Abstract
The seven Mycobacterium tuberculosis whiB-like genes encode small proteins postulated to be transcriptional regulators. A systematic real-time reverse transcription-PCR analysis following exposure to antibiotics and a variety of growth and in vitro stress conditions indicates differential, and in some cases dramatic, transcription modulations for the different M. tuberculosis whiB family members. This information together with biochemical analyses of the whiB1 to whiB7 gene products will be important for understanding the biology of this novel family of proteins in mycobacteria and related actinomycetes.
Upon infection, Mycobacterium tuberculosis has the ability to adapt to many different environments within the host organism. Tubercle bacilli are able to avoid immune system detection and persist inside the host for decades. Many of the conditions to which the bacteria are exposed, such as the acidic environment within the phagolysosomal compartment, are harsh (14). In the lung, there is a recruitment of activated macrophages to the infection site, and these, along with other immune cells, contain the infection by forming a tuberculous granuloma (49). M. tuberculosis is adept at surviving within the hypoxic and fatty acid-rich granulomatous environment (8), a facet critical to its ability to persist despite immune pressure (25, 28). Moreover, tubercle bacilli can survive for long periods of exposure to low-nutrient conditions, such as may occur within a granuloma (4, 34), as well as temperature and oxidative stress which may act to disrupt the M. tuberculosis cell membrane. The bacterium also has a remarkable tolerance to a wide range of antibiotics (12, 26). To survive, the organism must sense and respond to exogenous stress conditions. Hence, differential expression of transcriptional regulators, which control sets of genes that respond to environmental stimuli, is an important mechanism of stress survival.
A family of genes which may be key transcriptional regulators in M. tuberculosis is the whiB gene family. The whiB-like genes are exclusive to the actinomycetes, such as Mycobacterium and Streptomyces spp., and are absent from all other organisms studied thus far (9, 44). whiB was identified in Streptomyces coelicolor as an essential gene for sporulation of aerial hyphae (9), and early sporulation genes were identified by morphological studies of a collection of mutants unable to form normal gray spore pigment (17). Because their aerial mycelium remained white upon prolonged incubation, these mutants were designated whi mutants. S. coelicolor whiB encodes an 87-amino-acid polypeptide with attributes suggesting that it may be a DNA binding protein (9). S. coelicolor WhiB and its related homologues, including S. coelicolor WhiD, each contain four cysteine residues, a feature common in metal-coordinating DNA binding proteins (33). Indeed, a recent study has found that Sc WhiD binds a [4Fe-4S] cluster under anaerobic conditions similar to the situation with Escherichia coli DNA binding regulators SoxR and Fnr (6, 10, 18). These data suggest that WhiB family members may be involved in DNA binding and in interactions with other proteins leading to transcriptional activation. The M. tuberculosis genome contains seven whiB-like genes (whiB1 to whiB7) (5, 27, 44). Determining the conditions under which the whiB family of genes are induced or repressed is an important step in understanding their possible function and role in the regulation of gene expression in M. tuberculosis under different environmental stimuli. We conducted a comprehensive analysis of the expression of the seven whiB-like genes in M. tuberculosis under various growth conditions and in response to several stresses using quantitative real-time reverse transcription-PCR (RT-PCR). Important stimuli examined were antibiotic exposure, nutrient deprivation, acid, ethanol, detergent, oxidative, heat, and low-iron stresses.
M. tuberculosis strain CDC1551 was grown in roller bottles at 37°C in Middlebrook 7H9 liquid medium supplemented with 10% ADC (albumin-dextrose-salt complex; Sigma), 0.5% glycerol, and 0.05% Tween 80. Cultures were grown to an optical density at 600 nm (OD600) of 0.4, divided into 50-ml aliquots, returned to the incubator for 2 h to equilibrate, and then treated with various stress conditions for 1.5 h. Stress conditions were as follows: 0.05% and 0.1% sodium dodecyl sulfate (SDS) (detergent stress), 5 mM diamide (oxidative stress), 1 mM cumene hydroperoxide (oxidative stress), pH 4.5 (acid stress), phosphate-buffered saline (PBS) (nutrient starvation), 2.5% and 5% ethanol, and 42°C (heat shock). Antibiotic stresses were carried out with the compounds for 3.5 h using high-end critical concentrations as follows: 50 μg/ml cycloserine, 12 μg/ml ethambutol, 0.75 μg/ml isoniazid, 7.5 μg/ml streptomycin, and 50 μg/ml kanamycin. The effect of acid stress was examined by washing the cultures twice with PBS, resuspending of cells in complete 7H9 medium at pH 4.5 (acidified with HCl), and incubating for 1.5 h. For nutrient starvation conditions, bacteria were washed as described above and then resuspended and incubated in PBS for 1.5 h. The cellular response to low-iron stress was studied by washing the cultures twice with PBS, resuspending of cells in GAST (glycerol-alanine-salt) media with and without iron, and incubating for 6 h and 24 h. Cultures were also grown and collected at different growth phases, when the OD600 values reached 0.2, 0.5, 0.8, 0.91, 1.9, and 3.0 and 2 and 5 days after the OD600 values reached 3.0. RNA was isolated as described previously (47) and treated with DNase I (Invitrogen), and RNA (1 μg) was reverse transcribed using iScript (Bio-Rad). Quantitative real-time RT-PCR was performed with cDNA corresponding to 50 ng RNA using a Bio-Rad iQ SYBR green Supermix kit. Relative expression levels were calculated using the sigA transcript for normalization. The average relative expression levels and the standard deviations were determined for four independent experiments.
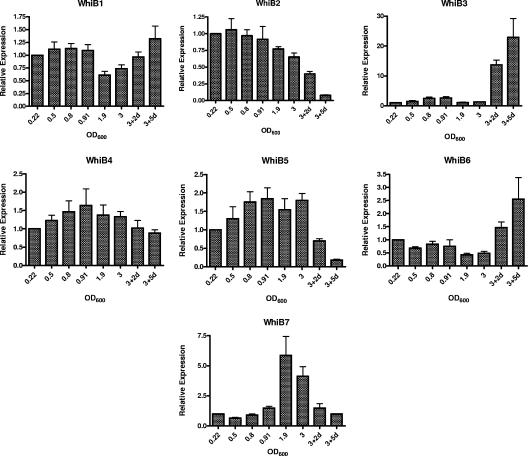
We first examined the expression of the M. tuberculosis whiB family of genes from the early exponential phase to the late stationary phase of growth. Figure 1 shows the relative expression levels of each whiB-like gene at the various growth phases. Transcript levels of whiB1 and whiB4 remained relatively constant throughout the different phases of growth. whiB2 expression was constant through early to late-exponential-phase growth, with a gradual decline to almost undetectable levels in late stationary phases. whiB3 showed a gradual increase of 2.5-fold in late exponential phase and then a decrease to control levels in early stationary phase and a dramatic increase in late stationary phases. whiB5 levels increased nearly twofold in late exponential and mid-stationary phases, with a decline to very low levels in late stationary phases. There was a decline in whiB6 expression from control levels to early stationary phase and from this point a gradual increase in late stationary phase to 2.5-fold over control levels. whiB7 expression increased in late exponential phase to over fivefold during early stationary phase with a decline to control levels in late stationary phases of growth.
FIG. 1.
Relative expression levels of whiB1 to whiB7 during various phases of growth. Total RNA was isolated from M. tuberculosis (CDC1551) cultures grown to the OD600 values shown. The OD600 values for the respective growth phases are as follows: early exponential (0.22), mid-late exponential (0.8 to 0.91), early stationary (1.9), late stationary (3.0), and very late stationary (2 days or 5 days after OD600 reaches 3.0). Real-time quantitative real-time RT-PCR was performed, and the relative expression level of each whiB was calculated using the sigA transcript for normalization for RNA amounts and the expression level at an OD600 of 0.22 as a control. Data shown are averages from four independent experiments.
Both an abundance of efflux pumps and an unusual cell envelope have been advanced as explanations for the high tolerance of M. tuberculosis to antibiotics (1, 19, 31). Recently, however, M. tuberculosis whiB7 was shown to govern a resistance mechanism to several well-known antituberculosis drugs (26). To further assess adaptive responses to antibiotics as a mechanism of antibiotic tolerance, we examined the effects of various drugs on the expression of the entire family of whiB-like genes. Ethambutol and isoniazid had minimal effects on most of the gene family, but increased transcript accumulation of whiB2 with both drugs (Fig. 2) and a slight decrease in whiB4 levels on treatment with isoniazid were observed (Table 1). Cycloserine, another cell wall-acting agent, also stimulated an increase in whiB2 transcript levels (Fig. 2) and slight decreases in whiB5, whiB6, and whiB7 (Table 1). As shown in Fig. 2, streptomycin induced whiB3 (10-fold), whiB6 (3-fold), and whiB7 (70-fold), while whiB2 and whiB5 were repressed (Table 1). Kanamycin induced whiB7 over 70-fold, while whiB2, whiB3, and whiB6 increased 2- to 3.5-fold (Fig. 2). Thus, cell wall-active agents (isoniazid, ethambutol, and cycloserine) stimulated whiB2 transcription, and ribosomally acting aminoglycosides (streptomycin and kanamycin) led to the induction of whiB7.
FIG. 2.
Relative expression levels of whiB1 to whiB7 following antibiotic stresses. Total RNA was isolated from M. tuberculosis (CDC1551) cultures (OD600, 0.4) after treatment with ethambutol, isoniazid, cycloserine, streptomycin, and kanamycin for 3.5 h. Real-time quantitative RT-PCR was performed, and the relative expression level of each whiB gene was calculated using the sigA transcript for normalization for RNA amounts and untreated cultures as a control. Data shown are averages from four independent experiments.
TABLE 1.
M. tuberculosis whiB-like genes displaying significant (>2-fold) downregulation under growth and stress conditions
| Gene | Growth and/or stress condition | Relative expression level |
|---|---|---|
| whiB1 | 0.05% SDS | 0.35 |
| 42°C | 0.29 | |
| whiB2 | Late stationary phase (2 days after OD600 reached 3.0) | 0.40 |
| Late stationary phase (5 days after OD600 reached 3.0) | 0.08 | |
| 0.05% SDS | 0.39 | |
| 42°C | 0.19 | |
| Streptomycin | 0.51 | |
| whiB3 | Low iron | 0.40 |
| whiB4 | 0.05% SDS | 0.27 |
| 5% ethanol | 0.47 | |
| whiB5 | Late stationary phase (5 days after OD600 reached 3.0) | 0.18 |
| 0.05% SDS | 0.31 | |
| 2.5% ethanol | 0.47 | |
| 5 mM diamide | 0.41 | |
| Cycloserine | 0.55 | |
| Streptomycin | 0.44 | |
| whiB6 | Early stationary phase (OD600, 1.9) | 0.42 |
| Stationary phase (OD600, 3.0) | 0.48 | |
| Cycloserine | 0.51 | |
| whiB7 | 2.5% ethanol | 0.42 |
| Cycloserine | 0.50 |
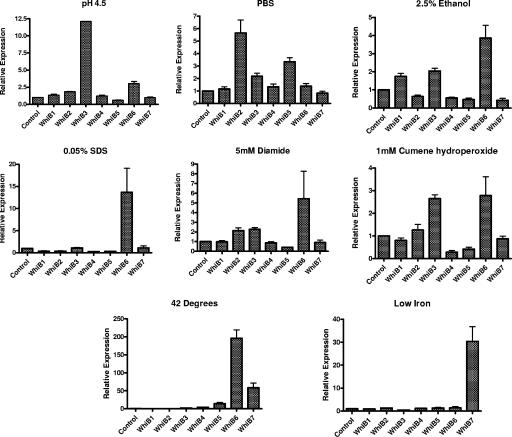
The phagocytosis of M. tuberculosis by macrophages in the lung is an early event during infection (16). The phagolysosomes of activated macrophages acidify, and the pH may drop below 6.0 (14, 41, 42), while in certain settings, pathogenic bacteria may interrupt the acidification process (16, 32). Previous transcriptional profiling studies of in vivo-grown M. tuberculosis have shown that whiB4 is upregulated (36). In this study, we examined the effect of acid pH (pH 4.5) on the expression of the whiB genes. We found that after 1.5 h of exposure, whiB3 and whiB6 were induced 12-fold and 3-fold, respectively, compared to untreated controls. whiB2 was induced less than twofold. (Fig. 3). The levels for the other whiB genes did not vary significantly under these conditions.
FIG. 3.
Relative expression levels of whiB1 to whiB7 under stress conditions. Total RNA was isolated from M. tuberculosis (CDC1551) cultures grown to an OD600 of 0.4 and subjected to various stresses as described in the text for 1.5 h or 6 h for low-iron stress. Real-time RT-PCR was performed, and the relative expression level of each whiB was calculated using the sigA transcript for normalization for RNA amounts and untreated cultures as a control. Data shown are averages from four independent experiments.
M. tuberculosis may be exposed to low-nutrient concentrations within host granulomatous lesions (4, 34). For a model of starvation, we washed M. tuberculosis cultures with PBS and incubated them for 1.5 h in PBS (43). Under these culture conditions, we found whiB2, whiB3, and whiB5 to be induced about fivefold, twofold, and threefold, respectively, over the untreated controls (Fig. 3). No appreciable variations were observed in the levels for the other members of this gene family under the conditions tested.
Because ethanol is a protein denaturant and causes membrane disruption (3), we used exposure to ethanol as a stimulus to assess the M. tuberculosis responses to protein denaturation and membrane disruption. After 1.5 h of exposure to 2.5% ethanol, whiB1, whiB3, and whiB6 were induced two- to fourfold compared to controls (Fig. 3). A similar pattern of induction was seen with 5% ethanol (data not shown). Reductions in the levels for whiB4, whiB5, and whiB7 were also observed with 2.5% ethanol treatment (Table 1).
Treatment of M. tuberculosis with the detergent SDS has been used to mimic detergent stress, which M. tuberculosis may encounter in the form of surfactant early on during the process of infection (20, 23). As shown in Fig. 3, only whiB6 was significantly induced at exposures of 0.05% SDS; similar data were obtained for 0.1% SDS (data not shown). The other whiB genes (except whiB3 and whiB7) were significantly reduced with SDS treatment (Table 1).
In order to survive the burst of reactive oxygen species in activated macrophages, M. tuberculosis induces a number of stress response genes to protect itself from the damaging effects of these agents (24, 42, 51). In addition, mycobacteria synthesize compounds like mycothiol, which may offer further protection under such hostile conditions (29, 30, 38). The thiol oxidant diamide has been used extensively to study the effects of changes in the oxidation state of thiols (22). In this study, we examined the effect of diamide treatment on the expression of the whiB family of genes. We observed that whiB2, whiB3, and whiB6 were induced two- to fivefold with exposure to 5 mM diamide for 1.5 h, whereas whiB5 levels were observed to drop (Fig. 3 and Table 1). We also examined the effect of another oxidizing agent, cumene hydroperoxide, which mimics the alkyl peroxides produced by metabolism of unsaturated fatty acids and nucleic acids (2). On exposure of cells to 1 mM cumene hydroperoxide for 1.5 h, we observed >2-fold induction in the levels of whiB3 and whiB6 (Fig. 3).
The heat shock response is an adaptive pathway involved in the survival of cells exposed to a sudden increase in ambient temperature, and heat shock proteins are induced in both the host and the pathogen during the process of infection (45). We investigated the expression of the whiB-like genes after exposure to a temperature of 42°C for 1.5 h. Heat stress led to an increase in the expression levels of whiB3 (2-fold), whiB4 (4-fold), whiB5 (14-fold), whiB6 (196-fold), and whiB7 (58-fold). Expression levels of whiB1 and whiB2 were observed to drop significantly following heat shock (Table 1).
Iron is an essential nutrient for most pathogens, and iron limitation at the site of infection is an important mechanism of host defense (11, 40, 50). This phenomenon was simulated in vitro by washing M. tuberculosis cultures with PBS and resuspending of cells in GAST media with and without iron. We examined the expression of the whiB-like genes after 6 and 24 h of exposure to conditions of low iron. Levels of whiB7 were induced 30-fold and levels of whiB3 reduced 2.5-fold at the 6-h time point (Table 1). No effect was observed at 24 h subsequent to iron starvation (data not shown).
The WhiB-like proteins are putative transcription factors involved in the regulation of significant cellular processes, such as cell division, pathogenesis, and response to oxidative stress (13, 15, 21, 28, 37, 46). While they are postulated to serve as DNA binding regulatory proteins based upon the presence of a helix-turn-helix motif (9), and there is evidence that M. tuberculosis WhiB3 binds to RNA polymerase sigma factor region 4.2 (46), formal proof that they bind DNA and modulate gene expression has not been established. Recently, Morris et al. demonstrated that exposure of M. tuberculosis to the antituberculous drug streptomycin, as well as macrolide and tetracycline antibiotics, led to a significant increase in transcription of whiB7 (26). Microarray analysis demonstrated that upon subinhibitory exposure to tetracycline, the expression of other genes was temporally dependent on the initial induction of whiB7, and hierarchical clustering indicated that 12 other genes were whiB7 dependent (26). Other M. tuberculosis whiB family members have also demonstrated conditional upregulation in response to environmental changes (4, 28, 39). These observations further support the notion that WhiB family member gene products may serve as DNA binding regulators which are responsive to exogenous conditions, including antimicrobial stress.
We further extended this theme by conducting a systematic assessment of the transcription of each of the seven M. tuberculosis whiB family members under standard growth conditions as well as exogenous stress conditions. Our results show that each of the whiB family members has a unique transcriptional response pattern. Moreover, we confirmed a dramatic induction of whiB7 with ribosomally active antimicrobial agents. Our study shows that in addition to whiB7, M. tuberculosis whiB2 is also responsive to antimicrobial stress. In contrast to whiB7 induction by ribosomally acting aminoglycoside agents, the expression of whiB2 was stimulated by exposure to isoniazid, ethambutol, and cycloserine, agents which inhibit cell wall biosynthesis in mycobacteria. M. tuberculosis whiB2 was also strongly stimulated by starvation and was downregulated by entry into late stationary phase. Earlier work on whiB2 focused on its Mycobacterium smegmatis orthologue, whmD, which was shown to be an essential gene for M. smegmatis viability and required for septum formation (15). Since septum formation is linked to new-cell-envelope biosynthesis, these observations associating whiB2 induction to envelope inhibitors are consistent with reports from other bacterial species (7, 35). The involvement of whiB2 in septation/cell division is also consistent with our results showing whiB2 expression to be decreased in late stationary phase, when cell division rates decline.
While whiB2 was downregulated in late stationary phase, our study revealed that M. tuberculosis whiB3 expression was dramatically upregulated by entry into late stationary phase. This is of biological relevance since in vitro stationary-phase expression patterns could be relevant to chronic infection in vivo. In addition, whiB3 transcription was induced more than 10-fold by acid-stress, a condition potentially related to that of in vitro-grown cultures in late stationary phase when culture medium becomes acidified. WhiB3 has been shown to interact with domain 4.2 of the M. tuberculosis principal sigma factor and to be required for full virulence in animal models of tuberculosis (46). In the study of whiB3 genetic deletion by Steyn et al., it is noteworthy that the greatest degree of virulence attenuation was seen in guinea pigs infected with whiB3 mutants of Mycobacterium bovis. Since guinea pigs develop necrotic granulomatous lesions which are rich in fatty acids (48), it is possible that whiB3 functions as a sensor of acid stress, as may be seen in granulomatous lesions, and that its expression is required for mycobacterial survival in this inhospitable environment.
M. tuberculosis whiB6 was the paralogue which displayed the greatest degree of stress modulation in our study. Like whiB3, whiB6 was strongly induced in late stationary phase, although in contrast to that of whiB3, its expression was only modestly increased by acid stress. However, whiB6 showed strong upregulation in response to SDS stress (∼12-fold), heat shock (∼200-fold), ethanol exposure (∼4-fold), and oxidative stress due to diamide (∼5-fold) or cumene hydroperoxide (∼3-fold). Neither M. tuberculosis whiB6 nor orthologous family members related to it in Streptomyces have been studied to date. Our study suggests that this particular family member is strongly responsive to a wide variety of stress conditions and may play a key role in generalized stress responses by mycobacteria.
Acknowledgments
The support of a Heiser Foundation postdoctoral fellowship grant to T.R.R. and funding from NIH grants AI51668, AI36973, AI37856, and AI43846 are gratefully acknowledged.
REFERENCES
- 1.Ainsa, J. A., M. C. Blokpoel, I. Otal, D. B. Young, K. A. De Smet, and C. Martin. 1998. Molecular cloning and characterization of Tap, a putative multidrug efflux pump present in Mycobacterium fortuitum and Mycobacterium tuberculosis. J. Bacteriol. 180:5836-5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antelmann, H., S. Engelmann, R. Schmid, and M. Hecker. 1996. General and oxidative stress responses in Bacillus subtilis: cloning, expression, and mutation of the alkyl hydroperoxide reductase operon. J. Bacteriol. 178:6571-6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bermudez, L. E., L. S. Young, J. Martinelli, and M. Petrofsky. 1993. Exposure to ethanol up-regulates the expression of Mycobacterium avium complex proteins associated with bacterial virulence. J Infect. Dis. 168:961-968. [DOI] [PubMed] [Google Scholar]
- 4.Betts, J. C., P. T. Lukey, L. C. Robb, R. A. McAdam, and K. Duncan. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717-731. [DOI] [PubMed] [Google Scholar]
- 5.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 6.Crack, J., J. Green, and A. J. Thomson. 2004. Mechanism of oxygen sensing by the bacterial transcription factor fumarate-nitrate reduction (FNR). J. Biol. Chem. 279:9278-9286. [DOI] [PubMed] [Google Scholar]
- 7.Dalhoff, A., T. Nasu, and K. Okamoto. 2003. Target affinities of faropenem to and its impact on the morphology of gram-positive and gram-negative bacteria. Chemotherapy 49:172-183. [DOI] [PubMed] [Google Scholar]
- 8.Dannenberg, A. M., Jr. 1993. Immunopathogenesis of pulmonary tuberculosis. Hosp. Pract. (Off. Ed.) 28:51-58. [DOI] [PubMed] [Google Scholar]
- 9.Davis, N. K., and K. F. Chater. 1992. The Streptomyces coelicolor whiB gene encodes a small transcription factor-like protein dispensable for growth but essential for sporulation. Mol. Gen. Genet. 232:351-358. [DOI] [PubMed] [Google Scholar]
- 10.Demple, B. 2002. Signal transduction by nitric oxide in cellular stress responses. Mol. Cell. Biochem. 234-235:11-18. [PubMed] [Google Scholar]
- 11.De Voss, J. J., K. Rutter, B. G. Schroeder, and C. E. Barry III. 1999. Iron acquisition and metabolism by mycobacteria. J. Bacteriol. 181:4443-4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doucet-Populaire, F., K. Buriankova, J. Weiser, and J. L. Pernodet. 2002. Natural and acquired macrolide resistance in mycobacteria. Curr. Drug Targets Infect. Disord. 2:355-370. [DOI] [PubMed] [Google Scholar]
- 13.Dubnau, E., J. Chan, V. P. Mohan, and I. Smith. 2005. Responses of Mycobacterium tuberculosis to growth in the mouse lung. Infect. Immun. 73:3754-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fisher, M. A., B. B. Plikaytis, and T. M. Shinnick. 2002. Microarray analysis of the Mycobacterium tuberculosis transcriptional response to the acidic conditions found in phagosomes. J. Bacteriol. 184:4025-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gomez, J. E., and W. R. Bishai. 2000. whmD is an essential mycobacterial gene required for proper septation and cell division. Proc. Natl. Acad. Sci. USA 97:8554-8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hackam, D. J., O. D. Rotstein, W. Zhang, S. Gruenheid, P. Gros, and S. Grinstein. 1998. Host resistance to intracellular infection: mutation of natural resistance-associated macrophage protein 1 (Nramp1) impairs phagosomal acidification. J. Exp. Med. 188:351-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hopwood, D. A., H. Wildermuth, and H. M. Palmer. 1970. Mutants of Streptomyces coelicolor defective in sporulation. J Gen. Microbiol. 61:397-408. [DOI] [PubMed] [Google Scholar]
- 18.Jakimowicz, P., M. R. Cheesman, W. R. Bishai, K. F. Chater, A. J. Thomson, and M. J. Buttner. 2005. Evidence that the Streptomyces developmental protein WhiD, a member of the WhiB family, binds a [4Fe-4S] cluster. J. Biol. Chem. 280:8309-8315. [DOI] [PubMed] [Google Scholar]
- 19.Jarlier, V., and H. Nikaido. 1994. Mycobacterial cell wall: structure and role in natural resistance to antibiotics. FEMS Microbiol. Lett. 123:11-18. [DOI] [PubMed] [Google Scholar]
- 20.Johansson, J., and T. Curstedt. 1997. Molecular structures and interactions of pulmonary surfactant components. Eur. J. Biochem. 244:675-693. [DOI] [PubMed] [Google Scholar]
- 21.Kim, T. H., J. S. Park, H. J. Kim, Y. Kim, P. Kim, and H. S. Lee. 2005. The whcE gene of Corynebacterium glutamicum is important for survival following heat and oxidative stress. Biochem. Biophys. Res. Commun. 337:757-764. [DOI] [PubMed] [Google Scholar]
- 22.Kosower, N. S., and E. M. Kosower. 1995. Diamide: an oxidant probe for thiols. Methods Enzymol. 251:123-133. [DOI] [PubMed] [Google Scholar]
- 23.Manganelli, R., E. Dubnau, S. Tyagi, F. R. Kramer, and I. Smith. 1999. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol. Microbiol. 31:715-724. [DOI] [PubMed] [Google Scholar]
- 24.Manganelli, R., M. I. Voskuil, G. K. Schoolnik, and I. Smith. 2001. The Mycobacterium tuberculosis ECF sigma factor sigmaE: role in global gene expression and survival in macrophages. Mol. Microbiol. 41:423-437. [DOI] [PubMed] [Google Scholar]
- 25.McKinney, J. D., K. Honer zu Bentrup, E. J. Munoz-Elias, A. Miczak, B. Chen, W. T. Chan, D. Swenson, J. C. Sacchettini, W. R. Jacobs, Jr., and D. G. Russell. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406:735-738. [DOI] [PubMed] [Google Scholar]
- 26.Morris, R. P., L. Nguyen, J. Gatfield, K. Visconti, K. Nguyen, D. Schnappinger, S. Ehrt, Y. Liu, L. Heifets, J. Pieters, G. Schoolnik, and C. J. Thompson. 2005. Ancestral antibiotic resistance in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 102:12200-12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mulder, N. J., H. Zappe, and L. M. Steyn. 1999. Characterization of a Mycobacterium tuberculosis homologue of the Streptomyces coelicolor whiB gene. Tuber. Lung Dis. 79:299-308. [DOI] [PubMed] [Google Scholar]
- 28.Muttucumaru, D. G., G. Roberts, J. Hinds, R. A. Stabler, and T. Parish. 2004. Gene expression profile of Mycobacterium tuberculosis in a non-replicating state. Tuberculosis (Edinburgh) 84:239-246. [DOI] [PubMed] [Google Scholar]
- 29.Newton, G. L., K. Arnold, M. S. Price, C. Sherrill, S. B. Delcardayre, Y. Aharonowitz, G. Cohen, J. Davies, R. C. Fahey, and C. Davis. 1996. Distribution of thiols in microorganisms: mycothiol is a major thiol in most actinomycetes. J. Bacteriol. 178:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newton, G. L., M. D. Unson, S. J. Anderberg, J. A. Aguilera, N. N. Oh, S. B. delCardayre, Y. Av-Gay, and R. C. Fahey. 1999. Characterization of Mycobacterium smegmatis mutants defective in 1-d-myo-inosityl-2-amino-2-deoxy-alpha-d-glucopyranoside and mycothiol biosynthesis. Biochem. Biophys. Res. Commun. 255:239-244. [DOI] [PubMed] [Google Scholar]
- 31.Nguyen, L., S. Chinnapapagari, and C. J. Thompson. 2005. FbpA-dependent biosynthesis of trehalose dimycolate is required for the intrinsic multidrug resistance, cell wall structure, and colonial morphology of Mycobacterium smegmatis. J. Bacteriol. 187:6603-6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oh, Y. K., C. Alpuche-Aranda, E. Berthiaume, T. Jinks, S. I. Miller, and J. A. Swanson. 1996. Rapid and complete fusion of macrophage lysosomes with phagosomes containing Salmonella typhimurium. Infect. Immun. 64:3877-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Halloran, T. V. 1993. Transition metals in control of gene expression. Science 261:715-725. [DOI] [PubMed] [Google Scholar]
- 34.Parish, T. 2003. Starvation survival response of Mycobacterium tuberculosis. J. Bacteriol. 185:6702-6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pinho, M. G., and J. Errington. 2005. Recruitment of penicillin-binding protein PBP2 to the division site of Staphylococcus aureus is dependent on its transpeptidation substrates. Mol. Microbiol. 55:799-807. [DOI] [PubMed] [Google Scholar]
- 36.Rachman, H., M. Strong, U. Schaible, J. Schuchhardt, K. Hagens, H. Mollenkopf, D. Eisenberg, and S. Kaufmann. 2006. Mycobacterium tuberculosis gene expression profiling within the context of protein networks. Microbes Infect. 8:747-757. [DOI] [PubMed] [Google Scholar]
- 37.Ramakrishnan, L., N. A. Federspiel, and S. Falkow. 2000. Granuloma-specific expression of Mycobacterium virulence proteins from the glycine-rich PE-PGRS family. Science 288:1436-1439. [DOI] [PubMed] [Google Scholar]
- 38.Rawat, M., G. L. Newton, M. Ko, G. J. Martinez, R. C. Fahey, and Y. Av-Gay. 2002. Mycothiol-deficient Mycobacterium smegmatis mutants are hypersensitive to alkylating agents, free radicals, and antibiotics. Antimicrob. Agents Chemother. 46:3348-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rickman, L., C. Scott, D. M. Hunt, T. Hutchinson, M. C. Menendez, R. Whalan, J. Hinds, M. J. Colston, J. Green, and R. S. Buxton. 2005. A member of the cAMP receptor protein family of transcription regulators in Mycobacterium tuberculosis is required for virulence in mice and controls transcription of the rpfA gene coding for a resuscitation promoting factor. Mol. Microbiol. 56:1274-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodriguez, G. M., B. Gold, M. Gomez, O. Dussurget, and I. Smith. 1999. Identification and characterization of two divergently transcribed iron regulated genes in Mycobacterium tuberculosis. Tuber. Lung Dis. 79:287-298. [DOI] [PubMed] [Google Scholar]
- 41.Saviola, B., S. C. Woolwine, and W. R. Bishai. 2003. Isolation of acid-inducible genes of Mycobacterium tuberculosis with the use of recombinase-based in vivo expression technology. Infect. Immun. 71:1379-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schnappinger, D., S. Ehrt, M. I. Voskuil, Y. Liu, J. A. Mangan, I. M. Monahan, G. Dolganov, B. Efron, P. D. Butcher, C. Nathan, and G. K. Schoolnik. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198:693-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Silberbach, M., M. Schafer, A. T. Huser, J. Kalinowski, A. Puhler, R. Kramer, and A. Burkovski. 2005. Adaptation of Corynebacterium glutamicum to ammonium limitation: a global analysis using transcriptome and proteome techniques. Appl. Environ. Microbiol. 71:2391-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soliveri, J. A., J. Gomez, W. R. Bishai, and K. F. Chater. 2000. Multiple paralogous genes related to the Streptomyces coelicolor developmental regulatory gene whiB are present in Streptomyces and other actinomycetes. Microbiology 146:333-343. [DOI] [PubMed] [Google Scholar]
- 45.Stewart, G. R., L. Wernisch, R. Stabler, J. A. Mangan, J. Hinds, K. G. Laing, D. B. Young, and P. D. Butcher. 2002. Dissection of the heat-shock response in Mycobacterium tuberculosis using mutants and microarrays. Microbiology 148:3129-3138. [DOI] [PubMed] [Google Scholar]
- 46.Steyn, A. J., D. M. Collins, M. K. Hondalus, W. R. Jacobs, Jr., R. P. Kawakami, and B. R. Bloom. 2002. Mycobacterium tuberculosis WhiB3 interacts with RpoV to affect host survival but is dispensable for in vivo growth. Proc. Natl. Acad. Sci. USA 99:3147-3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun, R., P. J. Converse, C. Ko, S. Tyagi, N. E. Morrison, and W. R. Bishai. 2004. Mycobacterium tuberculosis ECF sigma factor sigC is required for lethality in mice and for the conditional expression of a defined gene set. Mol. Microbiol. 52:25-38. [DOI] [PubMed] [Google Scholar]
- 48.Turner, O. C., R. J. Basaraba, and I. M. Orme. 2003. Immunopathogenesis of pulmonary granulomas in the guinea pig after infection with Mycobacterium tuberculosis. Infect. Immun. 71:864-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ulrichs, T., and S. H. Kaufmann. 2006. New insights into the function of granulomas in human tuberculosis. J. Pathol. 208:261-269. [DOI] [PubMed] [Google Scholar]
- 50.Wong, D. K., B. Y. Lee, M. A. Horwitz, and B. W. Gibson. 1999. Identification of Fur, aconitase, and other proteins expressed by Mycobacterium tuberculosis under conditions of low and high concentrations of iron by combined two-dimensional gel electrophoresis and mass spectrometry. Infect. Immun. 67:327-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zahrt, T. C., and V. Deretic. 2002. Reactive nitrogen and oxygen intermediates and bacterial defenses: unusual adaptations in Mycobacterium tuberculosis. Antioxid. Redox Signal 4:141-159. [DOI] [PubMed] [Google Scholar]