Abstract
We have developed a sensitive and quantitative assay using a DNA microarray for the detection of hepatitis B virus (HBV) DNA in serum. Fluorescently labeled target cDNA prepared from cloned HBV DNA or serum HBV DNA was hybridized to capture DNA on a slide. A linear relationship was obtained between the intensities of the array spot and the amount of the cloned or serum HBV DNA, indicating the quantitative accuracy of this assay system. In addition, there was a significant correlation between the number of molecules of serum HBV DNA determined by the DNA microarray and that determined by a branched-DNA assay (n = 21, r = 0.89). Given these results, we conclude that the DNA microarray assay system may be useful as a diagnostic technique in the clinical laboratory.
The hepatitis B virus (HBV) is a partially double-stranded DNA virus that infects the human liver (14). The detection and quantification of HBV DNA are clinically useful because they allow for estimation of HBV replication and responsiveness to therapy. Several assay systems have been developed for the detection of HBV DNA in serum. The PCR is the most sensitive assay, detecting as low as 10−5 pg of HBV DNA (12). Hybridization techniques such as slot blot hybridization (1, 21), cross-linking assay (13), or branched-DNA (bDNA) signal amplification assay (6, 19) are quantitative, but these are relatively insensitive and complex assays. There are several other potentially quantitative and sensitive methods for detection of HBV DNA, including the transcription-mediated amplification assay (from 5 × 103 to 5 × 108 copies/ml) (11), Hybrid Capture II assay (Digene; from 8 × 103 to 1.7 × 109 copies/ml) (7), and the HBV COBAS MONITOR (Roche; from 2 × 102 to 2 × 105 copies/ml) (5).
Microarray technology allows the quantitative and simultaneous analysis of hundreds to thousands of genes (16, 17, 18). This would allow for high-throughput screening for multiple pathogens in large numbers of samples. This technology utilizes a capture DNA that is fixed on a slide. Fluorescently labeled target cDNAs are prepared from samples, and these are hybridized to the capture DNA. Because of its potential advantages, in the present studies, we developed a microarray technique for the detection and quantification of HBV DNA in sera from patients.
HBV DNA fragments including a portion of the core (nucleotides [nt] 2248 to 2449; HB200), almost the entire core region (nt 1961 to 2449; HB500), the core to the pre-S1 gene (nt 1961 to 2961; HB1000), or the complete HBV DNA of pBlue-HBadr4 (HB3200) (4) were amplified by PCR with specific primers. The amplified PCR fragments were used as the capture DNA on a microarray analysis slide (Fig. 1A). Approximately 1 kb each of firefly and Renilla luciferase cDNA was also amplified by PCR and used for positive and negative controls, respectively (8). These fragments were spotted onto glass slides as described previously (2, 18). The HBV DNA from serum samples (100 μl) was precipitated with polyethylene glycol and processed with a DNA Extractor kit (Wako Pure Chemical Industries, Osaka, Japan) as described previously (10). The extracted sample DNA was mixed with 2.0 pg of the firefly luciferase DNA fragment as a positive control and labeled with cyanine-5-dUTP according to a procedure described previously (15). Hybridization on the slides was performed at 65°C for 1 h. The slides were washed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.03% sodium dodecyl sulfate at 65°C for 5 min, 1× SSC at 65°C for 5 min, and 0.2× SSC at room temperature for 5 min.
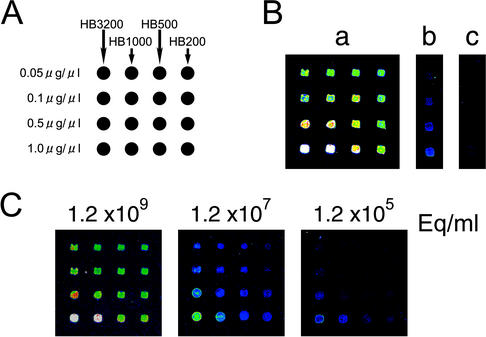
FIG. 1.
(A) DNA microarray matrix of HBV DNA spots. HBV DNAs with different sizes and concentrations were spotted on the slide. HB3200, the full HBV genome; HB1000, the core to the pre-S1 gene (nt 1961 to 2961); HB500, the whole core region (nt 1961 to 2449); HB200, a part of the core (nt 2248 to 2449). The diameter of each spot is 250 μm, and the estimated volume loaded for each spot is 10 nl. (B) Typical results from an array of HBV DNA spots captured on a ScanArray 5000 instrument. (a) HBV DNA isolated from a patient's serum and 2.0 pg of the firefly luciferase gene were labeled with Cy5 and hybridized onto the spotted DNA. The longer capture DNA (3.2 kb) generated a higher fluorescence intensity, as did higher concentrations of capture DNA (1.0 μg/μl). (b) Firefly luciferase DNA spots were used as positive controls. A total of 2.0 pg (1.7 × 106 copies) of firefly luciferase probe DNA generated a weak but clear signal. (c) The Renilla luciferase gene used as negative control did not generate a signal. (C) One of 21 serum samples was serially diluted (1.2 × 109 eq/ml, 1.2 × 107 eq/ml, and 1.2 × 105 eq/ml) and used in the DNA microarray analysis.
The glass slides were scanned on a ScanArray 5000 instrument (General Scanning, Watertown, Mass.), and image analysis was carried out with QuantArray software (General Scanning). To standardize each experiment, HBV DNA fluorescence intensities were normalized by the intensities of the positive control gene (2.0 pg [1.7 × 106 copies] of firefly luciferase). The intensity of the positive control, spotted at a concentration of 0.1 μg/μl, was set at 103 counts. Based on this standard, the relative counts with 0.1 μg of HB1000/μl as capture DNA were determined (Fig. 1A and B).
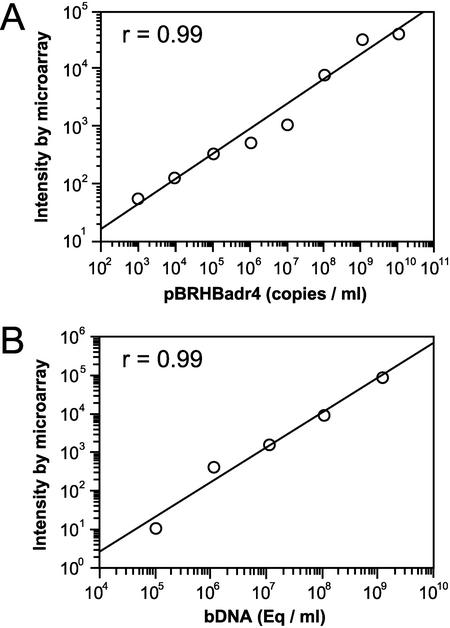
Results from a typical array with a patient's serum sample are shown in Fig. 1B-a. Use of a longer capture DNA (HB3200) or a higher concentration of spotted capture DNA (1.0 μg/μl) resulted in higher fluorescence intensity (Fig. 1B-a). The positive control of the firefly luciferase gene generated a relatively weak but clear signal as shown in Fig. 1B-b. As seen in Fig. 1B-c, the negative Renilla luciferase control did not produce any obvious signal. Figure 2A shows that there was a linear correlation between the various concentrations of pBRHBadr4 target DNA and the signal intensities (r = 0.99) and that the detection range for the microarray was from 103 to 1010 copies/ml.
FIG. 2.
(A) The fluorescence intensity from DNA microarray analysis as a function of serial dilutions from 103 to 1010 copies of plasmid pBRHBadr4 (r = 0.99, P < 0.01)/ml. The intensity of the HB1000 HBV DNA spot (spotted at a concentration of 0.1 μg/μl) was used to calculate the absolute intensity of HBV DNA. The intensity was normalized to the standard firefly luciferase gene of 103 counts in each assay. (B) Fluorescence intensity from DNA microarray analysis as a function of serial dilutions from 1.2 × 105 to 1.2 × 109 eq/ml of one patient's serum (r = 0.99, P < 0.01).
Serial dilutions of a patient's serum with a known viral titer (1.2 × 109 eq/ml as determined by the bDNA assay) were made in normal serum and evaluated in the microarray assay. Figure 2B shows that there was a strong linear correlation between the concentrations of the HBV DNA and the fluorescence intensities obtained from the microarray assay (r = 0.99). The patterns from HBV DNA spots with diluted serum samples (1.2 × 109 eq/ml, 1.2 × 107 eq/ml, and 1.2 × 105 eq/ml) are shown in Fig. 1C. The lower detection limit of HBV DNA in serum was 1.2 × 105 eq/ml, which is 2 log units higher than that with cloned DNA. However, the assay was at least as sensitive as the conventional bDNA assay (6).
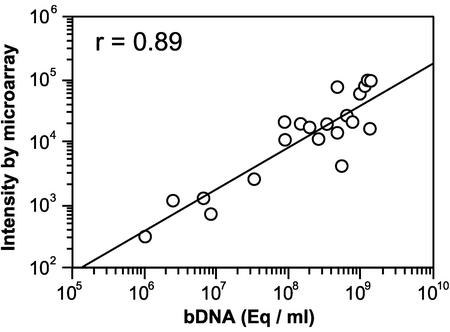
Twenty-five patients' sera from HBV carriers (positive for HBs antigen over 6 months) were analyzed with the DNA microarray assay, and the results were compared with those obtained with the bDNA assay. All patients were positive for HBe antigen and diagnosed as having chronic hepatitis. HBV DNA was detected in 21 patients' sera (84%). Four patients' sera were negative for HBV DNA. These sera were also negative according to the bDNA assay. Within the detection range from 106 to 109 eq/ml, there was a good correlation between the two assay systems (n = 21, r = 0.89) (Fig. 3). Six serum samples that were HBs antigen negative (one from a healthy volunteer, three from infectious mononucleosis patients, and two from patients with chronic hepatitis C) were also examined by this assay. No specific signal intensities were obtained from the samples (data not shown). Finally, the reproducibility and accuracy of this assay system were evaluated by repeated measurement, and the within-run coefficient of validation was 5.3%, while the between-run coefficient of validation was 2.6%.
FIG. 3.
Comparison of the HBV DNA levels in 21 serum samples determined by DNA microarray analysis and bDNA assays. There was a significant correlation between the results of these assays (r = 0.89, P < 0.01).
The cDNA microarray assay could be applied to both basic scientific and clinical research (16, 17, 18). For example, in addition to gene expression profiling, cDNA microarray technology has been applied to the detection of single-nucleotide pleomorphisms of the human genome and to genotyping of viruses such as the human immunodeficiency virus (20). In this study, we used DNA microarray technology to develop a sensitive and quantitative method for detecting HBV DNA in human serum samples.
Although hybridization techniques, including the microarray system, are less sensitive than are amplification-based assays like PCR or the recently developed transcription-mediated amplification assay, a DNA microarray assay may be more useful as a diagnostic tool. One advantage is that point mutations or deletions of the virus genome could be detected by the DNA microarray assay by using the appropriate sets of the viral DNA fragments as capture DNA (3). For example, evaluating patient samples for the YMDD mutation or mutations in the pre-C region of the polymerase gene in the HBV genome would provide important clinical information for monitoring interferon therapy and the prognosis of patients (9). Moreover, our system can be applied to the diagnosis of other DNA virus infections that cause hepatitis, such as those with Epstein-Barr virus, cytomegalovirus, and herpes simplex virus. In fact, these viral DNAs could be spotted together on a microarray slide and assayed in a single reaction.
It took about 12 h to complete our assay, i.e., 5 h for the HBV DNA extraction and 7 h for the labeling and hybridization. Modifying the DNA extraction procedure could reduce this time. Also, the sensitivity of the assay could be enhanced by increasing the amount of capture DNA on the slide or by pretreatment of the sample DNA. Finally, although the microarray assay system that we have described is preliminary and could be modified for optimal performance, it is clear that cDNA microarray technology could be applied to other pathogens and that it will be a useful diagnostic method in the clinical laboratory.
REFERENCES
- 1.Berninger, M., M. Hammer, B. Hoyer, and J. L. Gerin. 1982. An assay for the detection of the DNA genome of hepatitis B virus in serum. J. Med. Virol. 9:57-68. [DOI] [PubMed] [Google Scholar]
- 2.DeRisi, J., L. Penland, P. O. Brown, M. L. Bittner, P. S. Meltzer, M. Ray, Y. Chen, Y. A. Su, and J. M. Trent. 1996. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat. Genet. 14:457-460. [DOI] [PubMed] [Google Scholar]
- 3.Favis, R., J. P. Day, N. P. Gerry, C. Phelan, S. Narod, and F. Barany. 2000. Universal DNA array detection of small insertions and deletions in BRCA1 and BRCA2. Nat. Biotechnol. 18:561-564. [DOI] [PubMed] [Google Scholar]
- 4.Fujiyama, A., A. Miyanohara, C. Nozaki, T. Yoneyama, N. Ohtomo, and K. Matsubara. 1983. Cloning and structural analyses of hepatitis B virus DNAs, subtype adr. Nucleic Acids Res. 11:4601-4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerken, G., J. Gomes, P. Lampertico, M. Colombo, T. Rothaar, M. Trippler, and G. Colucci. 1998. Clinical evaluation and applications of the Amplicor HBV Monitor test, a quantitative HBV DNA PCR assay. J. Virol. Methods 74:155-165. [DOI] [PubMed] [Google Scholar]
- 6.Hendricks, D. A., B. J. Stowe, B. S. Hoo, J. Kolberg, B. D. Irvine, P. D. Neuwald, M. S. Urdea, and R. P. Perrillo. 1995. Quantitation of HBV DNA in human serum using a branched DNA (bDNA) signal amplification assay. Am. J. Clin. Pathol. 104:537-546. [DOI] [PubMed] [Google Scholar]
- 7.Ho, S. K., T. M. Chan, I. K. Cheng, and K. N. Lai. 1999. Comparison of the second-generation Digene Hybrid Capture assay with the branched-DNA assay for measurement of hepatitis B virus DNA in serum. J. Clin. Microbiol. 37:2461-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Honda, M., S. Kaneko, E. Matsushita, K. Kobayashi, G. A. Abell, and S. M. Lemon. 2000. Cell cycle regulation of hepatitis C virus internal ribosomal entry site-directed translation. Gastroenterology 118:152-162. [DOI] [PubMed] [Google Scholar]
- 9.Hunt, C. M., J. M. McGill, M. I. Allen, and L. D. Condreay. 2000. Clinical relevance of hepatitis B viral mutations. Hepatology 31:1037-1044. [DOI] [PubMed] [Google Scholar]
- 10.Ishizawa, M., Y. Kobayashi, T. Miyamura, and S. Matsuura. 1991. Simple procedure of DNA isolation from human serum. Nucleic Acids Res. 19:5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamisango, K., C. Kamogawa, M. Sumi, S. Goto, A. Hirao, F. Gonzales, K. Yasuda, and S. Iino. 1999. Quantitative detection of hepatitis B virus by transcription-mediated amplification and hybridization protection assay. J. Clin. Microbiol. 37:310-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaneko, S., S. M. Feinstone, and R. H. Miller. 1989. Rapid and sensitive method for the detection of serum hepatitis B virus DNA using the polymerase chain reaction technique. J. Clin. Microbiol. 27:1930-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai, V. C. H., R. Guan, M. L. Wood, S. K. Lo, M. F. Yuen, and C. L. Lai. 1999. Nucleic acid-based cross-linking assay for detection and quantification of hepatitis B virus DNA. J. Clin. Microbiol. 37:161-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahoney, F. J. 1999. Update on diagnosis, management, and prevention of hepatitis B virus infection. Clin. Microbiol. Rev. 12:351-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pollack, J. R., C. M. Perou, A. A. Alizadeh, M. B. Eisen, A. Pergamenschikov, C. F. Williams, S. S. Jeffrey, D. Botstein, and P. O. Brown. 1999. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat. Genet. 23:41-46. [DOI] [PubMed] [Google Scholar]
- 16.Schena, M., D. Shalon, R. W. Davis, and P. O. Brown. 1995. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270:467-470. [DOI] [PubMed] [Google Scholar]
- 17.Schena, M., D. Shalon, R. Heller, A. Chai, P. O. Brown, and R. W. Davis. 1996. Parallel human genome analysis: microarray-based expression monitoring of 1000 genes. Proc. Natl. Acad. Sci. USA 93:10614-10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shalon, D., S. J. Smith, and P. O. Brown. 1996. A DNA microarray system for analyzing complex DNA samples using two-color fluorescent probe hybridization. Genome Res. 6:639-645. [DOI] [PubMed] [Google Scholar]
- 19.Urdea, M. S., T. Horn, T. J. Fultz, M. Anderson, J. A. Running, S. Hamren, D. Ahle, and C. A. Chang. 1991. Branched DNA amplification multimers for the sensitive, direct detection of human hepatitis viruses. Nucleic Acids Symp. Ser. 24:197-200. [PubMed] [Google Scholar]
- 20.Vahey, M., M. E. Nau, S. Barrick, J. D. Cooley, R. Sawyer, A. A. Sleeker, P. Vickerman, S. Bloor, B. Larder, N. L. Michael, and S. Wegner. 1999. Performance of the Affymetrix Gene Chip HIV PRT 440 platform for antiretroviral drug resistance genotyping of human immunodeficiency virus type 1 clades and viral isolates with length polymorphisms. J. Clin. Microbiol. 37:2533-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walter, E., H. E. Blum, W. B. Offensperger, C. Zeschnigk, S. Offensperger, and W. Gerok. 1987. Spot-blot hybridization assay for the detection of hepatitis B virus DNA in serum: factors determining its sensitivity and specificity. Hepatology 7:557-562. [DOI] [PubMed] [Google Scholar]