Abstract
We investigated ertapenem-susceptible and -resistant extended-spectrum β-lactamase-producing Enterobacter cloacae isolates obtained from the same patient. Gene transcription of OmpD and OmpF was diminished in the ertapenem-resistant isolate. An efflux pump inhibitor decreased the MICs of ertapenem in the resistant strain, suggesting a potential role of efflux pumps in ertapenem resistance.
Ertapenem is typically active in vitro against the Enterobacteriaceae (12). Elevated ertapenem MICs in Klebsiella pneumoniae have been associated with extended-spectrum β-lactamase or AmpC production and deficiency in the expression of outer membrane proteins OmpK35 and OmpK36 (3, 4). In this report, we have studied the antibiotic resistance mechanisms in a clinical isolate of Enterobacter cloacae exhibiting ertapenem resistance and compared them to those of a prior ertapenem-susceptible isolate from the same patient.
A 55-year-old male developed bacteremia due to E. cloacae (strain ES24), thought to be secondary to an infected dialysis catheter. The catheter was removed, and imipenem and amikacin were administered for 3 weeks. The patient did not receive ertapenem. Two weeks after discontinuing the antibiotics, blood cultures again grew E. cloacae (strain ER24), thought to be related to an infection of a new dialysis catheter.
The MICs of various antibiotics were determined by Etest on Mueller-Hinton agar, with and without 40 μg/ml phenylalanyl arginyl β-naphthylamide (PAβN) (Sigma, St. Louis, MO), an antibiotic efflux pump inhibitor. Characterization of the strains by analytical isoelectric focusing; detection and sequencing of blaCTX-M, blaAmpC, blaTEM and blaSHV genes; assessment of nitrocefin hydrolysis by total cell lysate; assessment of β-lactamase induction; plasmid profile analysis; and pulsed-field gel electrophoresis were performed by previously described methods (9, 11, 13-15).
Real-time reverse transcriptase PCR (RT-PCR) was performed to determine the expression of ompD and ompF porin genes and the acrB efflux pump gene relative to the rpoB housekeeping gene. Total RNA was isolated using the RNeasy kit (QIAGEN Inc., Valencia, CA) and treated with RNase-free DNase I. The concentration of the RNA was determined spectrophotometrically. One hundred twenty-five micrograms of RNA was reverse transcribed into single-stranded cDNA using the High Capacity cDNA Archive kit (Applied Biosystems, Foster City, CA). The cDNAs were subsequently quantified by real-time PCR amplification with primers specific to the outer membrane protein (OMP) and efflux pump genes (Table 1) using the ABI 7300 RealTime PCR system (Applied Biosystems, Foster City, CA) with initial incubations of 50°C for 2 min and 95°C for 12 min, followed by 40 cycles of 15 s at 95°C and 60 s at 60°C. Expression level results were standardized relative to the transcription levels of rpoB (housekeeping gene) for each isolate. The relative change in gene expression for the ompD, ompF, and acrB genes in the ER24 strain was calculated as the ratio of ES24 (reference) to ER24 (target) using the 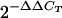 method, where CT is the cycle threshold (8).
method, where CT is the cycle threshold (8).
TABLE 1.
Primers used in real-time RT-PCR
| Primera | Sequence (5′ to 3′)b | Annealing temp (°C) | Product size (bp) |
|---|---|---|---|
| rpoB (F) | CAGCCGCGAYCAGGTTGACTACA | 60 | 63 |
| rpoB (R) | GACGCACCGACGGATACCACCTG | ||
| ompD (F) | AGAATTTGGCGGCGACACCTAC | 60 | 145 |
| ompD (R) | TGCGCTACCGTTTTTACCCTGATA | ||
| ompF (F) | CCGGCGTGAGCGATAACTTCTTCT | 60 | 136 |
| ompF (R) | AACGCATCGGTACGCTCATTTTTG | ||
| acrB (F) | TCCGGGCGCAGAGAACAAGGTAGA | 60 | 100 |
| acrB (R) | TGCGGGCAGGTTAAAGGCGAAGAC |
F, forward primers; R, reverse primers.
Y = C or T.
Strain ES24 was susceptible to carbapenems (ertapenem MIC, 2 μg/ml; imipenem and meropenem MICs, 0.25 μg/ml), whereas ER24 had higher carbapenem MICs (ertapenem, >32 μg/ml; imipenem, 4 μg/ml; and meropenem, 6 μg/ml). In the ES24 strain, the PAβN had minimal effect on the MIC of all antibiotics except ciprofloxacin (MIC decreased from >32 to 2 μg/ml (Table 2). In the ER24 strain, the presence of PAβN decreased the MICs of ciprofloxacin (from >32 to 3 μg/ml), meropenem (from 6 to 0.75 μg/ml), and ertapenem (from >32 to 12 μg/ml) (Table 2).
TABLE 2.
Effect of efflux pump inhibitor PAβN on various antibiotic susceptibilities
| E. cloacae strain | MIC (μg/ml)
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ceftazidime
|
Cefotaxime
|
Cefepime
|
Ertapenem
|
Imipenem
|
Meropenem
|
Ciprofloxacin
|
||||||||
| −PAβN | +PAβN | −PAβN | +PAβN | −PAβN | +PAβN | −PAβN | +PAβN | −PAβN | +PAβN | −PAβN | +PAβN | −PAβN | +PAβN | |
| ATCC 13047 | 3 | 2 | 2 | 1.5 | 0.19 | 0.5 | 0.064 | 0.064 | 0.19 | 0.19 | 0.032 | 0.032 | 0.023 | 0.012 |
| ES24 | >256 | >256 | 24 | 16 | 16 | 16 | 2 | 2 | 0.25 | 0.25 | 0.25 | 0.25 | >32 | 2 |
| ER24 | >256 | >256 | >256 | 64 | 24 | 24 | >32 | 12 | 4 | 2 | 6 | 0.75 | >32 | 3 |
By analytical isoelectric focusing, both ES24 and ER24 had four bands with isoelectric points of 5.4, 7.0, 7.6, and ≥9. Each strain harbored blaTEM-1, blaSHV-7, blaSHV-30, and blaMIR (14). The profiles of plasmid DNA isolated from ES24 and ER24 were identical, and the pulsed-field gel electrophoresis patterns of the two strains were indistinguishable.
The beta-lactamases of both ES24 and ER24 led to relatively little hydrolytic activity of nitrocefin, when in the uninduced state (hydrolytic activities of 10 and 20 μM/s/mg total protein, respectively). However, when induced by the presence of cefoxitin, hydrolytic activity increased substantially to 290 and 140 μM/s/mg total protein, respectively. The transcription levels of the ompD and ompF genes were 11 and 55 times lower, respectively, in the ertapenem-resistant ER24 isolate compared to the susceptible ES24 (Table 3). The transcription levels of the acrB gene were equivalent in the two strains (1.2 times higher in ER24 than ES24).
TABLE 3.
RT-PCR analysis of acrB, ompD, and ompF expression
| Gene | Strain | Relative expressiona |
|---|---|---|
| ompF | ES24 | 1 |
| ER24 | 0.018 | |
| ompD | ES24 | 1 |
| ER24 | 0.088 | |
| acrB | ES24 | 1 |
| ER24 | 1.2 |
Relative expression is calculated as 2−ΔΔCT, where ΔΔCT = CT target − CT reference. The target is ER24, and the reference is ES24.
In this report, we investigated an ertapenem-resistant E. cloacae blood culture isolate, compared to an earlier ertapenem-susceptible isolate from the same patient. Although there was slightly higher β-lactamase production by the ertapenem-resistant isolate, there were no differences in the number or type of β-lactamases between the two strains. However, the ertapenem-resistant isolate had decreased amounts of RNA transcripts of the ompF and ompD genes compared to the ertapenem-susceptible isolate. A notable feature of our study is that we used RT-PCR to determine the expression levels of the porin transcripts. This method potentially eliminates some of the difficulties associated with subjective interpretation of outer membrane protein profiles.
It has been previously shown that reduced outer membrane permeability and high-level cephalosporinase production can act in combination in clinical isolates of E. cloacae to confer carbapenem resistance (1, 5, 6). No phenotypic evidence of efflux pump overexpression in ertapenem-resistant strains has been found (4). However, when we used PAβN, an efflux pump inhibitor, we found that the ciprofloxacin, ertapenem, and meropenem MICs decreased substantially. Ciprofloxacin and meropenem are known substrates of efflux pumps (10), but ertapenem is not known as an efflux pump substrate. Several previous studies have investigated the role efflux pumps (notably the AcrAB pumps) play in antibiotic resistance in Enterobactericaeae (2, 7). We could not detect any difference in the expression level of AcrB between the ertapenem-susceptible and -resistant strains. Based on these results, we speculate that there is an additional unknown efflux pump influencing ertapenem resistance.
It must be recognized that while the PAβN effect supports the existence of an efflux pump, it does not necessarily indicate pump overexpression. Efflux pump-mediated resistance in absolute terms is determined in part by the permeability of the outer membrane. The decreased amounts of RNA transcripts of the ompF and ompD genes suggest that the outer membrane of the ertapenem-resistant isolate was less permeable than that of the susceptible strain—potentially this could lead to higher resistance provided by a pump without necessarily requiring overexpression per se. Therefore, although certainly possible, we have not been able to show direct evidence to actually support pump overexpression. Further investigations into the nature of the purported efflux pump and its expression are ongoing.
Acknowledgments
Jennifer Adams and Anna Gushchin are thanked for their laboratory assistance.
Dóra Szabó was supported by a Hungarian State Eötvös Fellowship (Ministry of Education of the Hungarian Republic, Budapest, Hungary), and Robert A. Bonomo was supported by the Department of Veterans Affairs Merit Review Program and NIH grant R01AI063517-01.
REFERENCES
- 1.Bush, K., S. K. Tanaka, D. P. Bonner, and R. B. Sykes. 1985. Resistance caused by decreased penetration of β-lactam antibiotics into Enterobacter cloacae. Antimicrob. Agents Chemother. 27:555-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chollet, R., J. Chevalier, C. Bollet, J.-M. Pages, and A. Davin-Regli. 2004. RamA is an alternate activator of the multidrug resistance cascade in Enterobacter aerogenes. Antimicrob. Agents Chemother. 48:2518-2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jacoby, G., P. Han, and J. Tran. 1997. Comparative in vitro activities of carbapenem L-749,345 and other antimicrobials against multiresistant gram-negative clinical pathogens. Antimicrob. Agents Chemother. 41:1830-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jacoby, G. A., D. M. Mills, and N. Chow. 2004. Role of β-lactamases and porins in resistance to ertapenem and other β-lactams in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48:3203-3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee, E. H., E. Collatz, J. Trias, and L. Gutmann. 1992. Diffusion of beta-lactam antibiotics into proteoliposomes reconstituted with outer membranes of isogenic imipenem-susceptible and -resistant strains of Enterobacter cloacae. J. Gen. Microbiol. 138:2347-2351. [DOI] [PubMed] [Google Scholar]
- 6.Lee, E. H., M. H. Nicolas, M. D. Kitzis, G. Pialoux, E. Collatz, and L. Gutmann. 1991. Association of two resistance mechanisms in a clinical isolate of Enterobacter cloacae with high-level resistance to imipenem. Antimicrob. Agents Chemother. 35:1093-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Linde, H. J., F. Notka, C. Irtenkauf, J. Decker, J. Wild, H. H. Niller, P. Heisig, and N. Lehn. 2002. Increase in MICs of ciprofloxacin in vivo in two closely related clinical isolates of Enterobacter cloacae. J. Antimicrob. Chemother. 49:625-630. [DOI] [PubMed] [Google Scholar]
-
8.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the
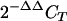 method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar] - 9.Mahlen, S. D., S. S. Morrow, B. Abdalhamid, and N. D. Hanson. 2003. Analyses of ampC gene expression in Serratia marcescens reveal new regulatory properties. J. Antimicrob. Chemother. 51:791-802. [DOI] [PubMed] [Google Scholar]
- 10.Pages, J. M., M. Masi, and J. Barbe. 2005. Inhibitors of efflux pumps in Gram-negative bacteria. Trends Mol. Med. 11:382-389. [DOI] [PubMed] [Google Scholar]
- 11.Paterson, D. L., K. M. Hujer, A. M. Hujer, B. Yeiser, M. D. Bonomo, L. B. Rice, R. A. Bonomo, and the International Klebsiella Study Group. 2003. Extended-spectrum β-lactamases in Klebsiella pneumoniae bloodstream isolates from seven countries: dominance and widespread prevalence of SHV- and CTX-M-type β-lactamases. Antimicrob. Agents Chemother. 47:3554-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paterson, D. L., F. Rossi, F. Baquero, P. R. Hsueh, G. L. Woods, V. Satishchandran, T. A. Snyder, C. M. Harvey, H. Teppler, M. J. Dinubile, and J. W. Chow. 2005. In vitro susceptibilities of aerobic and facultative Gram-negative bacilli isolated from patients with intra-abdominal infections worldwide: the 2003 Study for Monitoring Antimicrobial Resistance Trends (SMART). J. Antimicrob. Chemother. 55:965-973. [DOI] [PubMed] [Google Scholar]
- 13.Sanders, C. C., and W. E. Sanders, Jr. 1986. Type I beta-lactamases of gram-negative bacteria: interactions with beta-lactam antibiotics. J Infect. Dis. 154:792-800. [DOI] [PubMed] [Google Scholar]
- 14.Szabó, D., R. A. Bonomo, F. Silveira, A. W. Pasculle, C. Baxter, P. K. Linden, A. M. Hujer, K. M. Hujer, K. Deeley, and D. L. Paterson. 2005. SHV-type extended-spectrum beta-lactamase production is associated with reduced cefepime susceptibility in Enterobacter cloacae. J. Clin. Microbiol. 43:5058-5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]


