Abstract
We compared the population structure and drug resistance patterns of the Mycobacterium tuberculosis strains currently circulating in the Beijing area of China. One hundred thirteen of 123 strains belonged to the Beijing family genotypes defined by spoligotyping. The Beijing genotype strains were further subdivided into old and modern sublineages on the basis of NTF locus analysis. A stronger association with resistance to the more recently introduced antituberculosis drugs has been observed for old versus modern strains of the Beijing genotype, suggesting that its different sublineages may differ in their mechanisms of adaptation to drug selective pressure.
In some areas of the world, the increased rate of multidrug-resistant (MDR) tuberculosis (TB) appears to be linked to a disequilibrium in the local population structure of Mycobacterium tuberculosis, manifested as a predominance of particular genetic lineages and sublineages. For example, strains of the Beijing genotype are endemically prevalent in eastern Asia, South Africa, and northern Eurasia (reviewed in references 1 and 9), and new unexpected routes of their transmission are being uncovered (7). This genotype was first identified in M. tuberculosis strains isolated in the Beijing area of China, for which it was named (23). Currently, these strains attract great attention worldwide because they demonstrate important pathogenic features (15, 24) and association with drug resistance (4, 9).
China is one of the MDR TB hot spots, along with Russia and India (6). The increased rate of drug-resistant and MDR strains of M. tuberculosis remains a serious problem of TB control in China (14, 20). In the present study, we investigated the current population structure of M. tuberculosis in the Beijing area of China and compared it with drug resistance patterns in order to gain new insights into the evolution of drug-resistant TB.
One hundred twenty-three M. tuberculosis isolates were recovered from 123 adult pulmonary TB patients admitted to the Beijing Chest Hospital in 2002 to 2005. No epidemiological connection of these patients could be detected by standard investigation. For each patient, only the first available isolate was included in this study. Löwenstein-Jensen medium was used for cultivation of isolates. Testing of susceptibility to rifampin (RIF), isoniazid (INH), streptomycin (STR), ethambutol (EMB), and pyrazinamide (PZA) was done by the method of absolute concentration as previously described (3).
DNA from cultured cells was extracted as described by van Embden et al. (22). Strain differentiation was performed by spoligotyping (11). A PCR approach was used to determine a possible IS6110 insertion(s) in the NTF region of the M. tuberculosis Beijing genotype strains (18). Three NTF variants of the Beijing strains are distinguished on the basis of the presence or absence of the IS6110 sequence, thus providing a rough subdivision within this genotype. The W branch prevalent in the United States harbors two head-to-tail IS6110 insertions separated by a 556-bp noncoding spacer (13). Most Beijing strains worldwide harbor only one IS6110 insertion (1, 13); we previously defined them as the NTF::IS6110 “modern” branch (16). Finally, the Beijing strains without an IS6110 insertion in the NTF region (12, 13) were previously defined as atypical (12, 17), ancient/primordial (16), or ancestral (17, 19); here, we assigned these strains to an “old” sublineage of the Beijing genotype.
Odds ratios and P values, were calculated with EpiCalc software (8).
Results and discussion.
A total of 123 M. tuberculosis strains were included in this study. They comprised 40 pansusceptible and 83 resistant isolates randomly selected among those isolated between 2002 and 2005 in Beijing, China. A majority of the resistant strains were multidrug resistant; the distribution of the drug resistance profiles of all of the strains is shown in Table 1.
TABLE 1.
Drug resistance profiles of all of the M. tuberculosis strains studied and of modern versus old Beijing genotype strains
| Drug resistance profile (no. of strains)a | No. (%) of strains
|
P value | Odds ratio (95% confidence interval) | |
|---|---|---|---|---|
| Old Beijing sublineage (n = 27) | Modern Beijing sublineage (n = 86) | |||
| Sensitive (40) | 6 (22.2) | 30 (34.9) | 0.2 | 1.88 (0.68, 5.15) |
| H (2) | 2 (2.3) | |||
| R (5) | 1 (3.7) | 2 (2.3) | ||
| SH (6) | 5 (5.8) | |||
| SR (6) | 3 (11.1) | 3 (3.5) | 0.1 | 3.46 (0.66, 18.25) |
| HR (16) | 5 (18.5) | 10 (11.6) | 0.4 | 1.73 (0.53, 5.59) |
| HZ (1) | 1 (1.2) | |||
| RZ (2) | 2 (2.3) | |||
| RE (1) | 1 (1.2) | |||
| SHR (25) | 6 (22.2) | 17 (19.8) | 0.8 | 1.16 (0.41, 3.32) |
| SHE (1) | 1 (1.2) | |||
| SRE (1) | 1 (1.2) | |||
| HRZ (11) | 6 (22.2) | 5 (5.8) | 0.01 | 4.63 (1.29, 16.65) |
| HRE (1) | 1 (1.2) | |||
| SHRE (5) | 5 (5.8) | |||
S, STR; H, INH; R, RIF; E, EMB; Z, PZA.
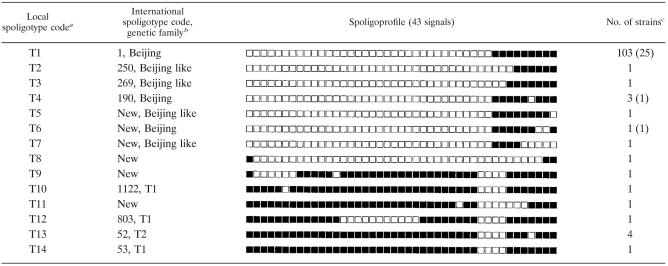
All strains were subjected to spoligotyping in order to assess their genetic relatedness. This genotyping method subdivided the 123 strains into 14 types (Table 2). The largest cluster (profile T1, Table 2) had a nine-signal spoligoprofile (signals 35 to 43) known to be a characteristic signature of the Beijing genotype (1, 9, 12). Additionally, eight strains (types T2 to T7) had abridged Beijing-like profiles with some of the nine signals absent. Thus, 113 (91.9%) of the 123 strains in this study belonged the Beijing family genotypes. The remaining spoligoprofiles (T8 to T14) were compared against the global spoligotype database SpolDB 4.0 (2) and the spoligotype signatures of the different M. tuberculosis families (2, 5). Accordingly, the respective codes (types and families) were assigned where available (Table 2).
TABLE 2.
Spoligotyping profiles of M. tuberculosis strains from Beijing, China
A further rough subdivision within the Beijing genotype was performed by analysis of the NTF region. It revealed that 27 strains had an intact NTF region (old Beijing genotype sublineage) while 86 strains had one IS6110 direct insertion on the right side of the NTF region (modern Beijing genotype sublineage). In our sample, old Beijing strains constituted 24% of the isolates. Previously, this rate was reported to be 5% in Russia (17), 14% in Hong Kong, and 25% in Vietnam (12). Mokrousov et al. (16) hypothesized that primary dispersal of the Beijing genotype strains took place in China and had been driven by Neolithic Proto-Sino-Tibetan farmers, whereas introduction of these strains into northern Eurasia was historically recent and might be associated with expansion of the Mongol empire in the 13th to the 15th centuries. In this view, it seems that the ratio of the old (ancient [16]) versus modern Beijing sublineages is a specific feature reflecting how “old” or “young,” as a whole, a Beijing strain population circulating in a given geographic area is, although it may be influenced by TB control measures such as therapy and vaccination.
The Beijing genotype strains constituted the majority of our collection (91.9%), and we further compared the distribution of drug resistance patterns within the Beijing genotype sample for old versus modern Beijing genotype sublineages. Interestingly, the spectrum of resistance patterns was much wider in the modern versus old Beijing group although this could mainly be due to the higher number of isolates in the modern group (Table 1). Our comparison revealed that resistance to RIF and PZA, as well as MDR and HRZ patterns, was statistically significantly associated with old Beijing strains (Tables 1 and 3). In the case of INH, this association was not significant while STR resistance was very weakly associated with modern Beijing strains. Similar observations on STR and INH resistance and multidrug resistance in modern (typical) and old (atypical) Beijing genotype strains have been made by Kremer (12) for strains from Hong Kong and Vietnam. Anti-TB chemotherapy was only introduced some 60 years ago (10), and neither old nor modern Beijing strains were subjected to the selective pressure of the drugs before then. A comparison of the order of implementation of the particular drugs used for anti-TB therapy with P values in Table 3 revealed an enigmatic and somewhat quantitative association pattern. It shows that resistance to the historically more recently introduced drugs (RIF and PZA) is associated with old Beijing strains and furthermore is the most prominent for the most recently implemented drug, PZA. Contrarily, this difference between old and modern Beijing sublineages is smoothed and even reversed for INH and STR, respectively, which were introduced earlier (Table 3).
TABLE 3.
Drug resistance in old versus modern Beijing genotype strains
| Drug resistancea | No. (%) of strains
|
P value | Odds ratio (95% confidence interval) | |
|---|---|---|---|---|
| Old Beijing sublineage (n = 27) | Modern Beijing sublineage (n = 86) | |||
| S (1944) | 9 (33.3) | 32 (37.2) | 0.7 | 0.84 (0.34, 2.10) |
| H (1952) | 17 (63.0) | 47 (54.7) | 0.5 | 1.41 (0.58, 3.43) |
| R (1970s) | 21 (77.8) | 47 (54.7) | 0.03 | 2.90 (1.07, 7.91) |
| E (1960s) | 9 (10.5) | |||
| Z (1980s) | 6 (22.2) | 8 (9.3) | 0.08 | 2.79 (0.87, 8.91) |
| Any drug resistance | 21 (77.8) | 56 (65.1) | 0.2 | 1.88 (0.68, 5.15) |
| MDR | 17 (63.0) | 38 (44.2) | 0.09 | 2.15 (0.88, 5.23) |
In parentheses is the time of introduction of the particular drug for anti-TB therapy (10). S, STR; H, INH; R, RIF; E, EMB, Z, PZA; MDR, resistance to at least RIF and INH.
It may be that different mechanisms including second-order selection acting on DNA repair systems (19, 21) lie behind mycobacterial adaptation and, in particular, behind the development of drug resistance in different sublineages of the Beijing genotype. The observed variation of the drug resistance patterns in the M. tuberculosis Beijing strain populations in different countries may be explained by different ratios of modern to old Beijing genotype subpopulations in the local populations of M. tuberculosis. Perhaps the prevalence of the modern Beijing strains in the currently circulating global population of this genotype results not from drug-driven selection but from a real situation prior to the advent of anti-TB therapy 60 years ago, an evolutionarily negligible period of time. It seems reasonable to assume that old (ancestral [17, 19], ancient [16], or atypical [12]) strains were also less numerous than modern “progeny” (typical [12]) strains in that population of the Beijing genotype. Alternatively, the current worldwide dissemination of the modern Beijing genotype strains may be due to their hypervirulent features rather than increased mutability leading to the rapid acquisition of drug resistance, although further studies of this effect are undoubtedly required.
Acknowledgments
I.M. and W.W.J. contributed equally to this study.
REFERENCES
- 1.Bifani, P. J., B. Mathema, N. E. Kurepina, and B. N. Kreiswirth. 2002. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 10:45-52. [DOI] [PubMed] [Google Scholar]
- 2.Brudey, K., J. R. Driscoll, L. Rigouts, W. M. Prodinger, A. Gori, S. A. Al-Hajoj, C. Allix, L. Aristimuno, J. Arora, V. Baumanis, L. Binder, P. Cafrune, A. Cataldi, S. Cheong, R. Diel, C. Ellermeier, J. T. Evans, M. Fauville-Dufaux, S. Ferdinand, D. Garcia de Viedma, C. Garzelli, L. Gazzola, H. M. Gomes, M. C. Gutierrez, P. M. Hawkey, P. D. van Helden, G. V. Kadival, B. N. Kreiswirth, K. Kremer, M. Kubin, S. P. Kulkarni, B. Liens, T. Lillebaek, H. M. Ly, C. Martin, I. Mokrousov, O. Narvskaya, Y. F. Ngeow, L. Naumann, S. Niemann, I. Parwati, M. Z. Rahim, V. Rasolofo-Razanamparany, T. Rasolonavalona, M. L. Rossetti, S. Rusch-Gerdes, A. Sajduda, S. Samper, I. Shemyakin, U. B. Singh, A. Somoskovi, R. Skuce, D. van Soolingen, E. M. Streicher, P. N. Suffys, E. Tortoli, T. Tracevska, V. Vincent, T. C. Victor, R. Warren, S. F. Yap, K. Zaman, F. Portaels, N. Rastogi, and C. Sola. 2006. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol. 6:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chinese Anti-tuberculosis Association. 1996. The laboratory science procedure of diagnostic bacteriology in tuberculosis. Zhong Guo Fang Lao Za Zhi 18:28-31. [Google Scholar]
- 4.Cox, H. S., T. Kubica, D. Doshetov, Y. Kebede, S. Rüsch-Gerdess, and S. Niemann. 2005. The Beijing genotype and drug resistant tuberculosis in the Aral Sea region of Central Asia. Respir. Res. 6:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferdinand, S., G. Valetudie, C. Sola, and N. Rastogi. 2004. Data mining of Mycobacterium tuberculosis complex genotyping results using mycobacterial interspersed repetitive units validates the clonal structure of spoligotyping-defined families. Res. Microbiol. 155:647-654. [DOI] [PubMed] [Google Scholar]
- 6.Floyd, K., L. Blanc, and M. Raviglione. 2002. Resources required for global tuberculosis control. Science 295:2040-2041. [DOI] [PubMed] [Google Scholar]
- 7.Garcia de Viedma, D., F. Chaves, and J. Inigo for the Tuberculosis Molecular Epidemiology Study Group. 2006. New route of importation of Mycobacterium tuberculosis Beijing genotype. Emerg. Infect. Dis. 12:169-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilman, J., and M. Myatt. 1998. EpiCalc 2000. Version 1.02. Brixton Books, London, United Kingdom.
- 9.Glynn, J. R., J. Whiteley, P. J. Bifani, K. Kremer, and D. van Soolingen. 2002. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerg. Infect. Dis. 8:843-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iseman, M. D. 2002. Tuberculosis therapy: past, present and future. Eur. Respir. J. 36:87s-94s. [DOI] [PubMed] [Google Scholar]
- 11.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. D. A. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kremer, K. 2005. Genetic markers for Mycobacterium tuberculosis: characterization and spread of the Beijing genotype. Ph.D. thesis. University of Amsterdam, Blaricum, The Netherlands.
- 13.Kurepina, N. E., S. Sreevatsan, B. B. Plikaytis, P. J. Bifani, N. D. Connell, R. J. Donnelly, D. van Soolingen, J. M. Musser, and B. N. Kreiswirth. 1998. Characterization of the phylogenetic distribution and chromosomal insertion sites of five IS6110 elements in Mycobacterium tuberculosis: non-random integration in the dnaA-dnaN region. Tuberc. Lung Dis. 79:31-42. [DOI] [PubMed] [Google Scholar]
- 14.Liu, J. J., H. Y. Yao, and E. Y. Liu. 2005. Analysis of factors affecting the epidemiology of tuberculosis in China. Int. J. Tuberc. Lung Dis. 9:450-454. [PubMed] [Google Scholar]
- 15.Lopez, B., D. Aguilar, H. Orozco, M. Burger, C. Espitia, V. Ritacco, L. Barrera, K. Kremer, R. Hernandez-Pando, K. Huygen, and D. van Soolingen. 2003. A marked difference in pathogenesis and immune response induced by different Mycobacterium tuberculosis genotypes. Clin. Exp. Immunol. 133:30-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mokrousov, I., H. M. Ly, T. Otten, N. N. Lan, B. Vyshnevskyi, S. Hoffner, and O. Narvskaya. 2005. Origin and primary dispersal of the Mycobacterium tuberculosis Beijing genotype: clues from human phylogeography. Genome Res. 15:1357-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mokrousov, I., O. Narvskaya, T. Otten, A. Vyazovaya, E. Limeschenko, L. Steklova, and B. Vyshnevskyi. 2002. Phylogenetic reconstruction within Mycobacterium tuberculosis Beijing genotype in northwestern Russia. Res. Microbiol. 153:629-637. [DOI] [PubMed] [Google Scholar]
- 18.Plikaytis, B. B., J. L. Marden, J. Crawford, C. L. Woodley, W. R. Butler, and T. M. Shinnick. 1994. Multiplex PCR assay specific for the multidrug resistant strain W of Mycobacterium tuberculosis. J. Clin. Microbiol. 32:1542-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rad, M. E., P. Bifani, C. Martin, K. Kremer, S. Samper, J. Rauzier, B. Kreiswirth, J. Blazquez, M. Jouan, D. van Soolingen, and B. Gicquel. 2003. Mutations in putative mutator genes of Mycobacterium tuberculosis strains of the W-Beijing family. Emerg. Infect. Dis. 9:838-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang, S., and S. B. Squire. 2005. What lessons can be drawn from tuberculosis (TB) control in China in the 1990s? An analysis from a health system perspective. Health Policy 72:93-104. [DOI] [PubMed] [Google Scholar]
- 21.Tenaillon, O., F. Taddei, M. Radman, and I. Matic. 2001. Second-order selection in bacterial evolution: selection acting on mutation and recombination rates in the course of adaptation. Res. Microbiol. 152:11-16. [DOI] [PubMed] [Google Scholar]
- 22.van Embden, J. D. A., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnik, and P. M. Small. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Soolingen, D., L. Qian, P. E. W. de Haas, J. T. Douglas, H. Traore, F. Portaels, Z. Quing, D. Enkhasaikan, P. Nymadawa, and J. D. A. van Embden. 1995. Predominance of a single genotype of Mycobacterium tuberculosis in countries of East Asia. J. Clin. Microbiol. 33:3234-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang, M., J. Cong, Z. Yang, B. Samten, and P. F. Barnes. 1999. Enhanced capacity of a widespread strain of Mycobacterium tuberculosis to grow in human macrophages. J. Infect. Dis. 179:1213-1217. [DOI] [PubMed] [Google Scholar]