Abstract
The spread of extended-spectrum-β-lactamase (ESBL)-producing organisms, particularly those harboring the CTX-M-type enzymes, both in the hospital and in the community, is difficult to discontinue due to the successful mobilization and evolution of the genetic elements harboring ESBL genes and coresistance rates in these isolates. The activities of tigecycline against 285 non-clonally related isolates (172 from Escherichia coli, 84 from Klebsiella spp., 20 from Enterobacter spp., 5 from Salmonella spp., and 4 from Citrobacter spp.) expressing well-characterized ESBLs and recovered in our hospital and its community area of influence were comparatively assessed (CLSI microdilution). Susceptibility rates for meropenem, imipenem, tigecycline, amikacin, and piperacillin-tazobactam were 100%, 100%, 97.5%, 93.3%, and 93%, respectively. Tigecycline (mode MIC, 0.5 μg/ml; MIC90, 1 μg/ml) was 4- to 256-fold more active than doxycycline and minocycline (mode MIC range, 2 to 128 μg/ml). CTX-Ms were the most frequent ESBLs (61.4%), 65.8% in community and 58.6% in nosocomial isolates. CTX-M-9 (22%), CTX-M-14 (15.8%), and CTX-M-10 (14%) were the most represented derivatives. SHV and TEM variants constituted 22.8% and 15.8% of the ESBLs, respectively. Overall coresistance rates were as follows: gentamicin, 27.4%; tobramycin, 27.4%; amikacin, 6.7%; and chloramphenicol, 29.1%. Sulfonamide (61.7%), trimethoprim (52.3%), streptomycin (50.5%), and ciprofloxacin (37.2%) resistance levels were significantly (P < 0.001) associated with CTX-M-9 producers. No tigecycline resistance was observed, although seven Klebsiella pneumoniae isolates exhibited intermediate MICs (4 μg/ml). Tigecycline, lacking cross-resistance with other compounds, could represent an opportunity to reduce the intensity of selection for ESBL-producing organisms derived from the use of other antimicrobial agents. However, this in vitro promise requires support from clinical studies.
Increasing rates of extended-spectrum-β-lactamase (ESBL)-producing Enterobacteriaceae isolates from either the community or the hospital setting have been observed (13, 26). In the last few years, CTX-M-type enzymes appear to be among the most widespread variants of the ESBL group even though there has been no decline in the incidence of ESBL TEM and SHV derivatives (2, 16, 25). ESBL dispersion models encompass mainly complex mobilization processes involving the dissemination of resistance determinants through transferable elements among either many different or a few selected bacterial clones. These clones may then become prevalent and successfully spread along diverse environments. The high prevalence of CTX-Ms in both the community and the hospital environments is suggestive of a dynamic flow of genes coding for these enzymes (26).
Continuous selective pressure exerted by β-lactam compounds is a risk factor for selection of resistant ESBL-producing strains (12, 27). The associated coresistance of these isolates to unrelated antimicrobials, such as aminoglycosides, tetracyclines, chloramphenicol, trimethoprim, sulfonamides, and quinolones, may play an important role in the spread and maintenance of these isolates. However, this associated coresistance also limits therapeutic options (12, 29). In this scenario, alternative antimicrobial compounds are needed to cover infections in which these isolates are increasingly involved.
Tigecycline is a glycylcycline derivative of minocycline with in vitro bacteriostatic activity. It has a broad antibacterial spectrum including gram-positive, gram-negative, atypical, and anaerobic bacteria (11, 20, 22). Tigecycline circumvents efflux and ribosomal protection, the two most frequent genetic mechanisms of tetracycline resistance (6). It is also unaffected by the presence of coresistance to unrelated antimicrobials, such as β-lactams, aminoglycosides, and quinolones (16). However, it is vulnerable to chromosomally mediated efflux in Pseudomonas aeruginosa and Proteeae as well as to the Tet(X)-degrading enzyme found in Bacteroides spp. (16, 21). In addition, reduced levels of susceptibility have been described not only for clinical isolates of Klebsiella pneumoniae and Enterobacter spp. due to an elevated expression of AcrAB efflux pumps (28) but also for Staphylococcus aureus due to alterations in the expression of a so-called MATE efflux pump (19). Emerging tigecycline resistance in standard laboratory selection experiments is rare (mutant selection frequency, <10−9) (3).
Tigecycline is indicated in complicated intra-abdominal and severe skin and soft-tissue infections (4, 22). In vitro studies may support coverage of multiresistant gram-positive cocci, Enterobacteriaceae, pan-resistant Acinetobacter spp., and anaerobes (1, 14, 16). We conducted this study for a detailed evaluation of tigecycline against well-characterized ESBL-producing strains. The comparative in vitro activities of clinically available tetracyclines and that of tigecycline as well as the impact of unrelated antimicrobial coresistance on tigecycline activity were also analyzed.
(These results were presented in part at the 45th Interscience Conference on Antimicrobial Agents and Chemotherapy, 16 to 19 December 2005, Washington, D.C.)
MATERIALS AND METHODS
Bacterial strains.
A total of 285 ESBL-producing Enterobacteriaceae isolates (61% from hospitalized and 39% from community patients) recovered from June 1989 through January 2004 were studied. Escherichia coli isolates from fecal carriers collected in 2003 and constituting 4.2% and 14.4% of the nosocomial and community isolates, respectively, were included (30). One non-clonally related isolate per subject was studied. The bacterial species used in this study were E. coli (n = 172), Klebsiella pneumoniae (n = 75), Klebsiella oxytoca (n = 9), Enterobacter cloacae (n = 16), Enterobacter aerogenes (n = 3), Enterobacter gergoviae (n = 1), Citrobacter freundii (n = 3), Citrobacter amalonaticus (n = 1), and Salmonella spp. (n = 5). Clinical specimens were obtained from urine (n = 141), fecal (n = 39), respiratory (n = 33), wound (n = 23), blood (n = 21), catheter (n = 7), intra-abdominal (n = 7), rectal (n = 4), skin (n = 4), and other (n = 6) sources. Pulsed-field gel electrophoresis with XbaI (Roche GmbH, Mannheim, Germany) as a restriction enzyme was used to study the genetic relatedness of the ESBL-producing isolates (10, 30).
Antimicrobial agents.
The antimicrobials tested were amoxicillin-clavulanate, piperacillin alone or in combination with a fixed concentration of 4 μg of tazobactam per ml, cefotaxime, ceftazidime, aztreonam, cefepime, imipenem, meropenem, ciprofloxacin, tetracycline, doxycycline, minocycline, and tigecycline. All compounds were provided by their respective manufacturers or obtained commercially from Sigma (St. Louis, Mo.). Coresistance was also studied by the disk diffusion method (7) using commercial disks (Oxoid Ltd., Basingstoke, Hampshire, United Kingdom) containing 30 μg of amikacin, 10 μg of gentamicin, 10 μg of tobramycin, 30 μg of kanamycin, 10 μg of streptomycin, 300 μg of sulfonamide, 5 μg of trimethoprim, 30 μg of tetracycline, 30 μg of chloramphenicol, 30 μg of nalidixic acid, and 5 μg of ciprofloxacin.
Antimicrobial susceptibility testing.
MICs were determined by the standard microdilution method (8) using Mueller-Hinton broth (Oxoid Ltd., Basingstoke, Hampshire, United Kingdom) and interpreted according to CLSI (formerly NCCLS) guidelines (9). The CLSI recommends that all confirmed E. coli, K. pneumoniae, K. oxytoca, and Proteus mirabilis ESBL-producing strains should be reported as resistant to all penicillins, cephalosporins, and aztreonam. This rule was applied to all ESBL-producing strains irrespective of species identification. The recommendation included in the U.S. FDA (Food and Drug Administration) package insert was used for tigecycline (susceptible, ≤2 μg/ml; resistant, ≥8 μg/ml) (31). The E. coli ATCC 25922 and 35218 strains and the K. pneumoniae ATCC 700603 strain were included as controls in each susceptibility test.
ESBL characterization.
The ESBLs were characterized by isoelectric focusing, amplification of bla genes by PCR using specific primers for TEM, SHV, or CTX-M ESBL types, and sequencing, as previously described (10, 18, 30).
Statistical analysis.
Statistical significance for comparison of proportions was calculated by the chi-square test (P values of <0.05 were considered statistically significant).
RESULTS
ESBL types among Enterobacteriaceae isolates.
Considering the two most represented species, 70.9% of E. coli ESBL-producing isolates harbored a CTX-M variant, while 16.9% and 12.2% harbored an SHV and a TEM derivative, respectively. In the case of K. pneumoniae, the rates were 34.7%, 44.0%, and 21.3% for CTX-M, SHV, and TEM variants, respectively. CTX-M enzymes (71.0%) were also the most prevalent in the remaining bacterial species, followed by TEM (21.0%) and SHV (7.9%) derivatives.
The 175 CTX-M derivatives, constituting the most numerous ESBL group (61.4%), were distributed as follows: CTX-M-9, 22%; CTX-M-14, 15.8%; and CTX-M-10, 14%. A group named “other CTX-M type” included CTX-M-1-like enzymes (10.2%) and CTX-M-2 (0.4%). The CTX-M group was followed in incidence by the SHV-type (22.8%) and the TEM-type (15.8%) enzymes.
The CTX-M group constituted 65.8% of the ESBLs found in the community and 58.6% of those found in the hospital setting. Within the SHV ESBL type, 20.7% and 24.1% of the isolates harbored these enzymes in the community and in the hospital environment, respectively. Similarly, extended-spectrum TEM derivatives were slightly less represented among community (13.5%) than among hospital (17.3%) isolates.
Susceptibility profiles.
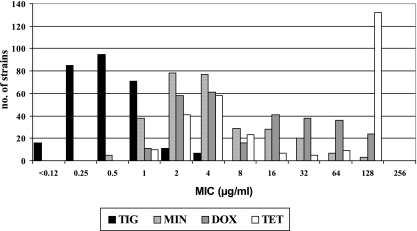
Comparative antimicrobial susceptibilities of ESBL-producing Enterobacteriaceae isolates are shown in Table 1. When a tigecycline susceptibility breakpoint of 2 μg/ml was used (31), 97.5% of the isolates were fully susceptible to this compound. This percentage decreased to 93.7% when the EUCAST susceptibility breakpoint (1 μg/ml) was used (http://www.eucast.org). Comparative in vitro activities of different tetracyclines are shown in Fig. 1. The intrinsic activity of tigecycline (mode MIC, 0.5 μg/ml) was 4- to 256-fold greater than those of tetracycline compounds (range of mode MICs, 2 to 128 μg/ml). In the case of tigecycline, only seven isolates (2.4%) displayed an intermediate MIC of 4 μg/ml and they were all K. pneumoniae isolates harboring a heterogeneous group of ESBLs (five SHV-type, one CTX-M-type, and one TEM-type enzyme). No full resistance to tigecycline was observed in any of the isolates tested.
TABLE 1.
Comparative in vitro activities of tigecycline against 285 extended-spectrum-β-lactamase-producing Enterobacteriaceae isolates
| Antimicrobial | MIC (μg/ml)
|
% of fully susceptible isolates | |||
|---|---|---|---|---|---|
| Range | 50% | 90% | Mode | ||
| Amoxicillin-clavulanate | ≤4/2-128/64 | 8/4 | 16/8 | ≤4/2 | 81a |
| Piperacillin-tazobactam | ≤0.5/4-≥128/4 | 16/4 | 16/4 | 16/4 | 93a |
| Cefotaxime | 0.5-512 | 256 | 256 | 256 | 0b |
| Ceftazidime | 0.12-256 | 4 | 256 | 256 | 0b |
| Cefepime | 1-64 | 64 | 64 | 64 | 0b |
| Aztreonam | 0.5-256 | 64 | 128 | 128 | 0b |
| Imipenem | ≤0.06-2 | 0.5 | 0.5 | 0.5 | 100 |
| Meropenem | ≤0.5-2 | ≤0.5 | 1 | ≤0.5 | 100 |
| Ciprofloxacin | ≤0.12-8 | 1 | 8 | ≤0.12 | 64.9a |
| Tetracycline | 1-128 | 32 | 128 | 128 | 38.2a |
| Doxycycline | 1-128 | 8 | 64 | 4 | 45.6a |
| Minocycline | 0.5-128 | 4 | 32 | 2 | 69.5a |
| Tigecycline | 0.12-4 | 0.5 | 1 | 0.5 | 97.5c |
FIG. 1.
Comparative in vitro activities of tetracyclines against extended-spectrum-β-lactamase-producing Enterobacteriaceae. TIG, tigecycline; MIN, minocycline; DOX, doxycycline; TET, tetracycline.
Coresistance among ESBL-producing Enterobacteriaceae isolates.
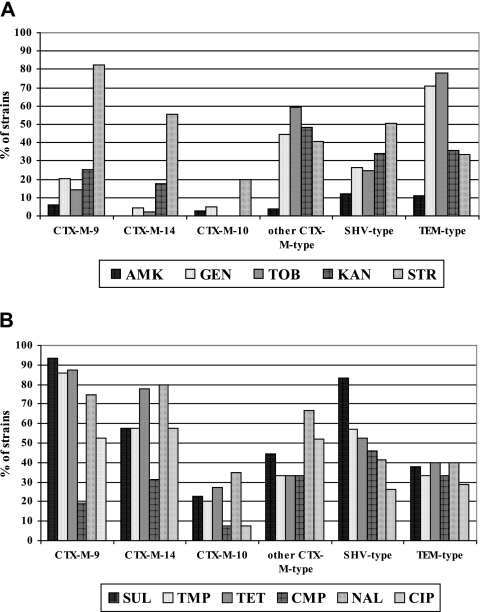
Overall full resistance rates (intermediate plus resistant isolates) to aminoglycoside compounds were as follows: amikacin, 6.7%; gentamicin and tobramycin, 27.4%; kanamycin, 26.3%; and streptomycin, 50.5% (Fig. 2A). In the case of quinolone compounds, resistance to nalidixic acid was 56.1% and resistance to ciprofloxacin was 37.2%. Resistances to chloramphenicol, sulfonamides, trimethoprim, and tetracycline were 29.1%, 61.7%, 52.3%, and 61.8%, respectively (Fig. 2B). Streptomycin, trimethoprim, sulfonamide, nalidixic acid, and ciprofloxacin resistances were significantly (P < 0.01) associated with CTX-M-9 producers.
FIG. 2.
Coresistances among the Enterobacteriaceae isolates of the different ESBL types. (A) Levels for amikacin (AMK), gentamicin (GEN), tobramycin (TOB), kanamycin (KAN), and streptomycin (STR). (B) Levels for sulfonamides (SUL), trimethoprim (TMP), tetracycline (TET), chloramphenicol (CMP), nalidixic acid (NAL), and ciprofloxacin (CIP).
Among CTX-M-9-producing isolates, 6.3% were resistant to amikacin, 20.6% to gentamicin, 14.3% to tobramycin, and 25.4% to kanamycin. Extremely high percentages of isolates harboring this enzyme were coresistant to sulfonamides (93.6%), tetracycline (87.3%), trimethoprim (85.7%), and streptomycin (82.5%). A resistance rate of 19% was found for chloramphenicol, while coresistances to nalidixic acid were as high as 74.6% and 52.4% in the case of ciprofloxacin. Interestingly, CTX-M-14-harboring isolates were all susceptible to amikacin and only 4.4% and 2.2% were resistant to gentamicin and tobramycin, respectively. However, kanamycin resistance was 17.8%, rising to 55.5% for streptomycin. Both sulfonamides and trimethoprim displayed resistance rates of 57.8%, while tetracycline and chloramphenicol resistance figures were 77.8% and 31.1%, respectively. The highest resistance rate corresponded to nalidixic acid (80%), with a significantly high percentage of isolates being coresistant to ciprofloxacin (57.8%). For strains harboring CTX-M-10, percentages of coresistance were significantly lower than those in the previous cases. Nevertheless, high coresistances to nalidixic acid (35%), tetracycline (27.5%), and trimethoprim (20%) were still observed. For the so-called “other CTX-M type” group, resistances to amikacin, gentamicin and tobramycin were 3.7%, 44.4%, and 59.2%, respectively. In the case of kanamycin and streptomycin, the corresponding resistance rates were 48.1% and 40.7%, respectively. Nearly 45% of the isolates were resistant to sulfonamides, while trimethoprim, tetracycline, and chloramphenicol had resistance rates of 33.3%. For nalidixic acid and ciprofloxacin, 66.7% and 51.8% of the isolates were resistant, respectively.
In the case of SHV-type ESBL isolates, resistance rates were as follows: amikacin, 2.3%; gentamicin, 26.1%; tobramycin, 24.6%; kanamycin, 33.8%; and streptomycin, 50.8%. A high resistance rate corresponded to sulfonamides (83.1%), but rates were lower for tetracycline, trimethoprim, chloramphenicol, nalidixic acid, and ciprofloxacin (52.3%, 56.9%, 46.1%, 41.5%, and 26.1%, respectively). The highest percentages of resistance to gentamicin (71.1%) and tobramycin (77.8%) were observed among TEM ESBL-producing Enterobacteriaceae. Among these isolates, 11.1% were resistant to amikacin, 35.5% to kanamycin, and 33.3% to streptomycin. Identical percentages of resistance of 33.3% were observed with chloramphenicol and trimethoprim. Similarly, both tetracycline and nalidixic acid percentages of resistance were 40%, while 28.9% of the isolates were resistant to ciprofloxacin.
DISCUSSION
The presence of ESBLs in the community and hospital settings has previously been documented, particularly the expanding group of CTX-M enzymes (2, 25, 30). Although with less impact, SHV and TEM ESBLs still represent a threat in the hospital environment (25). The distribution of ESBL types in our collection is similar to that reported in other studies including hospitalized patients or community ESBL fecal carriers (13, 18, 27, 30).
Dissemination of CTX-M-coding genes among different Enterobacteriaceae isolates and environments would seem to support a variety of mechanisms of dispersion. Coresistance, frequently linked to resistance genes found in bla gene surroundings, is also important for maintenance and dispersion of ESBL-producing organisms (2, 15, 18, 25). The multidrug resistance found in isolates harboring CTX-M-9 and CTX-M-14 is a matter of concern. Resistance rates over 80% for nalidixic acid and over 50% for ciprofloxacin were observed in both groups. Although quinolone resistance has been linked to the presence of qnr elements in CTX-M-producing variants, particularly when harbored in the class 1 integron-bearing CR1 (Common Region 1) (23), the presence of qnr was ruled out and discarded from our collection (L. Poirel, R. Cantón, and P. Nordmann, unpublished data). The high levels of streptomycin and sulfonamide resistance in these isolates are related to the presence of streptomycin and sulfonamide resistance gene cassettes linked to class 1 integrons (18). The significantly low coresistance levels found in CTX-M-10-harboring isolates are of note. In our environment, the blaCTX-M-10 gene is surrounded by a unique structure that is not accompanied by resistant cassettes (24). On the other hand, despite no apparent linkage of blaSHV or blaTEM genes to integrons (18), a substantial proportion of isolates with these ESBL variants was also resistant to aminoglycosides and sulfonamides. Almost 30% of these isolates were also resistant to ciprofloxacin.
It is well known that the use of expanded-spectrum cephalosporins in the hospital setting influences the selection of ESBL-producing organisms (17, 25). In addition, the use of aminoglycosides increases the risk of hospitalized patients being infected or colonized with this type of isolate, as does the use of quinolones in the community setting (25). Obviously, the combined use of all these drugs implies additive increases in selective pressure. Previous courses with unrelated antimicrobials and the influence of a coselection event in the onset of ESBLs were not analyzed in our study.
Coresistance, such as that found in a significant proportion of ESBL-producing isolates from our study and from others (29), precludes the empirical use of most of the antimicrobial families, particularly in the case of severe infections. Carbapenems represent the first option when these infections are caused by ESBL producers (17), but the viability of this option might be weakened in the presence of strains producing metallo-β-lactamases (12). The efficacy of the combination of a β-lactam and a β-lactamase inhibitor is unclear, as treatment failures have been reported. In vitro data supports the assumption that tigecycline can also be considered an alternative for infections involving multiresistant ESBL-producing isolates (1, 14). The possibility of gut colonization with ESBL-producing organisms selected by previous courses of therapy or prophylaxis with unrelated antimicrobials and a history of previous hospitalization permit only limited therapeutic options, such as carbapenems and tigecycline, if a subsequent infection develops (16, 22, 32).
According to our study and other reports (1, 20), tigecycline is not affected in multiresistant ESBL-producing Enterobacteriaceae. Significantly, tigecycline has recently been advocated as one of the few options with in vitro activity against certain metallo-β-lactamase-producing gram-negative pathogens (12) and particularly in the case of carbapenemase-producing K. pneumoniae isolates (5). However, tigecycline has to cope with intrinsic resistance in Proteeae and an overall decreased activity in Serratia spp. and some K. pneumoniae strains and Enterobacter spp. (16, 28). In our study, a tigecycline MIC90 of 1 μg/ml was observed and no fully resistant isolates were observed. This is in agreement with the findings of Biedenbach et al. (1) with a smaller collection of ESBL producers, which included only E. coli and K. pneumoniae isolates and in which ESBLs were not characterized. As in this study, a small number of isolates, all of them from K. pneumoniae, displayed nonsusceptible tigecycline MICs of 4 μg/ml. Serratia spp. and Proteeae were not included in our work. Our study confirms previous reports that demonstrate that tigecycline assures in vitro coverage of most ESBL-producing enteric bacilli and is not involved, at least at present, in the coresistance phenomenon. The introduction of a drug such as tigecycline, lacking cross-resistance with other selective compounds, could be an opportunity to reduce the intensity of selection for ESBL-producing organisms derived from the use of other antimicrobial agents. However, this in vitro promise will require support from clinical studies.
Acknowledgments
We thank Felisa Almaraz and Azucena Rollán for their excellent technical assistance.
María García-Castillo, Ângela Novais, and Aránzazu Valverde were funded by fellowships from the European Commission (LSHM-CT-2003-503335), Ministerio de Ciencia y Tecnología (SAF 2003-09285), and Fondo de Investigaciones Sanitarias (PI020943-2002), respectively. This work was partially supported by an unrestricted grant from Wyeth Farma S.A., Spain, and the Microbial Sciences Foundation, Madrid, Spain.
REFERENCES
- 1.Biedenbach, D. J., M. L. Beach, and R. N. Jones. 2001. In vitro antimicrobial activity of GAR-936 tested against antibiotic-resistant gram-positive blood stream infection isolates and strains producing extended-spectrum β-lactamases. Diagn. Microbiol. Infect. Dis. 40:173-177. [DOI] [PubMed] [Google Scholar]
- 2.Bonnet, R. 2004. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouchillon, S. K., D. J. Hoban, B. M. Johnson, T. M. Stevens, M. J. Dowzicky, D. H. Wu, and P. A. Bradford. 2005. In vitro evaluation of tigecycline and comparative agents in 3,049 clinical isolates: 2001 to 2002. Diagn. Microbiol. Infect. Dis. 51:291-295. [DOI] [PubMed] [Google Scholar]
- 4.Bradford, P. A., D. T. Weaver-Sands, and P. J. Petersen. 2005. In vitro activity of tigecycline against isolates from patients enrolled in phase 3 clinical trials of treatment for complicated skin and skin-structure infections and complicated intra-abdominal infections. Clin. Infect. Dis. 41(Suppl. 5):S315-S332. [DOI] [PubMed] [Google Scholar]
- 5.Bratu, S., P. Tolaney, U. Karamudi, J. Quale, M. Mooty, S. Nichani, and D. Landman. 2005. Carbapenemase-producing Klebsiella pneumoniae in Brooklyn, N.Y.: molecular epidemiology and in vitro activity of polymyxin B and other agents. J. Antimicrob. Chemother. 56:128-132. [DOI] [PubMed] [Google Scholar]
- 6.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial disk susceptibility tests, 9th ed. Approved standard M2-A9. Clinical Laboratory and Standards Institute, Wayne, Pa.
- 8.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility test for bacteria that grow aerobically, 7th ed. Approved standard M7-A7. Clinical Laboratory and Standards Institute, Wayne, Pa.
- 9.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing; 16th informational supplement. CLSI document M100-S16. Clinical and Laboratory Standards Institute, Wayne, Pa.
- 10.Coque, T. M., A. Oliver, J. C. Pérez-Díaz, F. Baquero, and R. Cantón. 2002. Genes encoding TEM-4, SHV-2, and CTX-M-10 extended-spectrum β-lactamases are carried by multiple Klebsiella pneumoniae clones in a single hospital (Madrid, 1989 to 2000). Antimicrob. Agents Chemother. 46:500-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fluit, A. C., A. Florijn, J. Verhoef, and D. Milatovic. 2005. Presence of tetracycline resistance determinants and susceptibility to tigecycline and minocycline. Antimicrob. Agents Chemother. 49:1636-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helfand, M. S., and R. A. Bonomo. 2005. Current challenges in antimicrobial chemotherapy: the impact of extended-spectrum-β-lactamases and metallo-β-lactamases on the treatment of resistant gram-negative pathogens. Curr. Opin. Pharmacol. 5:452-458. [DOI] [PubMed] [Google Scholar]
- 13.Hernández, J. R., L. Martínez-Martínez, R. Cantón, T. M. Coque, A. Pascual, and the Spanish Group for Nosocomial Infections (GEIH). 2005. Nationwide study of Escherichia coli and Klebsiella pneumoniae producing extended-spectrum β-lactamases in Spain. Antimicrob. Agents Chemother. 49:2122-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoban, D. J., S. K. Bouchillon, B. M. Johnson, J. L. Johnson, and M. J. Dowzicky. 2005. In vitro activity of tigecycline against 6,792 gram-negative and gram-positive clinical isolates from the global tigecycline evaluation and surveillance trial (TEST Program, 2004). Diagn. Microbiol. Infect. Dis. 52:215-227. [DOI] [PubMed] [Google Scholar]
- 15.Lartigue, M. F., L. Poirel, and P. Nordmann. 2004. Diversity of genetic environment of blaCTX-M genes. FEMS Microbiol. Lett. 234:201-207. [DOI] [PubMed] [Google Scholar]
- 16.Livermore, D. M. 2005. Tigecycline: what is it, and where should it be used? J. Antimicrob. Chemother. 56:611-614. [DOI] [PubMed] [Google Scholar]
- 17.Livermore, D. M., and P. M. Hawkey. 2005. CTX-M: changing the face of ESBLs in the United Kingdom. J. Antimicrob. Chemother. 56:451-454. [DOI] [PubMed] [Google Scholar]
- 18.Machado, E., R. Cantón, F. Baquero, J. C. Galán, A. Rollán, L. Peixe, and T. M. Coque. 2005. Integron content of extended-spectrum-β-lactamase-producing Escherichia coli strains over 12 years in a single hospital in Madrid, Spain. Antimicrob. Agents Chemother. 49:1823-1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McAleese, F., P. Petersen, A. Ruzin, P. M. Dunman, E. Murphy, S. J. Projan, and P. A. Bradford. 2005. A novel MATE family efflux pump contributes to the reduced susceptibility of laboratory-derived Staphylococcus aureus mutants to tigecycline. Antimicrob. Agents Chemother. 49:1865-1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milatovic, D., F. J. Schmitz, J. Verhoef, and A. C. Fluit. 2003. Activities of the glycylcycline tigecycline (GAR-936) against 1,924 recent European clinical bacterial isolates. Antimicrob. Agents Chemother. 47:400-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moore, I. F., D. W. Hughes, and G. D. Wright. 2005. Tigecycline is modified by the flavin-dependent monooxygenase TetX. Biochemistry 44:11829-11835. [DOI] [PubMed] [Google Scholar]
- 22.Nathwani, D. 2005. Tigecycline: clinical evidence and formulary positioning. Int. J. Antimicrob. Agents 25:185-192. [DOI] [PubMed] [Google Scholar]
- 23.Nordmann, P., and L. Poirel. 2005. Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae. J. Antimicrob. Chemother. 56:463-469. [DOI] [PubMed] [Google Scholar]
- 24.Oliver, A., T. M. Coque, D. Alonso, A. Valverde, F. Baquero, and R. Cantón. 2005. CTX-M-10 linked to a phage-related element is widely disseminated among Enterobacteriaceae in a Spanish hospital. Antimicrob. Agents Chemother. 49:1567-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paterson, D. L., and R. A. Bonomo. 2005. Extended-spectrum β-lactamases: a clinical update. Clin. Microbiol. Rev. 18:657-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pitout, J. D. D., P. Nordmann, K. B. Laupland, and L. Poirel. 2005. Emergence of Enterobacteriaceae producing extended-spectrum β-lactamases (ESBLs) in the community. J. Antimicrob. Chemother. 56:52-59. [DOI] [PubMed] [Google Scholar]
- 27.Rodríguez-Baño, J., M. D. Navarro, L. Romero, L. Martínez-Martínez, M. A. Muniain, E. J. Perea, R. Pérez-Cano, and A. Pascual. 2004. Epidemiology and clinical features of infections caused by extended-spectrum beta-lactamase-producing Escherichia coli in nonhospitalized patients. J. Clin. Microbiol. 42:1089-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruzin, A., M. A. Visalli, D. Keeney, and P. A. Bradford. 2005. Influence of transcriptional activator RamA on expression of multidrug efflux pump AcrAB and tigecycline susceptibility in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 49:1017-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwaber, M. J., S. Navon-Venezia, D. Schwartz, and Y. Carmeli. 2005. High levels of antimicrobial coresistance among extended-spectrum-β-lactamase-producing Enterobacteriaceae. Antimicrob. Agents Chemother. 49:2137-2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Valverde, A., T. M. Coque, M. P. Sánchez-Moreno, A. Rollán, F. Baquero, and R. Cantón. 2004. Dramatic increase in prevalence of fecal carriage of extended-spectrum-β-lactamase-producing Enterobacteriaceae during nonoutbreak situations in Spain. J. Clin. Microbiol. 42:4769-4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wyeth Pharmaceutics. 2005. Tygacil (tigecycline) for injection [package insert]. Wyeth Pharmaceuticals Inc., Philadelphia, Pa.
- 32.Zhanel, G. G., J. A. Karlowsky, E. Rubinstein, and D. J. Hoban. 2006. Tigecycline: a novel glycylcycline antibiotic. Expert Rev. Anti Infect. Ther. 4:9-25. [DOI] [PubMed] [Google Scholar]