and 44/403 (11%) INHr isolates (Table 2). These results definitively show that none of the common mutant kasA alleles are uniquely present in INHr M. tuberculosis. Thus, it appears unlikely that these alterations in kasA result in INH resistance, as has been reported previously (2, 40, 44).
We detected 51 mutations in inhA, in 3/608 (0.5%) INHs isolates and 48/403 (12%) INHr isolates. All promoter mutations (except the −17 G-to-T mutation) were confined to INHr isolates, with the C-to-T mutation at position −15 being the most abundant (Table 2). Two ORF mutations, I47T and I194T, were detected in INHs isolates; however, all other ORF mutations were only detected in INHr isolates. Importantly, inhA S94A mutants, which appear to alter binding of the INH-NADH adduct to InhA (45), were confined to INHr isolates. These results are consistent with the known structural interactions between INH and InhA and add further support to the role of inhA mutations in clinical INH resistance.
There were a total of 73 mutations detected in ahpC, in 19/608 (3%) INHs isolates and 54/403 (13%) INHr isolates. The most common mutation was found at position −46 in both INHs and INHr isolates (Table 2), confirming that this mutation is not associated with INH resistance (3, 27). The D73H mutation and the G-to-A mutation at position −9 were also detected in both susceptible and resistant isolates. The T5I mutation was the only ORF mutation detected and was present in only one INHr isolate. These results help to confirm a role for ahpC promoter mutations in INH resistance but suggest that mutations in the ahpC ORF make little or no contribution to INH resistance.
We detected a total of 30 ndh mutations, in 16/608 (3%) INHs isolates and 14/403 (3%) INHr isolates. The most common mutation, V18A, was detected in both INHs and INHr isolates. The R268H mutation was only detected in two INHr isolates. These results suggest that these ndh mutations play a small role, at most, in clinical INH resistance.
In order to limit any potential bias introduced by sampling or geographic origin of the isolates tested, we examined the prevalence of the mutations detected by gene in each country. The proportions were comparable for all countries with substantial (>30) sample sizes (data not shown).
Associations between mutations and drug resistance.
The large number of isolates analyzed in our study permitted us to test the statistical strength of reported associations between certain genes and INH resistance for the first time. We observed strong individual associations between any mutations in the katG, inhA, and ahpC genes and resistance to INH (P = 0.0001). Conversely, any mutations in the kasA gene were associated with susceptibility to INH (P = 0.012), while any mutations in ndh were not associated with INH resistance (P = 0.412) or susceptibility.
Logistic regression modeling was conducted using the five genes. The results of this analysis confirmed that any mutation in ndh was not associated with INH resistance or susceptibility (P = 0.68), while mutations in the other four genes were associated either with INH resistance (for katG, odds ratio [OR] = 214.2, standard deviation [SD] = 92.5, and P = 0.0001; for inhA, OR = 55.0, SD = 33.8, and P = 0.0001; and for ahpC, OR = 8.0, SD = 2.5, and P = 0.001) or with INH susceptibility (for kasA, OR = 0.5 [for resistance], SD = 0.2, and P = 0.036), with an area under the curve of 0.876. In order to address the potential clustering of samples within a country, the final logistic regression model was rerun, allowing for correlation within the country. The model did not alter significantly, supporting the representative nature of the overall sample. These results strongly implicate katG, ahpC, and inhA mutations in the mechanism of INH resistance in clinical isolates. The association between kasA mutations and INH susceptibility has no obvious biological explanation and will need to be explored in further studies.
We next examined the association between mutations in INH resistance-associated genes and resistance to other antituberculosis drugs. The development of MDR M. tuberculosis has long been attributed to the independent acquisition of resistance to individual drugs. However, INH resistance in itself (i.e., by any mechanism) appears to be associated with other types of drug resistance (9, 36). We desired to test the hypothesis that certain INH resistance-associated mutations might be more strongly associated with resistance to other drugs than other mutations. This type of unequal association might be observed if particular INH resistance-associated mutations also aided in the development of MDR. The fact that we had already observed this phenomenon in a previous study of embB306 mutations and drug-resistant M. tuberculosis (18) supported this line of analysis.
RIF resistance and STR resistance were associated with mutations in katG, inhA, and ahpC, both individually and in logistic regression models (P values were as follows: RIF, 0.0001, 0.0001, and 0.002, respectively; STR, 0.0001, 0.0001, and 0.005, respectively). RIF resistance was not associated with mutations in kasA (P = 0.69) or ndh (P = 0.07), and STR resistance was not associated with mutations in kasA (P = 0.68). Thus, each of the genes found to be associated with INH resistance was also associated with RIF resistance. This observation suggests that no individual INH resistance-associated mutation predisposes M. tuberculosis to the development of RIF resistance.
In contrast, resistance to EMB was associated with mutations in katG (OR = 5.10 [1.08]; P = 0.0001) and ahpC (OR = 2.16 [0.70]; P = 0.024) but not in inhA (P = 0.16), kasA (P = 0.127), or ndh (P = 0.422). This suggests that INHr katG mutants may be particularly prone to developing EMB resistance compared to other INHr isolates. We postulate that inhA mutations are not well tolerated by some EMB-resistant isolates.
Differences between mono-INHr and MDR isolates.
The genetic mechanisms responsible for INH resistance are commonly thought to be identical in mono-INHr and INHr MDR M. tuberculosis. However, it has been suggested that some drug resistance-associated mutations occur at higher frequencies in MDR M. tuberculosis than in mono-INHr clinical isolates (4, 19, 40). The size of our current investigation made it possible to rigorously test for differences between mono-INHr and MDR isolates for the first time.
We examined the proportion of INHr-associated mutations that could be detected in clinical isolates as a function of the total number of drugs to which each isolate was resistant. In this analysis, we only examined isolates that were resistant to INH or INH plus another drug. We also excluded from the analysis all mutations observed in INHs isolates. The results (Fig. 2) show that INH resistance-associated mutations were less common in mono-INHr isolates (61/101 [60%]) than in isolates that were resistant to INH plus one other drug (84/120 [70%]; P = 0.09) or isolates that were resistant to INH plus two or more drugs (148/177 [84%]; P = 0.001). Conversely, the proportion of isolates without any detectable mutation decreased as isolates became resistant to INH plus an increasing number of drugs. We repeated the analysis within each country, and the distributions observed were similar, limiting the possibility that bias may have been introduced by sampling or geographic origin (data not shown).
FIG. 2.
Distribution of INH resistance-associated mutations in drug-resistant clinical M. tuberculosis isolates. The proportion of M. tuberculosis isolates with detectable INH resistance-associated mutations and the number of mutations detected in each isolate are shown according to the drug susceptibility pattern of the M. tuberculosis isolate.
It has been suggested that Beijing clade isolates (which fall into principal genetic group 1 [PGG-1] [53]) with katG315 mutations are overrepresented among MDR M. tuberculosis isolates (4, 19). We determined the PGGs for 74% (295/398) of the mono-INHr and MDR isolates in the study (Table 3) to test for this potential confounder. The results were used to study the PGG distributions among all isolates and among the katG315 mutants (Table 3). We found that PGG-1 isolates were not overrepresented in the MDR isolates or in the sample limited to isolates with katG315 mutations. In fact, only 23% of the MDR isolates were in PGG-1, compared to 46% of the mono-INHr isolates. This difference remained when we analyzed our data by including only nonclustered isolates and one isolate from each cluster (to control for bias due to clustering) (Table 3).
TABLE 3.
Distribution of clinical isolates by PGG
| Study approach | Isolates included | Resistance pattern | No. of isolates in PGG/total no. of isolates (%)a
|
||
|---|---|---|---|---|---|
| PGG-1 | PGG-2 | PGG-3 | |||
| Counting all isolates | All | Mono-INHr | 31/68 (46) | 31/68 (46) | 6/68 (8) |
| All | INHr + resistance to ≥1 drug | 52/227 (23) | 142/227 (62) | 33/227 (15) | |
| katG315 mutants | Mono-INHr | 17/32 (53) | 14/32 (44) | 1/32 (3) | |
| katG315 mutants | INHr + resistance to ≥1 drug | 38/143 (27) | 90/143 (63) | 15/143 (10) | |
| Counting one isolate per clusterb | All | Mono-INHr | 30/65 (46) | 29/65 (45) | 6/65 (9) |
| All | INHr + resistance to ≥1 drug | 41/192 (21) | 122/192 (64) | 29/192 (15) | |
| katG315 mutants | Mono-INHr | 17/31 (55) | 13/31 (42) | 1/31 (3) | |
| katG315 mutants | INHr resistance to ≥1 drug | 27/110 (25) | 71/110 (64) | 12/110 (11) | |
Results based on 68/101(67%) and 227/297(76%) mono-INHr isolates and MDR isolates, respectively.
Results using all unique isolates plus one isolate from each cluster. In cases where a cluster contained isolates with different drug susceptibility profiles and/or mutations, each variation was counted as a new cluster.
The increase in detectable mutations in MDR isolates was almost entirely due to an increase in the proportion of katG315 mutations in MDR isolates (Fig. 3A). This observation did not change when the results were reanalyzed with a sample that included all unique isolates and only one isolate from each cluster (Fig. 3B). In contrast, we found that the proportion of isolates with inhA promoter mutations decreased as resistance increased (P = 0.57 for the difference between mono-INHr and resistance to INH plus one other drug; P = 0.011 for the difference between mono-INHr and resistance to INH plus two or more drugs); however, this trend was lost when the results were reanalyzed with a sample that included all unique isolates and only one isolate from each cluster, and a larger sample will be needed to make a conclusion about the significance of this finding. In contrast, no differences were observed when mono-INHr and MDR isolates were compared regarding the proportions of isolates with mutations in katG other than those in codon 315 and of ahpC and inhA ORF mutants. A similar analysis was performed to determine if MDR isolates were more likely to have multiple INH resistance-associated mutations than mono-INHr isolates (Fig. 2). No association was seen between mutation number and increasing drug resistance.
FIG. 3.
Distributions of mutations in specific genes. The proportions of isolates with INH resistance-associated mutations in the katG, inhA, and ahpC genes are shown according to the drug susceptibility patterns of the M. tuberculosis isolates. (A) Results using all isolates in the study; (B) results using all unique isolates plus one from each cluster. In cases where a cluster contained isolates with different drug susceptibility profiles and/or mutations, each variation was counted as a new cluster. *, P < 0.01; **, P = 0.034.
These results support the hypothesis that katG315 mutations maintain or increase the fitness of M. tuberculosis isolates as they evolve from INH monoresistance to MDR compared to other categories of INH resistance-associated mutations. They do not support the hypothesis that M. tuberculosis accumulates multiple mutations as it evolves to MDR. These results also suggest that inhA promoter mutations may attenuate M. tuberculosis isolates, decreasing the likelihood that these isolates result in clusters of MDR disease.
Interactions between mutations.
Totals of 1/608 (0%) INHs isolates and 80/403 (20%) INHr isolates contained more than one mutation in katG, inhA, ahpC, kasA, and ndh. This suggested that mutations in these genes might interact in a way that increases the drug resistance or general fitness of the isolate. In fact, an interaction between ahpC promoter mutations and mutations in katG that occur outside of the katG315 codon has been postulated previously (52, 54, 63). This hypothesis was based on the observation that ahpC mutations are not sufficient in themselves to confer INH resistance, yet these mutations could be found in isolates containing katG mutations at loci other than katG315. We reasoned that our study would be sufficiently powered to identify statistically significant interactions between certain mutations. Our results (Table 4) demonstrated a significant association between ahpC promoter mutants and mutations in katG that were not at position 315, in contrast to the case for mutations at katG315. A total of 9/221 (4%) katG315 mutants had mutations in the ahpC promoter, while 5/15 (33%) non-katG315 mutants had similar mutations in the ahpC promoter (P = 0.03). After removing mutants with mutations at ahpC position 46 (which are not associated with INH resistance), only 1/221 (0.5%) ahpC promoter mutants were found to have a mutation at katG315, while 4/16 (25%) had a non-katG315 mutation (P < 0.0001).
TABLE 4.
Frequency and distribution of multiple mutations in individual M. tuberculosis isolates
| Gene | Location of mutation | No. (%) of isolates with mutations in indicated gene location
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
katG
|
inhA
|
ahpC
|
kasA
|
ndh | |||||||||||
| All | Position 315 | Positions other than 315 | All | Promoter | ORF | All | Promoter | ORF | D73H | All | Position 269 | Position 312 | |||
| katG | All | 238 (100) | 221 (100) | 17 (100) | 5 (11) | 2 (5) | 3 (25) | 20 (27) | 14 (24) | 6 (43) | 5 (38) | 34 (24) | 29 (24) | 5 (26) | 9 (30) |
| Position 315 | 221 (93) | 221 (100) | 0 | 5 (11) | 2 (5) | 3 (25) | 15 (21) | 9 (15) | 6 (43) | 5 (38) | 34 (24) | 29 (24) | 5 (26) | 9 (30) | |
| Positions other than 315 | 17 (7) | 0 | 17 (100) | 0 | 0 | 0 | 5 (7) | 5 (8) | 0 | 0 | 0 | 0 | 0 | 0 | |
| inhA | All | 5 (2) | 5 (2) | 0 | 46 (100) | 39 (100) | 12 (100) | 6 (8) | 6 (10) | 0 | 0 | 5 (3) | 3 (2) | 2 (11) | 1 (3) |
| Promoter | 2 (1) | 2 (1) | 0 | 39 (85) | 39 (100) | 5 (42) | 6 (8) | 6 (10) | 0 | 0 | 3 (2) | 1 (1) | 2 (11) | 0 | |
| ORF | 3 (1) | 3 (1) | 0 | 12 (26) | 5 (13) | 12 (100) | 1 (1) | 1 (2) | 0 | 0 | 3 (2) | 2 (2) | 1 (5) | 1 (3) | |
| ahpC | All | 20 (8) | 15 (7) | 5 (29) | 6 (13) | 6 (15) | 1 (8) | 73 (100) | 59 (100) | 14 (100) | 13 (100) | 0 | 0 | 0 | 2 (7) |
| Promoter | 14 (6) | 9 (4) | 5 (29) | 6 (13) | 6 (15) | 0 | 59 (81) | 59 (100) | 0 | 0 | 0 | 0 | 0 | 1 (3) | |
| ORF | 6 (3) | 6 (3) | 0 | 0 | 0 | 1 (8) | 14 (19) | 0 | 14 (100) | 13 (100) | 0 | 0 | 0 | 1 (3) | |
| D73H | 5 (2) | 5 (2) | 0 | 0 | 0 | 0 | 13 (18) | 0 | 13 (93) | 13 (100) | 0 | 0 | 0 | 0 | |
| kasA | All | 34 (14) | 34 (15) | 0 | 5 (11) | 3 (8) | 3 (25) | 0 | 0 | 0 | 0 | 144 (100) | 123 (100) | 19 (100) | 0 |
| Position 269 | 29 (12) | 29 (13) | 0 | 3 (7) | 1 (3) | 2 (17) | 0 | 0 | 0 | 0 | 123 (85) | 123 (100) | 0 | 0 | |
| Position 312 | 5 (2) | 5 (2) | 0 | 2 (4) | 2 (5) | 1 (8) | 0 | 0 | 0 | 0 | 19 (13) | 0 | 19 (100) | 0 | |
| ndh | All | 9 (4) | 9 (4) | 0 | 1 (2) | 0 | 1 (8) | 2 (3) | 1 (2) | 1 (7) | 0 | 0 | 0 | 0 | 30 (100) |
We also observed a strong negative association between mutations in katG315 and mutations in the promoter region of inhA (P < 0.0088). This result suggests that this type of double mutant is selected against during strain evolution or clustering. Interestingly, we noted a very strong negative association between mutations in the promoter region of ahpC and mutations in kasA (P < 0.0001), mainly due to kasA269 mutations (P < 0.0006). In fact, these mutations were never found in the same INHs or INHr isolate, despite the high overall prevalence of these mutations in our sample. The significance of this observation is unclear. A weaker but still statistically significant positive association between mutations in ndh and mutations in kasA269 was also observed (P < 0.041).
DISCUSSION
This study provides new insights into the population genetics and evolution of drug-resistant M. tuberculosis while also confirming a number of previously held hypotheses. There are several reasons why this investigation represents a significant advance.
First, we tested a large number of drug-susceptible isolates for INH resistance-associated mutations, unlike many previous studies that mostly focused on INHr isolates. This approach enabled us to discover that the katG G316S, kasA D66N, ahpC −9 G-to-A, inhA −17 G-to-T, inhA I47T, and inhA I194T mutations, previously seen only in INHr M. tuberculosis isolates, could also be found in INHs isolates. It also confirmed that kasA269, kasA312, aphC −46, ahpC D73H, and ndh V18A mutations could be detected in INHs isolates. KasA is a protein involved in mycolic acid biosynthesis (50), and its role in resistance to INH is still controversial (26). These results provide strong evidence in clinical isolates that none of the above mutations are causally involved in INH resistance.
Second, we tested a much larger number of INHr M. tuberculosis isolates than any previous study. The large number of observations available for analysis enabled us to use statistical techniques to search for associations between mutations and drug resistance and to test for interactions between mutations. This type of analysis is likely to have been underpowered with the smaller data sets previously available. Our results show a strong statistical association between mutations in the katG, ahpC, and inhA genes and INH resistance. This confirms previously held assumptions about the roles of these genes (26, 49, 54). We also discovered a surprisingly strong inverse association between kasA mutations and INH resistance. Our results suggest that these mutations did not evolve because they conferred INH resistance. Perhaps this newly discovered association with INHs isolates will provide a clue to the roles of these mutations in M. tuberculosis evolution.
We found that katG, ahpC, and inhA mutations were associated with RIF resistance but that only katG mutations were associated with EMB resistance. These results do not directly implicate INH resistance mutations as a cause of resistance to either RIF or EMB. Rather, we believe that these results suggest that the type of mutation causing resistance to one drug can influence the probability of developing resistance to other drugs. Our results suggest that katG mutations provide a more favorable environment for the development of EMB resistance than do mutations in other genes. These results are consistent with our observation that inhA promoter mutations were inversely associated with katG mutations and that inhA promoter mutations occurred less frequently in highly MDR isolates than in mono-INHr isolates. We suggest below that this observation is due to a fitness advantage conferred by katG315 mutations and by a possible attenuating effect of inhA promoter mutations.
Third, we studied both mono-INHr and MDR isolates for INH resistance-associated mutations in sufficiently large numbers that it was possible to perform statistical comparisons between these groups. Prior studies, which focused principally on MDR isolates, appear to have overestimated the importance of katG mutations (and especially katG315 mutations) in causing INH monoresistance. Almost 56% of the mono-INHr isolates in our study did not have katG mutations, and 34% of the mono-INHr isolates did not have any previously identified mutation that was likely to have caused INH resistance. This suggests that many of the genetic causes of primary INH resistance remain to be discovered.
Our comparison between mono-INHr and MDR M. tuberculosis isolates also showed that katG315 mutations occurred with increasing frequency as clinical M. tuberculosis isolates became progressively more drug resistant. Thus, our results confirm the primary hypothesis of this investigation and corroborate previous reports (4, 19, 40). We speculate that two different mechanisms could account for this observation. One hypothesis suggests that katG mutations may have an important role as secondary mutations that develop after a (often unknown) primary mutation has caused an isolate to become INHr. Under this hypothesis, mutations in katG may be delayed until after an isolate has already become MDR. An alternate hypothesis suggests that katG315 mutations confer a survival advantage on INHr isolates. Under this hypothesis, katG315 mutants would be more likely to survive in a treated patient, to be transmitted, and to evolve into MDR strains because they have increased fitness. This second hypothesis is supported by our observation that inhA promoter mutations were observed with diminished frequency in highly MDR isolates when all clustered samples were included. Laboratory studies have suggested that expression of the inhA gene may be tightly regulated, as inhA cannot be overexpressed on a multicopy plasmid using its native promoter (5). If inhA overexpression occurs at a cost to the bacterium, then this mutation may be selected against compared to mutations in katG315. These results point to a continuing evolutionary process in MDR isolates and suggest that future MDR isolates may be even more fit and difficult to control than those currently encountered. These results also suggest that future INH analogues might select for MDR M. tuberculosis less frequently if they were able to circumvent katG315 resistance and prevent the selection of this mutation.
In addition to the insights that our analysis brings to the evolution of INH resistance, our results may also contribute to the design of genetic assays to detect INH resistance. We would have been able to detect INH resistance in 295 of 397 (74%) of the INHr isolates in our study by simply testing for mutations at katG315, inhA94, the promoter regions of inhA (spanning the positions between −19 and −2), and the ahpC promoter (except position −46). Only three INHs isolates would have been misclassified as INHr using this approach. Thus, these regions could be used as molecular markers for INH resistance, and they could be screened easily with only five assays. Our results also suggest that most of the positions evaluated in the ORFs of katG, kasA, inhA, ahpC, and ndh have little value in predicting resistance to INH.
This study has a number of potential limitations. We only analyzed the M. tuberculosis isolates for mutations that had been associated with INH resistance in previous studies. Additional mutations might have been discovered if we had completely sequenced each of the relevant genes. However, the SNPs targeted by this study included almost all SNPs identified in a large body of literature spanning ∼15 years. Furthermore, the frequency of katG mutations outside of codon 315 was sufficiently low to suggest that additional sequencing would have yielded only a small number of additional SNPs. It is very unlikely that the results would have changed any of the conclusions brought forth in this study. We were only able to detect mutations in 78% (309/397) of the INHr isolates in this study. It has been suggested that a number of other genes could be involved in INH resistance (44), and these additional genes were not examined here. However, few of these potential INH resistance genes contained mutations in M. tuberculosis isolates that did not also have mutations in katG, ahpC, inhA, kasA, or ndh, and most mutations were quite rare. Thus, it is unlikely that the inclusion of these genes would have changed the results of our study.
Future population genetic studies based on even larger sample sizes will have the power to examine mutations and other genetic variations among clinical isolates in fine detail, ultimately providing information that will aid in the development of improved treatments for tuberculosis.
Supplementary Material
Acknowledgments
We acknowledge the excellent technical support provided by Joyce Eskdale (UMDNJ), Tania Porras, Claudia Castro, and Wellman A. Ribón (from the Instituto Nacional de Salud, Colombia).
This work was supported by Public Health Service grants AI-46669 and AI-49352 from the National Institutes of Health, grant 176W009 from The Wellcome Trust, grants G26264M and 30987M from the Mexican Council of Science and Technology, and grant 6570-12-636-95 from COLCIENCIAS.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Alland, D., G. E. Kalkut, A. R. Moss, R. A. McAdam, J. A. Hahn, W. Bosworth, E. Drucker, and B. R. Bloom. 1994. Transmission of tuberculosis in New York City. An analysis by DNA fingerprinting and conventional epidemiologic methods. N. Engl. J. Med. 330:1710-1716. [DOI] [PubMed] [Google Scholar]
- 2.Alland, D., T. S. Whittam, M. B. Murray, M. D. Cave, M. H. Hazbón, K. Dix, M. Kokoris, A. Duesterhoeft, J. A. Eisen, C. M. Fraser, and R. D. Fleischmann. 2003. Modeling bacterial evolution with comparative-genome-based marker systems: application to Mycobacterium tuberculosis evolution and pathogenesis. J. Bacteriol. 185:3392-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, L. V., T. J. Brown, O. Maxwell, A. L. Gibson, Z. Fang, M. D. Yates, and F. A. Drobniewski. 2005. Molecular analysis of isoniazid-resistant Mycobacterium tuberculosis isolates from England and Wales reveals the phylogenetic significance of the ahpC −46A polymorphism. Antimicrob. Agents Chemother. 49:1455-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bakonyte, D., A. Baranauskaite, J. Cicenaite, A. Sosnovskaja, and P. Stakenas. 2003. Molecular characterization of isoniazid-resistant Mycobacterium tuberculosis clinical isolates in Lithuania. Antimicrob. Agents Chemother. 47:2009-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banerjee, A., E. Dubnau, A. Quemard, V. Balasubramanian, K. S. Um, T. Wilson, D. Collins, G. de Lisle, and W. R. Jacobs, Jr. 1994. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 263:227-230. [DOI] [PubMed] [Google Scholar]
- 6.Barnes, P. F., H. el-Hajj, S. Preston-Martin, M. D. Cave, B. E. Jones, M. Otaya, J. Pogoda, and K. D. Eisenach. 1996. Transmission of tuberculosis among the urban homeless. JAMA 275:305-307. [PubMed] [Google Scholar]
- 7.Bobadilla-del-Valle, M., A. Ponce-de-León, C. Arenas-Huertero, G. Vargas-Alarcon, M. Kato-Maeda, P. M. Small, P. Couary, G. M. Ruiz-Palacios, and J. Sifuentes-Osornio. 2001. rpoB gene mutations in rifampin-resistant Mycobacterium tuberculosis identified by polymerase chain reaction single-stranded conformational polymorphism. Emerg. Infect. Dis. 7:1010-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaves, F., F. Dronda, M. D. Cave, M. Alonso-Sanz, A. Gonzalez-Lopez, K. D. Eisenach, A. Ortega, L. Lopez-Cubero, I. Fernandez-Martin, S. Catalan, and J. H. Bates. 1997. A longitudinal study of transmission of tuberculosis in a large prison population. Am. J. Respir. Crit. Care Med. 155:719-725. [DOI] [PubMed] [Google Scholar]
- 9.Churchyard, G. J., E. L. Corbett, I. Kleinschmidt, D. Mulder, and K. M. De Cock. 2000. Drug-resistant tuberculosis in South African gold miners: incidence and associated factors. Int. J. Tuberc. Lung Dis. 4:433-440. [PubMed] [Google Scholar]
- 10.Cockerill, F. R., III, J. R. Uhl, Z. Temesgen, Y. Zhang, L. Stockman, G. D. Roberts, D. L. Williams, and B. C. Kline. 1995. Rapid identification of a point mutation of the Mycobacterium tuberculosis catalase-peroxidase (katG) gene associated with isoniazid resistance. J. Infect. Dis. 171:240-245. [DOI] [PubMed] [Google Scholar]
- 11.Dye, C., and M. A. Espinal. 2001. Will tuberculosis become resistant to all antibiotics? Proc. Biol. Sci. 268:45-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Escalante, P., S. Ramaswamy, H. Sanabria, H. Soini, X. Pan, O. Valiente-Castillo, and J. M. Musser. 1998. Genotypic characterization of drug-resistant Mycobacterium tuberculosis isolates from Peru. Tuber. Lung Dis. 79:111-118. [DOI] [PubMed] [Google Scholar]
- 13.Espinal, M. A., A. Laszlo, L. Simonsen, F. Boulahbal, S. J. Kim, A. Reniero, S. Hoffner, H. L. Rieder, N. Binkin, C. Dye, R. Williams, and M. C. Raviglione. 2001. Global trends in resistance to antituberculosis drugs. N. Engl. J. Med. 344:1294-1303. [DOI] [PubMed] [Google Scholar]
- 14.Fang, Z., C. Doig, A. Rayner, D. T. Kenna, B. Watt, and K. J. Forbes. 1999. Molecular evidence for heterogeneity of the multiple-drug-resistant Mycobacterium tuberculosis population in Scotland (1990 to 1997). J. Clin. Microbiol. 37:998-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frothingham, R., and W. A. Meeker-O'Connell. 1998. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology 144:1189-1196. [DOI] [PubMed] [Google Scholar]
- 16.García-García, M. L., A. Ponce de León, M. E. Jimenez-Corona, A. Jimenez-Corona, M. Palacios-Martinez, S. Balandrano-Campos, L. Ferreyra-Reyes, L. Juarez-Sandino, J. Sifuentes-Osornio, H. Olivera-Diaz, J. L. Valdespino-Gomez, and P. M. Small. 2000. Clinical consequences and transmissibility of drug-resistant tuberculosis in southern Mexico. Arch. Intern. Med. 160:630-636. [DOI] [PubMed] [Google Scholar]
- 17.Hazbón, M. H., and D. Alland. 2004. Hairpin primers for simplified single-nucleotide polymorphism analysis of Mycobacterium tuberculosis and other organisms. J. Clin. Microbiol. 42:1236-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hazbon, M. H., M. Bobadilla del Valle, M. I. Guerrero, M. Varma-Basil, I. Filliol, M. Cavatore, R. Colangeli, H. Safi, H. Billman-Jacobe, C. Lavender, J. Fyfe, L. Garcia-Garcia, A. Davidow, M. Brimacombe, C. I. Leon, T. Porras, M. Bose, F. Chaves, K. D. Eisenach, J. Sifuentes-Osornio, A. Ponce de Leon, M. D. Cave, and D. Alland. 2005. Role of embB codon 306 mutations in Mycobacterium tuberculosis revisited: a novel association with broad drug resistance and IS6110 clustering rather than ethambutol resistance. Antimicrob. Agents Chemother. 49:3794-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hillemann, D., T. Kubica, R. Agzamova, B. Venera, S. Rusch-Gerdes, and S. Niemann. 2005. Rifampicin and isoniazid resistance mutations in Mycobacterium tuberculosis strains isolated from patients in Kazakhstan. Int. J. Tuberc. Lung Dis. 9:1161-1167. [PubMed] [Google Scholar]
- 20.Johansen, I. S., V. O. Thomsen, M. Marjamaki, A. Sosnovskaja, and B. Lundgren. 2004. Rapid, automated, nonradiometric susceptibility testing of Mycobacterium tuberculosis complex to four first-line antituberculous drugs used in standard short-course chemotherapy. Diagn. Microbiol. Infect. Dis. 50:103-107. [DOI] [PubMed] [Google Scholar]
- 21.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kapetanaki, S. M., S. Chouchane, S. Yu, X. Zhao, R. S. Magliozzo, and J. P. Schelvis. 2005. Mycobacterium tuberculosis KatG(S315T) catalase-peroxidase retains all active site properties for proper catalytic function. Biochemistry 44:243-252. [DOI] [PubMed] [Google Scholar]
- 23.Kato-Maeda, M., J. Sifuentes-Osornio, M. Bobadilla-del-Valle, G. M. Ruiz-Palacios, and A. Ponce-de-Leon. 1999. Drug resistance among acid-fast bacilli. Lancet 353:1709. [DOI] [PubMed] [Google Scholar]
- 24.Kent, P. T., and G. P. Kubica. 1985. Public health mycobacteriology: a guide for the level III laboratory. Centers for Disease Control, Atlanta, Ga.
- 25.Kim, S. Y., Y. J. Park, W. I. Kim, S. H. Lee, C. Ludgerus Chang, S. J. Kang, and C. S. Kang. 2003. Molecular analysis of isoniazid resistance in Mycobacterium tuberculosis isolates recovered from South Korea. Diagn. Microbiol. Infect. Dis. 47:497-502. [DOI] [PubMed] [Google Scholar]
- 26.Larsen, M. H., C. Vilcheze, L. Kremer, G. S. Besra, L. Parsons, M. Salfinger, L. Heifets, M. H. Hazbon, D. Alland, J. C. Sacchettini, and W. R. Jacobs, Jr. 2002. Overexpression of inhA, but not kasA, confers resistance to isoniazid and ethionamide in Mycobacterium smegmatis, M. bovis BCG and M. tuberculosis. Mol. Microbiol. 46:453-466. [DOI] [PubMed] [Google Scholar]
- 27.Lavender, C., M. Globan, A. Sievers, H. Billman-Jacobe, and J. Fyfe. 2005. Molecular characterization of isoniazid-resistant Mycobacterium tuberculosis isolates collected in Australia. Antimicrob. Agents Chemother. 49:4068-4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, A. S., I. H. Lim, L. L. Tang, A. Telenti, and S. Y. Wong. 1999. Contribution of kasA analysis to detection of isoniazid-resistant Mycobacterium tuberculosis in Singapore. Antimicrob. Agents Chemother. 43:2087-2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, A. S., A. S. Teo, and S. Y. Wong. 2001. Novel mutations in ndh in isoniazid-resistant Mycobacterium tuberculosis isolates. Antimicrob. Agents Chemother. 45:2157-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, C. N., and L. B. Heifets. 1987. Determination of minimal inhibitory concentrations of antituberculosis drugs by radiometric and conventional methods. Am. Rev. Respir. Dis. 136:349-352. [DOI] [PubMed] [Google Scholar]
- 31.Marttila, H. J., H. Soini, E. Eerola, E. Vyshnevskaya, B. I. Vyshnevskiy, T. F. Otten, A. V. Vasilyef, and M. K. Viljanen. 1998. A Ser315Thr substitution in KatG is predominant in genetically heterogeneous multidrug-resistant Mycobacterium tuberculosis isolates originating from the St. Petersburg area in Russia. Antimicrob. Agents Chemother. 42:2443-2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Master, R. 1992. Mycobacteriology, p. 3.12.3-3.12.7. In H. D. Isenberg (ed.), Clinical microbiology procedures handbook, vol. 1. American Society for Microbiology, Washington, D.C. [Google Scholar]
- 33.Mdluli, K., R. A. Slayden, Y. Zhu, S. Ramaswamy, X. Pan, D. Mead, D. D. Crane, J. M. Musser, and C. E. Barry III. 1998. Inhibition of a Mycobacterium tuberculosis beta-ketoacyl ACP synthase by isoniazid. Science 280:1607-1610. [DOI] [PubMed] [Google Scholar]
- 34.Miesel, L., T. R. Weisbrod, J. A. Marcinkeviciene, R. Bittman, and W. R. Jacobs, Jr. 1998. NADH dehydrogenase defects confer isoniazid resistance and conditional lethality in Mycobacterium smegmatis. J. Bacteriol. 180:2459-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mokrousov, I., N. V. Bhanu, P. N. Suffys, G. V. Kadival, S. F. Yap, S. N. Cho, A. M. Jordaan, O. Narvskaya, U. B. Singh, H. M. Gomes, H. Lee, S. P. Kulkarni, K. C. Lim, B. K. Khan, D. van Soolingen, T. C. Victor, and L. M. Schouls. 2004. Multicenter evaluation of reverse line blot assay for detection of drug resistance in Mycobacterium tuberculosis clinical isolates. J. Microbiol. Methods 57:323-335. [DOI] [PubMed] [Google Scholar]
- 36.Moore, M., I. M. Onorato, E. McCray, and K. G. Castro. 1997. Trends in drug-resistant tuberculosis in the United States, 1993-1996. JAMA 278:833-837. [PubMed] [Google Scholar]
- 37.Musser, J. M. 1995. Antimicrobial agent resistance in mycobacteria: molecular genetic insights. Clin. Microbiol. Rev. 8:496-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Musser, J. M., V. Kapur, D. L. Williams, B. N. Kreiswirth, D. van Soolingen, and J. D. van Embden. 1996. Characterization of the catalase-peroxidase gene (katG) and inhA locus in isoniazid-resistant and -susceptible strains of Mycobacterium tuberculosis by automated DNA sequencing: restricted array of mutations associated with drug resistance. J. Infect. Dis. 173:196-202. [DOI] [PubMed] [Google Scholar]
- 39.Parsons, L. M., M. Salfinger, A. Clobridge, J. Dormandy, L. Mirabello, V. L. Polletta, A. Sanic, O. Sinyavskiy, S. C. Larsen, J. Driscoll, G. Zickas, and H. W. Taber. 2005. Phenotypic and molecular characterization of Mycobacterium tuberculosis isolates resistant to both isoniazid and ethambutol. Antimicrob. Agents Chemother. 49:2218-2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piatek, A. S., A. Telenti, M. R. Murray, H. El-Hajj, W. R. Jacobs, Jr., F. R. Kramer, and D. Alland. 2000. Genotypic analysis of Mycobacterium tuberculosis in two distinct populations using molecular beacons: implications for rapid susceptibility testing. Antimicrob. Agents Chemother. 44:103-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pym, A. S., B. Saint-Joanis, and S. T. Cole. 2002. Effect of katG mutations on the virulence of Mycobacterium tuberculosis and the implication for transmission in humans. Infect. Immun. 70:4955-4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramaswamy, S., and J. M. Musser. 1998. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber. Lung Dis. 79:3-29. [DOI] [PubMed] [Google Scholar]
- 43.Ramaswamy, S. V., S. J. Dou, A. Rendon, Z. Yang, M. D. Cave, and E. A. Graviss. 2004. Genotypic analysis of multidrug-resistant Mycobacterium tuberculosis isolates from Monterrey, Mexico. J. Med. Microbiol. 53:107-113. [DOI] [PubMed] [Google Scholar]
- 44.Ramaswamy, S. V., R. Reich, S. J. Dou, L. Jasperse, X. Pan, A. Wanger, T. Quitugua, and E. A. Graviss. 2003. Single nucleotide polymorphisms in genes associated with isoniazid resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 47:1241-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rawat, R., A. Whitty, and P. J. Tonge. 2003. The isoniazid-NAD adduct is a slow, tight-binding inhibitor of InhA, the Mycobacterium tuberculosis enoyl reductase: adduct affinity and drug resistance. Proc. Natl. Acad. Sci. USA 100:13881-13886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rouse, D. A., J. A. DeVito, Z. Li, H. Byer, and S. L. Morris. 1996. Site-directed mutagenesis of the katG gene of Mycobacterium tuberculosis: effects on catalase-peroxidase activities and isoniazid resistance. Mol. Microbiol. 22:583-592. [DOI] [PubMed] [Google Scholar]
- 47.Rouse, D. A., Z. Li, G. H. Bai, and S. L. Morris. 1995. Characterization of the katG and inhA genes of isoniazid-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 39:2472-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rozwarski, D. A., G. A. Grant, D. H. Barton, W. R. Jacobs, Jr., and J. C. Sacchettini. 1998. Modification of the NADH of the isoniazid target (InhA) from Mycobacterium tuberculosis. Science 279:98-102. [DOI] [PubMed] [Google Scholar]
- 49.Saint-Joanis, B., H. Souchon, M. Wilming, K. Johnsson, P. M. Alzari, and S. T. Cole. 1999. Use of site-directed mutagenesis to probe the structure, function and isoniazid activation of the catalase/peroxidase, KatG, from Mycobacterium tuberculosis. Biochem. J. 338:753-760. [PMC free article] [PubMed] [Google Scholar]
- 50.Schaeffer, M. L., G. Agnihotri, C. Volker, H. Kallender, P. J. Brennan, and J. T. Lonsdale. 2001. Purification and biochemical characterization of the Mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthases KasA and KasB. J. Biol. Chem. 276:47029-47037. [DOI] [PubMed] [Google Scholar]
- 51.Schroeder, E. K., L. A. Basso, D. S. Santos, and O. N. de Souza. 2005. Molecular dynamics simulation studies of the wild-type, I21V, and I16T mutants of isoniazid-resistant Mycobacterium tuberculosis enoyl reductase (InhA) in complex with NADH: toward the understanding of NADH-InhA different affinities. Biophys. J. 89:876-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sherman, D. R., K. Mdluli, M. J. Hickey, T. M. Arain, S. L. Morris, C. E. Barry III, and C. K. Stover. 1996. Compensatory ahpC gene expression in isoniazid-resistant Mycobacterium tuberculosis. Science 272:1641-1643. [DOI] [PubMed] [Google Scholar]
- 53.Sreevatsan, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sreevatsan, S., X. Pan, Y. Zhang, V. Deretic, and J. M. Musser. 1997. Analysis of the oxyR-ahpC region in isoniazid-resistant and -susceptible Mycobacterium tuberculosis complex organisms recovered from diseased humans and animals in diverse localities. Antimicrob. Agents Chemother. 41:600-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sreevatsan, S., K. E. Stockbauer, X. Pan, B. N. Kreiswirth, S. L. Moghazeh, W. R. Jacobs, Jr., A. Telenti, and J. M. Musser. 1997. Ethambutol resistance in Mycobacterium tuberculosis: critical role of embB mutations. Antimicrob. Agents Chemother. 41:1677-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van Rie, A., R. Warren, I. Mshanga, A. M. Jordaan, G. D. van der Spuy, M. Richardson, J. Simpson, R. P. Gie, D. A. Enarson, N. Beyers, P. D. van Helden, and T. C. Victor. 2001. Analysis for a limited number of gene codons can predict drug resistance of Mycobacterium tuberculosis in a high-incidence community. J. Clin. Microbiol. 39:636-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Soolingen, D., P. E. de Haas, H. R. van Doorn, E. Kuijper, H. Rinder, and M. W. Borgdorff. 2000. Mutations at amino acid position 315 of the katG gene are associated with high-level resistance to isoniazid, other drug resistance, and successful transmission of Mycobacterium tuberculosis in the Netherlands. J. Infect. Dis. 182:1788-1790. [DOI] [PubMed] [Google Scholar]
- 58.Vilcheze, C., T. R. Weisbrod, B. Chen, L. Kremer, M. H. Hazbon, F. Wang, D. Alland, J. C. Sacchettini, and W. R. Jacobs, Jr. 2005. Altered NADH/NAD+ ratio mediates coresistance to isoniazid and ethionamide in mycobacteria. Antimicrob. Agents Chemother. 49:708-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wei, C. J., B. Lei, J. M. Musser, and S. C. Tu. 2003. Isoniazid activation defects in recombinant Mycobacterium tuberculosis catalase-peroxidase (KatG) mutants evident in InhA inhibitor production. Antimicrob. Agents Chemother. 47:670-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weis, S. E., J. M. Pogoda, Z. Yang, M. D. Cave, C. Wallace, M. Kelley, and P. F. Barnes. 2002. Transmission dynamics of tuberculosis in Tarrant county, Texas. Am. J. Respir. Crit. Care Med. 166:36-42. [DOI] [PubMed] [Google Scholar]
- 61.WHO/IUATLD. 2000. Global Project on Anti-Tuberculous Drug Resistance Surveillance: antituberculous drug resistance in the world. Report 2: prevalence and trends. World Health Organization, Geneva, Switzerland.
- 62.Wilson, R. W., Z. Yang, M. Kelley, M. D. Cave, J. M. Pogoda, R. J. Wallace, Jr., J. P. Cegielski, D. F. Dunbar, D. Bergmire-Sweat, L. B. Elliott, and P. F. Barnes. 1999. Evidence from molecular fingerprinting of limited spread of drug-resistant tuberculosis in Texas. J. Clin. Microbiol. 37:3255-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wilson, T. M., and D. M. Collins. 1996. ahpC, a gene involved in isoniazid resistance of the Mycobacterium tuberculosis complex. Mol. Microbiol. 19:1025-1034. [DOI] [PubMed] [Google Scholar]
- 64.Zhang, M., J. Yue, Y. P. Yang, H. M. Zhang, J. Q. Lei, R. L. Jin, X. L. Zhang, and H. H. Wang. 2005. Detection of mutations associated with isoniazid resistance in Mycobacterium tuberculosis isolates from China. J. Clin. Microbiol. 43:5477-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang, Y., B. Heym, B. Allen, D. Young, and S. Cole. 1992. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature 358:591-593. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.