Abstract
Mutations in and around the catalytic site of the reverse transcriptase (RT) of human immunodeficiency virus type 1 (HIV-1) are associated with resistance to nucleoside RT inhibitors (NRTIs), whereas changes in the hydrophobic pocket of the RT are attributed to nonnucleoside RT inhibitor (NNRTI) resistance. In this study, we report a novel series of nonnucleoside inhibitors of HIV-1, exemplified by VRX-329747 and VRX-413638, which inhibit both NNRTI- and NRTI-resistant HIV-1 isolates. Enzymatic studies indicated that these compounds are HIV-1 RT inhibitors. Surprisingly, however, following prolonged (6 months) tissue culture selection, this series of nonnucleoside inhibitors did not select NNRTI-resistant mutations in HIV-1 RT. Rather, four mutations (M41L, A62T/V, V118I, and M184V) known to cause resistance to NRTIs and two additional novel mutations (S68N and G112S) adjacent to the catalytic site of the enzyme were selected. Although the M184V mutation appears to be the initial mutation to establish resistance, this mutation alone confers only a two- to fourfold decrease in susceptibility to VRX-329747 and VRX-413638. At least two additional mutations must accumulate for significant resistance. Moreover, while VRX-329747-selected viruses are resistant to lamivudine and emtricitabine due to the M184V mutation, they remain susceptible to zidovudine, stavudine, dideoxyinosine, abacavir, tenofovir, and efavirenz. These results directly demonstrate that VRX-329747 and VRX-413638 are novel nonnucleoside inhibitors of HIV-1 RT with the potential to augment current therapies.
Standard human immunodeficiency virus (HIV) therapies consist of combinations of nucleoside reverse transcriptase inhibitors (NRTIs), nonnucleoside reverse transcriptase inhibitors (NNRTIs), protease inhibitors, and a fusion inhibitor. Although they are generally effective and have resulted in reduced AIDS-related morbidity and mortality, none is curative. Treatment failures often occur when viruses arise that are resistant to one or more components of the combination therapy regimens. Cross-resistance within each class of HIV type 1 (HIV-1) drugs adds to the challenge of treating patients experiencing drug failure (7, 28). Upon failure of one drug regimen, options are often limited when selecting subsequent therapy. Thus, there is an urgent need for HIV drugs that target novel steps in the virus life cycle or that exhibit distinct resistance profiles (33).
The mechanisms of drug resistance are class specific for NRTIs and NNRTIs because they bind to HIV-1 reverse transcriptase (RT) in distinct manners and select completely different sets of resistance mutations. Mutations such as M184V, L74V, K65R, and thymidine analogue mutations, including M41L, D67N, K70R, L210W, T215Y/F, and K219Q, are responsible for resistance to the currently approved NRTIs. NNRTIs bind to a unique hydrophobic pocket located near the palm subdomain of HIV-1 RT, which interferes with cooperative movements between the finger and thumb subdomains, thereby inhibiting catalysis of the enzyme (16, 19, 22, 26). The predominant RT mutations in viruses isolated from NNRTI-treated patients are K103N and Y181C.
In this report, we describe a novel class of NNRTIs that are active against multiple NRTI- and NNRTI-resistant viruses. In culture, these compounds selected four mutations associated with NRTI resistance and two additional novel mutations located adjacent to the catalytic site of HIV-1 RT, giving a profile distinct from the resistance profiles of all current NNRTIs. The majority of approved NRTIs and efavirenz are fully effective against the selected resistant viruses, indicating that this new class of compounds has the potential to be used in combination with current antiretrovirals, even in patients who are experiencing failure with current therapies.
MATERIALS AND METHODS
Compounds.
The nonnucleosides VRX-329747 and VRX-413638 were purchased from ChemDiv, Inc. (San Diego, CA). Zidovudine (AZT), lamivudine (3TC), and stavudine (d4T) were purchased from Sigma-Aldrich (St. Louis, MO). Efavirenz, dideoxyinosine (ddI), and abacavir were obtained from the NIH AIDS Research and Reference Reagent Program. Tenofovir was obtained from Moravek Biochemicals, Inc. (Brea, CA). Emtricitabine was synthesized at Valeant Research and Development. All test compound stocks were prepared in dimethyl sulfoxide (DMSO).
Cell lines, plasmids, and virus isolates.
HeLa-JC53 cells were obtained from David Kabat at Oregon Health Sciences University (24). The HeLa-JC53-LTR-β-gal cell line was constructed by transduction of HeLa-JC53 cells with an HIV type 1 (HIV-1) vector carrying the long terminal repeat (LTR)-β-galactosidase. The HeLa-JC53-LTR-Luci cell line was likewise constructed by transduction with an HIV-1 vector carrying LTR-luciferase.
The HIV-1 molecular clones pNL4-3 and pNL4-3.Luc.R−E− were obtained from the NIH AIDS Research and Reference Reagent Program (8). A panel of HIV-1 isolates carrying single and double NNRTI resistance mutations (Table 1) was constructed based on the molecular clone pNL4-3.Luc.R−E−, using QuikChange II XL site-directed mutagenesis (Stratagene, La Jolla, CA) according to the manufacturer's protocol. A positive clone for each mutant was digested with restriction enzymes with unique recognition sites (ApaI and AgeI; nucleotides 2006 to 3490 in the NL4-3 strain), and this DNA fragment was reintroduced into pNL4-3.Luc.R−E−. The resulting mutant plasmids were sequenced throughout the insert to ensure that only the desired mutations were introduced into the plasmids.
TABLE 1.
Changes in EC50 values (relative to wt EC50) of VRX-329747, VRX-413638, and efavirenz against NNRTI-resistant HIV-1 molecular clonesa
| NNRTI resistance mutation(s) | Mean change (fold) in EC50 ± SDb
|
||
|---|---|---|---|
| VRX-329747 (wt EC50 = 156 ± 20.1 nM) | VRX-413638 (wt EC50 = 23 ± 4.5 nM) | Efavirenz (wt EC50 = 0.1 ± 0.04 nM) | |
| K103N | 0.6 ± 0.09 | 0.5 ± 0.15 | 27.1 ± 2.1* |
| Y181C | 2.9 ± 0.06 | 1.4 ± 0.67 | 1.2 ± 0.10 |
| Y188L | 0.6 ± 0.15 | 0.3 ± 0.05 | 392 ± 3.72* |
| G190S | 1.7 ± 1.04 | 1.7 ± 0.53 | 168 ± 6.38* |
| K103N, L100I | 3.7 ± 0.52* | 3.6 ± 0.45* | 13,016 ± 1,230* |
| K103N, V108I | 1.2 ± 0.13 | 1.2 ± 0.19 | 114 ± 4.45* |
| K103N, Y181C | 2.2 ± 0.10 | 1.2 ± 0.39 | 23 ± 1.9* |
| K103N, Y188L | 0.7 ± 0.35 | 0.3 ± 0.21 | >50,000* |
| K103N, P225H | 0.3 ± 0.29 | 0.2 ± 0.05 | 381 ± 136* |
| V106I, Y188L | 0.4 ± 0.05 | 0.3 ± 0.16 | 2035 ± 32.4* |
Molecular clones were built in reporter vector pNL4-3.Luc.R−E−, and EC50 values were determined using HeLa-JC-53 cells.
From two tests. *, EC50 was statistically significantly higher than that against wild-type virus (P < 0.05).
The panel of NRTI-resistant HIV-1 isolates (Table 2) consisted of 10 viruses obtained from the NIH AIDS Research and Reference Reagent Program (10, 17) and 2 viruses constructed in-house, while a panel of 15 HIV-1 isolates carrying a combination of NRTI and NNRTI resistance mutations (Table 3) was generated and tested by Monogram Biosciences Inc. (South San Francisco, CA) (23). HIV-2(NIH-Z) was obtained from ABI Advanced Biotechnologies Inc. (Columbia, MD).
TABLE 2.
Changes in EC50 values (relative to wt EC50) of VRX-413638, AZT, and FTC against NRTI-resistant HIV-1 isolates
| NRTI resistance mutation(s)a | Mean change (fold) in EC50 ± SDb
|
||
|---|---|---|---|
| VRX-413638 (wt EC50 = 27.1 ± 3.40 nM) | AZT (wt EC50 = 16.4 ± 2.80 nM) | FTC (wt EC50 = 42.5 ± 3.70 nM) | |
| K65R | 0.2 ± 0.03 | 0.6 ± 0.06 | 102.7 ± 7.16* |
| M41L, D67N, T69N, K70R, T215F, K219E | 0.7 ± 0.14 | 21.0 ± 7.66* | 13.5 ± 6.60* |
| M41L, E44D, D67N, T69D, V118I, L210W, T215Y | 0.7 ± 0.19 | 389.1 ± 143.78* | 25.8 ± 21.24* |
| V75I, F77L, F116Y, Q151M | 0.9 ± 0.08 | 43.9 ± 9.43* | 17.9 ± 6.99* |
| M184V | 3.0 ± 0.66* | 0.3 ± 0.03 | Max* |
| M41L, K70R, T215F, K219E | 1.4 ± 0.03 | 83.7 ± 21.37* | 9.4 ± 4.30* |
| K65R, K70R, V75I, F77L, F116Y, Q151M | 1.6 ± 0.65 | Max* | Max* |
| D67N, T69N, K70R, M184V, T215F, K219Q | 1.9 ± 0.01 | 4.8 ± 2.20* | Max* |
| M41L, D67N, V118I, M184V, L210W, T215Y | 4.4 ± 1.18* | 9.5 ± 0.91* | Max* |
| M41L, D67N, M184V, L210W, T215Y | 5.7 ± 1.05* | 13.2 ± 7.32* | Max* |
| M41L, E44D, D67N, T69D, V118I, M184V, L210W, T215Y | 11.2 ± 0.87* | 54.7 ± 8.68* | Max* |
| T69K, K70G, V75I, F77L, F116Y, Q151M, M184V | 15.7 ± 2.98* | 274.3 ± 33.35* | Max* |
Molecular clones with K65R and M184V mutations were in the HIV-1 NL4-3 format. All other viruses were in pNLPFB and were obtained from the U.S. NIH AIDS Research and Reference Reagent Repository.
EC50 values were determined using the reporter cell line HeLa-JC-53-LTR-Luci. Mean EC50 values ± SD were from two tests. Max; the change could not be determined because the EC50 was >10 μM; *, EC50 was statistically significantly higher than that against wild-type virus (P < 0.05).
TABLE 3.
Changes in EC50 values (relative to wt EC50) of VRX-413638 and efavirenz against clinical HIV-1 isolates containing both NRTI and NNRTI resistance mutationsa
| NRTI mutation(s) | NNRTI mutation(s) | Mean change (fold) in EC50
|
|
|---|---|---|---|
| VRX-413638 (wt EC50 = 36 nM) | Efavirenz (wt EC50 = 1.0 nM) | ||
| L74V | V106A, F227L | 0.4 | 630 |
| M41L, D67N, T69N, L74V, M184V, L210W, T215Y, K219E | K101P, Y188L, F227L | 0.4 | >2,500 |
| L210W, K219E | K101E, Y181C, G190A | 1.1 | 328 |
| M41L, D67N, T69N, L210W, T215Y | K103N, G190A | 1.3 | 443 |
| M41L, D67N, T69N, L74V, M184V, L210W, T215Y, K219E | L100I, K103N | 1.4 | >2,500 |
| M41L, T69N, L210W, T215Y | K103N, Y181V | 1.5 | 19 |
| M41L, D67N, L74V, M184V, L210W, T215Y, K219E | K101H, Y181C, G190S | 1.7 | >2,500 |
| M41L, D67N, T69N, V75I, M184V, L210W, T215Y, K219E | K101E, K103N, V108I | 2 | 1,912 |
| D67N, K70R, K219E | V106I, V108I, Y181C | 2.2 | 7 |
| M184V, T215Y | Y188L | 2.7 | 106 |
| D67N, T69N, K70R, M184V, T215Y, K219E | K103N | 3.3 | 34 |
| M41L, A62V, V75I, M184V, T215Y | K103N, V108I | 4.4 | 196 |
| D67N, K70R, M184V, T215Y, K219E | Y188L, G190A | 5.2 | 2,385 |
| L74V, M184V | K103N, V106A, F227L | 8 | 685 |
| M41L, A62V, M184V, T215Y, K219E | K101Q, V106A | 8.2 | 3 |
EC50 values were determined at Monogram Biosciences, Inc., using the company's PhenoScreen test. Replicate assays were not performed, so the SD is not available.
Wild-type (wt) and RT mutant HIV-1 stocks were generated by transfection of 293T cells, using FuGene6 (Roche Applied Science, Indianapolis, IN). To generate infectious luciferase-expressing virus stocks, a plasmid encoding the vesicular stomatitis virus glycoprotein (VSV-G) was cotransfected with pNL4-3.Luc.R−E−-derived plasmids (0.4:1 ratio) (8, 25). Supernatants containing virus were collected and aliquots frozen at 48 h posttransfection. Viral titers (tissue culture infective units/ml) of the stocks were determined using HeLa-JC53-LTR-β-gal cells as previously described by Kimpton and Emerman (18).
In vitro RT assay and enzyme kinetics.
HIV-1 RT was expressed in Escherichia coli and purified by the procedure of Boretto et al. (6). The expression plasmid p66RTB was a gift from B. Canard. In vitro RT reactions for 50% inhibitory concentration (IC50) measurements were carried out for 1 h at 25°C in the presence of 16 μg/ml poly(rA)/oligo(dT)18, 2 μM TTP (labeled with 0.5 μCi of α-33P), 1 nM RT, and 0 to 100 μM inhibitor in a buffer containing 50 mM Tris, pH 7.5, 5 mM MgCl2, 1 mM dithiothreitol, and 60 μg/ml bovine serum albumin. Reactions were quenched and precipitated by adding equal volumes of 20% trichloroacetic acid-1% sodium pyrophosphate. Radioactivity in the precipitated product was analyzed. The IC50 was defined as the concentration of inhibitor required to inhibit RT activity by 50%.
Steady-state kinetic assays for determining the mode of inhibition were carried out under the same conditions as those described above in the presence of 0 to 30 μM [α-33P]TTP and a fixed concentration of 13 μg/ml poly(rA)/oligo(dT)18 or 0 to 30 μg/ml poly(rA)/oligo(dT)18 and a fixed concentration of 10 μM [α-33P]TTP. Data were fitted to the following equations using a nonlinear global fitting program in Prism 4.0 (GraphPad Software, San Diego, CA): Competitive inhibition:
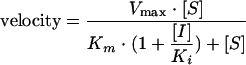 |
(1) |
Mixed-type inhibition:
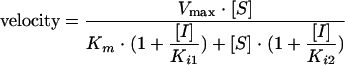 |
(2) |
Uncompetitive inhibition:
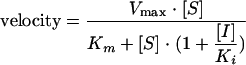 |
(3) |
In vitro anti-HIV-1 activity assays.
The antiviral activities of VRX-329747, VRX-413638, and current HIV-1 drugs were characterized against panels of NNRTI- and NRTI-resistant HIV-1 molecular clones and clinical isolates. HeLa-JC53 cells were used for VSV-G-pseudotyped pNL4-3.Luc.R−E−-derived virus infections, while HeLa-JC53-LTR-Luci cells (expressing luciferase under the control of the HIV-1 LTR) were used for infections of pNL4-3-derived viruses and HIV-2 (14, 24, 25, 30). Serial dilutions (1:4) of test compounds were performed in DMSO and then in culture medium (phenol red-free Dulbecco's modified Eagle's medium, 2% fetal bovine serum, and 1% penicillin-streptomycin) to yield a final DMSO concentration of 0.2% in the cell culture. Using 96-well solid white plates (Beckman Coulter, Fullerton, CA), cells (15,000 in 85 μl of culture medium) were mixed with 5 μl of virus (multiplicity of infection = 0.03) and 10 μl of the serially diluted compounds. All cultures were maintained at 37°C in 5% CO2 for 48 h. Following incubation, an equal volume (100 μl) of Bright-Glo luciferase reagent (Promega, Madison, WI) was added to each well, and chemiluminescence was read on an LJL Analyst system (LJL BioSystems, Sunnyvale, CA). Fifty percent effective concentration (EC50) values (defined as the concentration of inhibitor required to inhibit luciferase activity by 50%) were determined by nonlinear regression using XLfit4 or GraphPad Prism 4. One-way analysis of variance to compare mutant and wild-type virus EC50 values was performed with GraphPad Prism 4 after logarithmic transformation of the EC50 values.
In vitro cellular toxicity assay.
The cellular toxicities of VRX-329747 and VRX-413638 were determined against a panel of tissue culture cell lines (HeLa, MT-2, Jurkat, CEM, SupT1, and HepG2) by measuring cell viability and cellular ATP activity in the presence of the compounds. Cells (10,000 to 15,000/well) were cultured with each compound for 48 h at 37°C and 5% CO2. For the cell viability assay, following incubation, an equal volume of CellTiter 96 Aqueous (Promega, Madison, WI) was added to each culture well, and plates were incubated for 2 h at 37°C. Absorbance was read at 490 nm. The CC50 was defined as the concentration of inhibitor required to induce cell death by 50%. For the cellular ATP activity assay, an equal volume of CellTiter-Glo reagent (Promega, Madison, WI) was added to each well, and chemiluminescence was determined on an LJL Analyst system. For this assay, the CC50 was defined as the concentration of inhibitor required to decrease cellular ATP levels by 50%.
Selection and determination of VRX-329747 resistance mutations.
SupT1 cells (2 × 106 in 1 ml of RPMI 1640 containing 10% fetal bovine serum) were exposed to wt NL4-3 virus (multiplicity of infection = 0.05) for 3 h. The culture was subsequently maintained in 1 ml of growth medium containing 0.2 μM VRX-329747. Every 3 to 4 days, 100 μl of culture was placed into 900 μl of medium containing fresh drug and 9 × 105 SupT1 cells. Virus replication was monitored microscopically by observing the formation of syncytia. At each virus breakthrough (massive syncytium formation), the concentration of inhibitor was doubled. Culture medium and cell pellets from each breakthrough point were collected. Cellular DNA was purified with a Wizard Genomic DNA isolation kit (Promega, Madison, WI). The protease and RT coding regions of proviruses were amplified using high-fidelity PfuTurbo DNA polymerase (Stratagene, La Jolla, CA). The amplified fragments were cloned into the TOPO TA cloning vector (Invitrogen, Carlsbad, CA). The entire protease and RT coding sequences of breakthrough viruses at 0.4, 0.8, 1.6, 3.2, 6.4, 10, and 20 μM were determined by sequencing 10 to 25 individual recovered PCR clones for each point.
Construction of VRX-329747-resistant viruses.
The major patterns of HIV-1 RT mutations selected by VRX-329747 were engineered into the parental pNL4-3.Luc.R−E− molecular clone. This was achieved by exchanging a 1,485-bp ApaI/AgeI RT fragment (nucleotides 2006 to 3490 in NL4-3) from recovered mutant PCR clones with that from pNL4-3.Luc.R−E−. The resulting plasmids were sequenced throughout the insert. Virus stocks with the RT mutations (Table 4) were generated by transfection as described above.
TABLE 4.
Cross-resistance of VRX-329747-selected viruses to current HIV-1 drugs and RCs of viruses carrying VRX-329747-selected mutations in the absence of compounda
| Compound | Mean EC50 ± SD (nM) for wt | Mean change (fold) in EC50 ± SDb
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| M184V | V118I-M184V | M41L-V118I- M184V | A62T-V118I-M184V | M41L-S68N-G112S-M184V | M41L-A62V- S68N-G112S-M184V | M41L-A62V-S68N-G112S-V118I-M184V | G112S-M184V | ||
| VRX-329747 | 176.9 ± 1.6 | 3.5 ± 1.0* | 3.3 ± 0.6* | 6.5 ± 1.6* | 6.3 ± 1.0* | 10.3 ± 0.4* | Max* | 34.7 ± 18.6* | ND |
| VRX-413638 | 45.1 ± 0.5 | 3.9 ± 0.3* | 4.2 ± 0.8* | 9.8 ± 0.6* | 9.5 ± 4.2* | 36.5 ± 3.0* | Max* | Max* | ND |
| Efavirenz | 0.7 ± 0.1 | 0.7 ± 0.02 | 0.6 ± 0.01 | 0.6 ± 0.01 | 0.5 ± 0.05 | 0.2 ± 0.02 | 0.3 ± 0.02 | 0.2 ± 0.002 | ND |
| AZT | 60.8 ± 1.2 | 0.5 ± 0.05 | 0.4 ± 0.07 | 0.4 ± 0.04 | 0.5 ± 0.02 | 0.1 ± 0.01 | 0.1 ± 0.02 | 0.1 ± 0.03 | ND |
| ddI | 2,636.5 ± 841 | 2.4 ± 0.4 | 2.2 ± 0.9 | 1.7 ± 0.2 | 1.9 ± 0.9 | 1.4 ± 0.5 | 0.3 ± 0.2 | 0.7 ± 0.2 | ND |
| d4T | 1,221.0 ± 255 | 1.3 ± 0.2 | 1.3 ± 0.3 | 1.1 ± 0.3 | 1.1 ± 0.1 | 0.6 ± 0.2 | 0.4 ± 0.02 | 0.4 ± 0.1 | ND |
| Abacavir | 1,647.5 ± 126 | 4.7 ± 1.4* | 4.9 ± 1.0* | 2.6 ± 0.3* | 3.0 ± 0.7* | 1.4 ± 0.1 | 0.6 ± 0.2 | 0.7 ± 0.2 | ND |
| Tenofovir | 555.5 ± 122 | 1.0 ± 0.3 | 1.0 ± 0.04 | 1.0 ± 0.3 | 1.1 ± 0.1 | 0.5 ± 0.1 | 0.5 ± 0.3 | 0.4 ± 0.1 | ND |
| FTC | 76.7 ± 7.9 | Max* | Max* | Max* | Max* | Max* | Max* | Max* | ND |
| % wt RCc | 100 | 85.2 ± 7.9 | 119.9 ± 11.1 | 81.8 ± 4.7 | 121.6 ± 12.6 | 143.8 ± 10.2 | 15.9 ± 1.1 | 105.9 ± 12.1 | 0.2 ± 0.04 |
Molecular clones were built in the reporter vector pNL4-3.Luc.R−E, and EC50 values were determined using HeLa-JC-53 cells.
Mean EC50 values ± SD were from two tests. Max, could not be determined because the EC50 was >10 μM; ND, not determined because of the poor replication capacity of the virus. *, EC50 was statistically significantly higher than that against wild-type virus (P < 0.05).
Virus replication capacity was determined in the absence of inhibitor.
RCs of VRX-329747-selected viruses.
The replication capacities (RCs) of viruses carrying VRX-329747-selected RT mutations relative to that of wt virus were studied in the absence of inhibitor in a single-round infection assay. Transfection-generated, VSV-G-pseudotyped viruses (normalized to 50 ng/ml p24) were used to infect 1.5 × 104 HeLa-JC53 cells. Infected cells were cultured at 37°C and 5% CO2 for 48 h. Following incubation, an equal volume of Bright-Glo luciferase reagent was added to each well, and chemiluminescence was read on an LJL Analyst system. The RC of a mutant virus was defined as the percentage of luciferase activity relative to that of the wt virus control.
RESULTS
VRX-329747 is a specific HIV-1 RT inhibitor.
VRX-329747 {1-(4-nitro-phenyl)-2-oxo-2,5-dihydro-1H-pyrido[3,2-b]indole-3-carbonitrile} was discovered during a cell-based high-throughput screening of Valeant compound libraries (Fig. 1). It was a potent HIV-1 inhibitor (EC50 = 0.15 to 0.2 μM) and did not exhibit cellular toxicity against the HeLa, MT-2, Jurkat, HepG2, and SupT1 cell lines at concentrations up to 100 μM (the highest concentration tested) (data not shown). The compound did not inhibit hepatitis B virus or hepatitis C replicon replication (data not shown). In vitro enzymatic experiments further demonstrated that VRX-329747 inhibits HIV-1 RT activity (IC50 = 0.1 μM) (data not shown). In an effort to identify more active analogs, structure-activity relationship studies were performed and led to the identification of a methyl derivative, 5-methyl-1-(4-nitrophenyl)-2-oxo-2,5-dihydro-1H-pyrido[3,2-b]indole-3-carbonitrile (VRX-413638), which was a more potent inhibitor of HIV-1 RT (EC50 = 20 to 30 nM; CC50 > 100 μM). Taken together, these results indicate that this series of nonnucleoside compounds act as specific HIV-1 RT inhibitors.
FIG. 1.
Structures of VRX-329747 and VRX-413638.
VRX-329747 and VRX-413638 inhibit NNRTI-resistant HIV-1 molecular clones.
The anti-HIV activities of VRX-329747 and VRX-413638 were tested against a panel of NNRTI-resistant HIV-1 molecular clones (Table 1). The virus panel consisted of HIV-1 isolates carrying the most prevalent NNRTI resistance mutations observed in patients following NNRTI therapies (4, 5, 27). VRX-329747 was active against all viruses in the panel, with a change in EC50 of <4-fold relative to the EC50 against wt virus. More importantly, the activity profile of this compound differed substantially from that of efavirenz. For example, while VRX-329747 was equally active against wt virus and viruses carrying RT mutations K103N, Y188L, and G190S, efavirenz had 27-, 392-, and 168-fold less activity, respectively, against these mutant viruses. In addition, VRX-329747 retained very good activity against viruses with double mutations, such as K103N-Y188L, K103N-L100I, V106I-Y188L, and K103N-P225H mutants. This result demonstrates that VRX-329747 exhibits an activity profile distinct from those of current NNRTIs. VRX-413638 had a similar activity profile but was more potent than VRX-329747 against wt virus and the majority of NNRTI-resistant HIV-1 isolates tested (Table 1). For this reason, VRX-413638 was used in some of the studies described below.
VRX-413638 inhibits NRTI-resistant HIV-1 isolates.
We tested VRX-413638 against a panel of prototypic multi-NRTI-resistant viruses obtained from the NIH AIDS Research and Reference Reagent Program (10, 17). The panel included isolates with the RT mutations that occur most frequently in HIV-infected individuals following NRTI therapies. VRX-413638 was a potent inhibitor of 8 of the 12 viruses, exhibiting <4-fold increases in the EC50 value (EC50, <100 nM) relative to that for wt virus (Table 2). The compound was less potent against the remaining four viruses (change in EC50 ranging from 4.4- to 16-fold). Of these four viruses, only one contained an additional mutation associated with NNRTI resistance. The K103N mutation was found in a virus containing the M41L, D67N, M184V, L210W, and T215Y mutations. Since the K103N mutation alone (Table 1) or combined with other NRTI resistance mutations (Table 3) does not cause resistance to VRX-413638, we believe that the patterns of NRTI resistance mutations in these four isolates were responsible for their reduced susceptibility to VRX-413638. As shown below, the resistance mutations selected by VRX-329747 substantiate this hypothesis. Notably, VRX-413638 showed clear superiority over both AZT and FTC against this panel of viruses, including the Q151M mutant associated with multinucleoside resistance. AZT lost most of its activity (>4-fold) against 10 of the 12 viruses, and FTC was not active against any of these isolates.
VRX-413638 inhibits clinical HIV-1 isolates carrying both NRTI and NNRTI resistance mutations.
The activity of VRX-413638 was tested against a panel of 15 HIV-1 clinical isolates that carry both NRTI and NNRTI resistance mutations (Table 3). VRX-413638 exhibited nearly equal potencies (<4-fold change) against 11 of these 15 clinical isolates. It showed reduced activities (4- to 8.2-fold change) against the remaining four isolates, all of which carry the M184V mutation. In contrast, efavirenz exhibited reduced activities (>4-fold change) against 14 of the 15 isolates (Table 3). Although the statistical significance of changes in EC50 values could not be evaluated because replicate assays were not performed, the control values (EC50 against wt virus) in Table 3 are consistent with the values in Tables 1, 2, and 4. Taken together, these results indicate that VRX-413638 is active against the most prevalent NNRTI- and NRTI-resistant virus isolates.
VRX-329747 selects mutations surrounding the active site of RT.
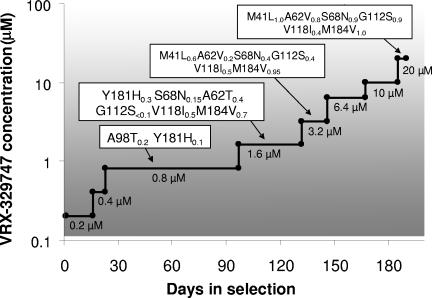
To confirm that this novel class of nonnucleosides represent true RT inhibitors, wild-type HIV-1 was cultured in cells in the presence of increasing concentrations of VRX-329747 to select resistant viruses. Starting at 0.2 μM, the compound concentration was doubled when virus replication resulted in apparent cytopathic effects in the cell culture. Over a 6-month period, the compound concentration was slowly increased to 20 μM (Fig. 2). While no consensus mutations were identified in the viral protease or integrase region (data not shown), a novel set of mutations was observed in the RT (Fig. 3). At low concentrations (0.4 to 0.8 μM), selected viruses contained A98T and Y181H mutations (in 20% and 10% of the recovered sequences, respectively). The remaining sequences recovered at these stages contained various transient RT mutations.
FIG. 2.
Time course of VRX-329747-resistant mutant selection. NL4-3 virus-infected SupT1 cells were cultured in the presence of VRX-329747. At each virus breakthrough point (massive syncytium formation), the compound concentration was doubled. Proviruses from each breakthrough point were sequenced. RT mutations and their prevalences (subscripts) in recovered pools are shown for 0.8, 1.6, 3.2, and 20 μM VRX-329747.
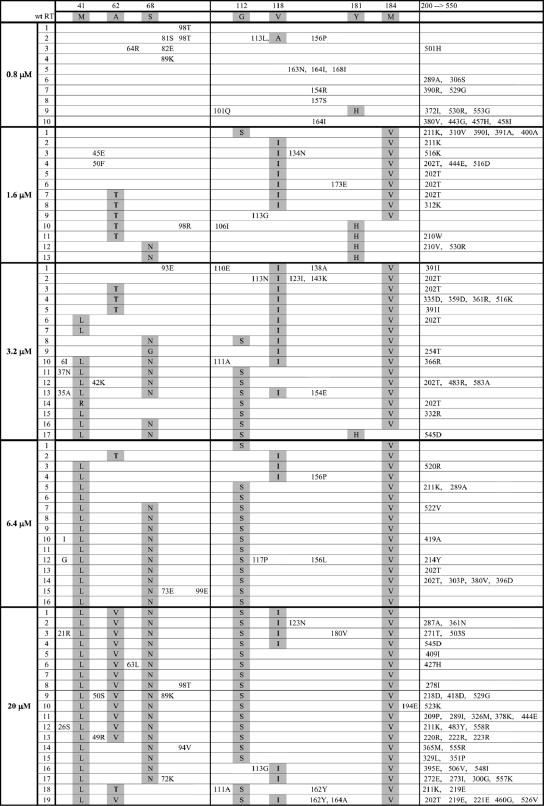
FIG. 3.
VRX-329747-selected mutations in HIV-1 RT. HIV-1 RT amino acids 1 to 550 are represented on the top line of the figure. Individual proviral clones recovered by PCR and characterized by sequence analysis are listed horizontally for 0.8, 1.6, 3.2, 6.4, and 20 μM VRX-329747. The dominant mutations M41L, A62T/V, S68N, G112S, V118I, and M184V are shaded.
Viral cytopathology emerged slowly when the concentration of VRX-329747 in the culture was increased from 0.4 μM to 0.8 μM. Virus breakthrough occurred after 70 days in culture at 0.8 μM (Fig. 2). Sequence analysis of the breakthrough viruses resistant to 1.6 μM VRX-329747 revealed that the majority (70%) contained the RT mutation M184V (Fig. 2). In addition, each of these M184V mutation-containing viruses carried at least two additional mutations (A62T, G112S, V118I, I202T, and/or R211K) (Fig. 3). None of the viruses recovered at this stage carried the M184V mutation alone. The V118I mutation was more prevalent (50%) than the Y181H mutation (30%). Approximately 40% of the viruses recovered at this stage contained the A62T mutation, 15% contained the S68N mutation, and fewer than 10% contained the G112S mutation.
After 40 days at 1.6 μM VRX-329747, the virus pool exhibited clear cytopathic effects and broke through subsequent passages more quickly than at lower inhibitor concentrations. The following four features were observed among the viruses selected at 3.2 μM versus 1.6 μM: (i) fewer than 6% contained the Y181H mutation; (ii) more than 50% contained the new mutation, M41L; (iii) greater percentages of viruses carried the M184V (95% versus 70%) and G112S (40% versus 10%) mutations; and (iv) all but one of the viruses contained combinations of at least three of the following mutations: M41L, A62T, S68N, G112S, V118I, M184V, and I202T. One virus carried a double V118I-M184V mutation but also had four additional nonconsensus mutations (Fig. 3 and data not shown).
At compound concentrations over 6.4 μM, the virus population became more homogeneous but never ceased evolving. Notably, at the highest concentration investigated (20 μM), an earlier and relatively minor A62T mutation-containing species was replaced by an A62V mutation-containing species (74%) (Fig. 2 and 3). Four of 19 recovered viruses carried a sextuple mutation core (M41L-A62V-S68N-G112S-V118I-M184V), while 10 contained quintuple mutation cores (M41L-A62V-S68N-G112S-M184V or M41L-A62V-G112S-V118I-M184V). Five viruses contained quadruple mutation cores (M41L-S68N-G112S-M184V, M41L-S68N-V118I-M184V, or M41L-A62T-G112S-M184V). Thus, at least four core mutations in the RT are required in response to the selection pressure exerted by 20 μM VRX-329747.
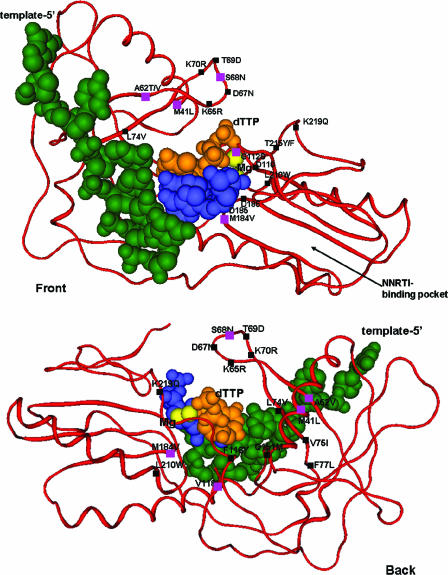
In summary, after 6 months of VRX-329747 selection, none of the mutations known to cause resistance to current NNRTIs was observed. Rather, the compound selected four NRTI resistance mutations, M41L, A62V, V118I, and M184V, as well as two novel mutations, S68N and G112S. All of these amino acid changes mapped to the active site of the RT, suggesting that this novel class of nonnucleosides bind to a site other than the classical NNRTI pocket (Fig. 4).
FIG. 4.
VRX-329747-selected mutations map near the HIV-1 RT catalytic complex (front and back views are shown). VRX-329747-selected (pink) and common NRTI (black) mutations are mapped on the complex structure of Huang et al. (16). The template, primer, dTTP, and Mg2+ are depicted in green, blue, orange, and yellow, respectively. The six mutations selected by VRX-329747 encompass the RT catalytic site.
Cross-resistance of VRX-329747-selected viruses to current NRTIs and efavirenz.
When the VRX-329747-selected mutations were engineered into wt HIV-1, they did indeed confer resistance to both VRX-329747 and VRX-413638 (Table 4). Notably, the M184V mutation alone resulted in only a fourfold increase in EC50 for both compounds. The addition of the V118I mutation to the M184V mutant resulted in no increase in resistance to either compound. At least two additional mutations to the M184V mutation were required to confer >5-fold resistance. More importantly, while the VRX-329747-selected M184V mutation-containing viruses were resistant to FTC and 3TC, they were all fully susceptible to other current NRTIs, including AZT, d4T, ddI, abacavir, and tenofovir, and to the NNRTI efavirenz (Table 4).
RCs of VRX-329747-selected viruses.
The RCs of viruses carrying VRX-329747-selected RT mutations relative to that of wt virus were studied in the absence of inhibitor in a single-round infection assay (Table 4). Most of the compound-selected viruses replicated at levels similar to that of wt virus under these conditions. Two viruses, however, showed reduced RCs. The first, carrying G112S and M184V mutations, replicated to only 0.2% of wt virus capacity (Table 4). In fact, this virus replicated so slowly in the absence of VRX-329747 that its susceptibility to the test compounds could not be determined. The other slow-growing virus was the most dominant quintuple mutant selected with 20 μM VRX-329747, the M41L-A62V-S68N-G112S-M184V mutant. This virus replicated to only 16% of the wt virus capacity in the absence of the compound (Table 4). These results strongly suggest that while mutations were selected to allow the virus to replicate at high VRX-329747 concentrations, the existence of at least some mutations caused the virus to lose replication capacity in the absence of the compound.
Kinetic analysis of RT inhibition.
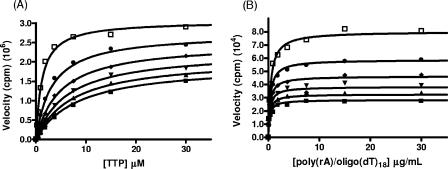
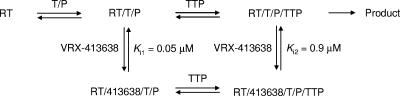
Because VRX-329747 selected resistance mutations are associated with resistance to nucleoside analogs, the relationship between VRX-413638 and the substrates of HIV-1 RT was explored by a steady-state kinetic analysis of the inhibition. Inhibition by VRX-413638 of synthesis on poly(rA)/oligo(dT)18 was examined in the presence of various concentrations of TTP or the template/primer. The steady-state velocity data were fitted to various inhibition equations using a nonlinear global curve-fitting program in Prism 4.0 (Graph Pad), and the fit to the different models was compared using the Akaike’s information criterion. As shown in Fig. 5, data for VRX-413638 inhibition against TTP were best fit to a mixed-type inhibition, giving a Km(TTP) of 1.2 ± 0.06 μM, a Ki1 of 0.051 ± 0.003 μM, and a Ki2 of 0.89 ± 0.07 μM (Fig. 6). Inhibition data with respect to the template/primer were best fit to an uncompetitive inhibition, with a Km[poly(rA)/oligo(dT)18] of 0.54 ± 0.03 μg/ml and Ki of 0.25 ± 0.01 μM.
FIG. 5.
Steady-state inhibition by VRX-413638 of HIV-1 RT synthesis on poly(A)/oligo(dT). (A) Inhibition with respect to TTP. Reactions were carried out as described in Materials and Methods, using 0 (□), 0.1 (•), 0.2 (⧫), 0.3 (▾), 0.4(▴), or 0.5 (▪) μM VRX-413638. Data were globally fit to a mixed-type inhibition pattern. (B) Inhibition with respect to poly(rA)/oligo(dT)18. Data are shown for 0 (□), 0.1 (•), 0.2 (⧫), 0.3 (▾), 0.4(▴), and 0.5 (▪) μM VRX-413638. Data were globally fit to an uncompetitive inhibition pattern.
FIG. 6.
Kinetic scheme of inhibition by VRX-413638. The inhibitor does not bind to RT but can bind after RT forms a complex with the template/primer (T/P). The inhibitor binds to the RT/T/P complex with a Ki of 0.05 μM or to the RT/T/P/TTP complex with a Ki of 0.9 μM, leading to the formation of an inactive complex.
Activity of VRX-413638 against HIV-2.
Using a HeLa-JC53-LTR-Luci reporter cell line that expresses luciferase under the control of the HIV-1 LTR, VRX-413638 was found to inhibit HIV-2(NIH-Z) with an EC50 of 1.5 μM (data not shown). This activity against HIV-2 was 22-fold weaker than that against HIV-1 but was still well below the CC50 of >100 μM for HeLa cells. The NNRTI efavirenz was not active against HIV-2 in this assay system at up to 20 μM (data not shown).
DISCUSSION
In this report, we have characterized a novel class of NNRTIs with potent activities against HIV-1. VRX-329747 seems to pose a higher genetic barrier against resistance development than the commercially available NNRTIs. It took 3 months for wt HIV-1 to break through 0.8 μM (∼4× the wt EC50) VRX-329747 pressure. At least three mutations were required for the virus to grow in the presence of 3.2 μM inhibitor. This contrasts with the case for current NNRTIs, which experience a significant loss in potency with just one amino acid change (K103N, Y181C, or Y188L) (2, 29). Although VRX-329747 is a nonnucleoside, it did not select mutations in the hydrophobic pocket of the HIV-1 RT, which is characteristic of other known NNRTIs. Rather, it selected mutations in or near the catalytic site (Fig. 4). Four of the selected mutations (M41L, A62V, V118I, and M184V) are involved in resistance to current NRTIs (13), while the other two mutations (S68N and G112S) are unique to this series.
The S68N and G112S mutations selected by VRX-329747 have not been associated with resistance to known NRTIs or NNRTIs. While the S68N mutation is novel, mutations of the neighboring residues, D67 and T69, have been described. The D67N mutation is a component of thymidine analogue mutations involved in AZT and d4T resistance (20), while the T69D mutation is involved in dideoxycytosine resistance (11). In addition, a two-serine insertion between T69 and K70 is associated with resistance to all NRTIs (9). These residues are located in the finger loop of the RT, which interacts with the thumb subdomain and affects enzyme catalysis. The G112S mutation is also in a region of RT that is important for catalysis. This residue abuts D113, a component of the dNTP-binding network, and is adjacent to D110, a catalytic residue of the aspartic acid triad (D110, D185, and D186) (15, 16). Although the nonnucleoside MSK-076 has been reported to select for G112E mutation in RT, this occurred only for HIV-2 (3). Selection of resistant HIV-1 with MSK-076 resulted in amino acid changes in the NNRTI pocket rather than at G112 (3). The replication capacity of the G112S-M184V mutant virus was quite low, suggesting that the G112S mutation may be deleterious to viral fitness (Table 4). The absence of emergence of the G112S mutation alone during the selection process also indicates that it is detrimental. If the G112S mutation does reduce the replication capacity, then other mutations may play a compensatory role. Further studies are required to determine the contributions of S68N and G112S mutations to viral resistance and fitness.
In addition to the S68N, G112S, and M184V mutations, VRX-329747 selected three other mutations, M41L, A62V, and V118I, which also exist in different multi-NRTI resistance complexes. It should be noted that in the presence of VRX-329747, these three mutations (M41L, A62V, and V118I) rarely emerged either alone or in any combination among themselves, suggesting that they represent secondary mutations and do not play a significant role by themselves in resistance to VRX-329747 (Fig. 3). Although these mutations are associated with resistance to NRTIs, viruses resulting from combining them with the M184V mutation and the two novel mutations, S68N and G112S, were fully susceptible to virtually all current NRTIs except for 3TC and FTC (Table 4 and data not shown). We believe that the specific combination of mutations governs the susceptibility to NRTIs or VRX-329747 so that viruses resistant to VRX-329747 are inhibited by NRTIs and viruses resistant to NRTIs can be inhibited by VRX-329747.
These novel NNRTIs may be useful in combination therapies with NRTIs, despite some potential issues with cross-resistance, for the following reasons: (i) VRX-329747 and VRX-413638 inhibited all important NNRTI-resistant virus isolates tested; (ii) VRX-413638 retained activity against many NRTI-resistant viruses containing M41L, A62V, V118I, and M184V mutations (Tables 2, 3, and 4); (iii) viruses resistant to VRX-329747 and VRX-413638 were still fully susceptible to inhibition by AZT, d4T, ddI, abacavir, tenofovir, and efavirenz (Table 4); (iv) the M184V mutation resulted in >100-fold resistance to 3TC and FTC but only 2- to 4-fold resistance to VRX-413638 (Tables 2 and 4), so mutants with this resistance mutation may still be susceptible to these inhibitors; and (v) the prevalence of the pattern of amino acid changes causing high-level resistance to these inhibitors is small in Stanford University's Drug Resistance Database (31). Of the 5,068 sequences containing mutations associated with NRTI resistance in this database, only 359, or 7%, contained the M184V mutation in combination with at least two additional VRX-329747-selected mutations.
Structural differences between the hydrophobic pockets of HIV-1 RT and HIV-2 RT render the approved NNRTIs inactive against HIV-2. Interestingly, VRX-413638 showed a weak but reasonable activity against HIV-2 (EC50 = 1 to 2 μM), presumably because this compound targets a conserved site distinct from the hydrophobic pocket. This result further supports the notion that this series of compounds and efavirenz bind to different sites of HIV RTs. In this regard, it will be interesting to determine the mutation profile selected by this series of compounds in HIV-2. Experiments to address this issue are being carried out.
It remains to be determined exactly where in the HIV-1 RT this novel series of NNRTIs binds. The selection of resistant viruses with amino acid changes surrounding the catalytic site initially suggested to us that this series binds to the catalytic site of HIV-1 RT, but this is not supported by the enzyme kinetic data. VRX-413638 was found to be a linear mixed-type inhibitor rather than a competitive inhibitor with respect to its nucleotide substrate, similar to many other NNRTIs (1, 12, 21). Unlike the other NNRTIs, however, VRX-329747 and VRX-413638 do not select for resistance mutations within the hydrophobic NNRTI pocket, and they retain activity against NNRTI-resistant mutants, suggesting that they do not bind within the NNRTI pocket either. Recently, another group described studies with an NNRTI, nucleotide competing RT inhibitor compound 1 (NcRTI-1), that is identical to VRX-413638. In agreement with our findings, they showed that the M184V mutation caused reduced susceptibility (M. Ehteshami, J. Deval, S. Barry, D. Jochmans, K. Hertogs, and M. Gotte, Abstr. 13th Conf. Retrovir. Opportunistic Infect., abstr. 47, 2006; D. Jochmans, H. Van Marck, M. Van Ginderen, I. De Baere, P. Dehertogh, A. Peeters, B. Kesteleyn, T. Pattery, P. McKenna, and K. Hertogs, Abstr. 13th Conf. Retrovir. Opportunistic Infect., abstr. 500, 2006). However, they concluded from their kinetic studies that VRX-413638, or NcRTI-1, was competitive with respect to deoxynucleoside triphosphates rather than a linear mixed-type inhibitor (D. Jochmans, B. Kesteleyn, B. Marchand, M. Gotte, T. Ivens, P. Dehertogh, A. Peeters, R. Pauwels, P. Wigerinck, and K. Hertgos, Abstr. 12 Conf. Retrovir. Opportunistic Infect., abstr. 156, 2005). Reliance on double-reciprocal plots rather than global curve fitting may account for our different conclusions. The finding that VRX-413638 is an uncompetitive inhibitor with respect to poly(rA)/oligo(dT)18 suggests that the binding site does not exist until after the formation of the HIV-1 RT/template-primer complex. This may explain why our attempts to cocrystallize VRX-413638 with HIV-1 RT to determine the binding site or to otherwise physically detect binding have been unsuccessful (data not shown). A kinetic scheme consistent with the data is shown in Fig. 6.
It is not known why mutations associated with NRTI resistance were selected by VRX-329747. The mechanism of resistance to this series of compounds clearly must be different than that to NRTIs since VRX-329747 is not incorporated into DNA. It is possible that these amino acid changes perturb the conformation of the binding site, resulting in a decreased affinity for the inhibitor. Alternatively, since HIV-1 RT in complex with NNRTIs is still capable of a low rate of polymerization (32), these changes may alter the conformation of the catalytic site to enable productive polymerization even when the complex is bound with VRX-329747. Additional studies with HIV-1 RT containing mutations associated with resistance to VRX-329747 may further elucidate the molecular interactions involved in enzyme inhibition.
In summary, our results demonstrate that VRX-329747 and VRX-413638 inhibit HIV-1 reverse transcription by a mechanism distinct from those of currently approved NNRTIs and NRTIs. This class of inhibitors exhibits the potential to be used in both naïve patients due to its high genetic barrier and in treatment-experienced patients because of its activity against both NNRTI- and NRTI-resistant viruses.
Acknowledgments
We thank Heli Walker and Dan Bellows for technical assistance, Shahul Nilar for structure modeling, and Weidong Zhong and Anneke Raney for critical reading of the manuscript. We also thank Yolanda Lie and Neil Parkin at Monogram Biosciences, Inc., for some of the phenotypic tests and Bruno Canard for the kind gift of plasmid p66RTB. Finally, we are indebted to David Ho for critical discussions.
REFERENCES
- 1.Althaus, I. W., J. J. Chou, A. J. Gonzales, M. R. Deibel, K. C. Chou, F. J. Kezdy, D. L. Romero, J. R. Palmer, R. C. Thomas, P. A. Aristoff, W. G. Tarpley, and F. Reusser. 1993. Kinetic studies with the non-nucleoside HIV-1 reverse transcriptase inhibitor U-88204E. Biochemistry 32:6548-6554. [DOI] [PubMed] [Google Scholar]
- 2.Antinori, A., M. Zaccarelli, A. Cingolani, F. Forbici, M. G. Rizzo, M. P. Trotta, S. Di Giambenedetto, P. Narciso, A. Ammassari, E. Girardi, A. De Luca, and C. F. Perno. 2002. Cross-resistance among nonnucleoside reverse transcriptase inhibitors limits recycling efavirenz after nevirapine failure. AIDS Res. Hum. Retrovir. 18:835-838. [DOI] [PubMed] [Google Scholar]
- 3.Auwerx, J., M. Stevens, A. R. Van Rompay, L. E. Bird, J. Ren, E. De Clercq, B. Oberg, D. K. Stammers, A. Karlsson, and J. Balzarini. 2004. The phenylmethylthiazolylthiourea nonnucleoside reverse transcriptase (RT) inhibitor MSK-076 selects for a resistance mutation in the active site of human immunodeficiency virus type 2 RT. J. Virol. 78:7427-7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bacheler, L., S. Jeffrey, G. Hanna, R. D'Aquila, L. Wallace, K. Logue, B. Cordova, K. Hertogs, B. Larder, R. Buckery, D. Baker, K. Gallagher, H. Scarnati, R. Tritch, and C. Rizzo. 2001. Genotypic correlates of phenotypic resistance to efavirenz in virus isolates from patients failing nonnucleoside reverse transcriptase inhibitor therapy. J. Virol. 75:4999-5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bacheler, L. T., E. D. Anton, P. Kudish, D. Baker, J. Bunville, K. Krakowski, L. Bolling, M. Aujay, X. V. Wang, D. Ellis, M. F. Becker, A. L. Lasut, H. J. George, D. R. Spalding, G. Hollis, and K. Abremski. 2000. Human immunodeficiency virus type 1 mutations selected in patients failing efavirenz combination therapy. Antimicrob. Agents Chemother. 44:2475-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boretto, J., S. Longhi, J. M. Navarro, B. Selmi, J. Sire, and B. Canard. 2001. An integrated system to study multiply substituted human immunodeficiency virus type 1 reverse transcriptase. Anal. Biochem. 292:139-147. [DOI] [PubMed] [Google Scholar]
- 7.Boyer, P. L., M. J. Currens, J. B. McMahon, M. R. Boyd, and S. H. Hughes. 1993. Analysis of nonnucleoside drug-resistant variants of human immunodeficiency virus type 1 reverse transcriptase. J. Virol. 67:2412-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 9.de Jong, J. J., J. Goudsmit, V. V. Lukashov, M. E. Hillebrand, E. Baan, R. Huismans, S. A. Danner, J. H. ten Veen, F. de Wolf, and S. Jurriaans. 1999. Insertion of two amino acids combined with changes in reverse transcriptase containing tyrosine-215 of HIV-1 resistant to multiple nucleoside analogs. AIDS 13:75-80. [DOI] [PubMed] [Google Scholar]
- 10.Dupnik, K., M. Gonzales, and R. W. Shafer. 2001. Most multidrug-resistant HIV-1 reverse transcriptase clones in plasma encode functional reverse transcriptase enzymes. Antivir. Ther. 6:42. [Google Scholar]
- 11.Fitzgibbon, J. E., R. M. Howell, C. A. Haberzettl, S. J. Sperber, D. J. Gocke, and D. T. Dubin. 1992. Human immunodeficiency virus type 1 pol gene mutations which cause decreased susceptibility to 2′,3′-dideoxycytidine. Antimicrob. Agents Chemother. 36:153-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fletcher, R. S., K. Syed, S. Mithani, G. I. Dmitrienko, and M. A. Parniak. 1995. Carboxanilide derivative non-nucleoside inhibitors of HIV-1 reverse transcriptase interact with different mechanistic forms of the enzyme. Biochemistry 34:4346-4353. [DOI] [PubMed] [Google Scholar]
- 13.Gallant, J. E., P. Z. Gerondelis, M. A. Wainberg, N. S. Shulman, R. H. Haubrich, M. St. Clair, E. R. Lanier, N. S. Hellmann, and D. D. Richman. 2003. Nucleoside and nucleotide analogue reverse transcriptase inhibitors: a clinical review of antiretroviral resistance. Antivir. Ther. 8:489-506. [PubMed] [Google Scholar]
- 14.Harrington, R., L. Wu, H. Pullen, and M. Emerman. 2000. Direct detection of infectious HIV-1 in blood using a centrifugation-indicator cell assay. J. Virol. Methods 88:111-115. [DOI] [PubMed] [Google Scholar]
- 15.Harris, D., N. Kaushik, P. K. Pandey, P. N. Yadav, and V. N. Pandey. 1998. Functional analysis of amino acid residues constituting the dNTP binding pocket of HIV-1 reverse transcriptase. J. Biol. Chem. 273:33624-33634. [DOI] [PubMed] [Google Scholar]
- 16.Huang, H., R. Chopra, G. L. Verdine, and S. C. Harrison. 1998. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282:1669-1675. [DOI] [PubMed] [Google Scholar]
- 17.Johnston, E., K. M. Dupnik, M. J. Gonzales, M. A. Winters, S. Y. Rhee, T. Imamichi, and R. W. Shafer. 2005. Panel of prototypical infectious molecular HIV-1 clones containing multiple nucleoside reverse transcriptase inhibitor resistance mutations. AIDS 19:731-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimpton, J., and M. Emerman. 1992. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J. Virol. 66:2232-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohlstaedt, L. A., J. Wang, J. M. Friedman, P. A. Rice, and T. A. Steitz. 1992. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science 256:1783-1790. [DOI] [PubMed] [Google Scholar]
- 20.Larder, B. A., and S. D. Kemp. 1989. Multiple mutations in HIV-1 reverse transcriptase confer high-level resistance to zidovudine (AZT). Science 246:1155-1158. [DOI] [PubMed] [Google Scholar]
- 21.Maga, G., D. Ubiali, R. Salvetti, M. Pregnolato, and S. Spadari. 2000. Selective interaction of the human immunodeficiency virus type 1 reverse transcriptase nonnucleoside inhibitor efavirenz and its thio-substituted analog with different enzyme-substrate complexes. Antimicrob. Agents Chemother. 44:1186-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pata, J. D., W. G. Stirtan, S. W. Goldstein, and T. A. Steitz. 2004. Structure of HIV-1 reverse transcriptase bound to an inhibitor active against mutant reverse transcriptases resistant to other nonnucleoside inhibitors. Proc. Natl. Acad. Sci. USA 101:10548-10553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petropoulos, C. J., N. T. Parkin, K. L. Limoli, Y. S. Lie, T. Wrin, W. Huang, H. Tian, D. Smith, G. A. Winslow, D. J. Capon, and J. M. Whitcomb. 2000. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 44:920-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Platt, E. J., K. Wehrly, S. E. Kuhmann, B. Chesebro, and D. Kabat. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 72:2855-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Popik, W., T. M. Alce, and W. C. Au. 2002. Human immunodeficiency virus type 1 uses lipid raft-colocalized CD4 and chemokine receptors for productive entry into CD4+ T cells. J. Virol. 76:4709-4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ren, J., C. E. Nichols, P. P. Chamberlain, K. L. Weaver, S. A. Short, and D. K. Stammers. 2004. Crystal structures of HIV-1 reverse transcriptases mutated at codons 100, 106 and 108 and mechanisms of resistance to non-nucleoside inhibitors. J. Mol. Biol. 336:569-578. [DOI] [PubMed] [Google Scholar]
- 27.Rhee, S. Y., T. Liu, J. Ravela, M. J. Gonzales, and R. W. Shafer. 2004. Distribution of human immunodeficiency virus type 1 protease and reverse transcriptase mutation patterns in 4,183 persons undergoing genotypic resistance testing. Antimicrob. Agents Chemother. 48:3122-3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richman, D. D. 1990. Zidovudine resistance of human immunodeficiency virus. Rev. Infect. Dis. 12(Suppl. 5):S507-S510. [DOI] [PubMed] [Google Scholar]
- 29.Richman, D. D., D. Havlir, J. Corbeil, D. Looney, C. Ignacio, S. A. Spector, J. Sullivan, S. Cheeseman, K. Barringer, D. Pauletti, et al. 1994. Nevirapine resistance mutations of human immunodeficiency virus type 1 selected during therapy. J. Virol. 68:1660-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roos, J. W., M. F. Maughan, Z. Liao, J. E. Hildreth, and J. E. Clements. 2000. LuSIV cells: a reporter cell line for the detection and quantitation of a single cycle of HIV and SIV replication. Virology 273:307-315. [DOI] [PubMed] [Google Scholar]
- 31.Shafer, R. W. 19 May 2005, posting date. NRTI mutation pattern & susceptibility. [Online.] http://hivdb.stanford.edu/pages/phenoSummary/Pheno.NRTI.Simple.html.
- 32.Spence, R. A., W. M. Kati, K. S. Anderson, and K. A. Johnson. 1995. Mechanism of inhibition of HIV-1 reverse transcriptase by nonnucleoside inhibitors. Science 267:988-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang, Z., R. Hamatake, and Z. Hong. 2004. Clinical utility of current NNRTIs and perspectives of new agents in this class under development. Antivir. Chem. Chemother. 15:121-134. [DOI] [PubMed] [Google Scholar]