Abstract
Dermcidin (DCD) is a recently described antimicrobial peptide, which is constitutively expressed in eccrine sweat glands and transported via sweat to the epidermal surface. By postsecretory proteolytic processing in sweat the dermcidin protein gives rise to several truncated DCD peptides which differ in length and net charge. In order to understand the mechanism of antimicrobial activity, we analyzed the spectrum of activity of several naturally processed dermcidin-derived peptides, the secondary structure in different solvents, and the ability of these peptides to interact with or permeabilize the bacterial membrane. Interestingly, although all naturally processed DCD peptides can adopt an α-helical conformation in solvents, they have a diverse and partially overlapping spectrum of activity against gram-positive and gram-negative bacteria. This indicates that the net charge and the secondary structure of the peptides are not important for the toxic activity. Furthermore, using carboxyfluorescein-loaded liposomes, membrane permeability studies and electron microscopy we investigated whether DCD peptides are able to permeabilize bacterial membranes. The data convincingly show that irrespective of charge the different DCD peptides are not able to permeabilize bacterial membranes. However, bacterial mutants lacking specific cell envelope modifications exhibited different susceptibilities to killing by DCD peptides than wild-type bacterial strains. Finally, immunoelectron microscopy studies indicated that DCD peptides are able to bind to the bacterial surface; however, signs of membrane perturbation were not observed. These studies indicate that DCD peptides do not exert their activity by permeabilizing bacterial membranes.
Antimicrobial peptides (AMPs) show a broad spectrum of antimicrobial activity against a wide range of pathogens, including bacteria, fungi, and enveloped viruses, and therefore play an important role in innate host defense (11, 15, 45). All AMPs are synthesized as proforms, which are subsequently processed into mature peptides of various lengths and structures (46). Although they differ significantly in their amino acid composition, most of them share similar features, such as a net positive charge and the ability to form amphipathic α-helix or β-sheet structures in which clusters of hydrophobic and hydrophilic amino acids are spatially separated. Therefore, it seems that these features are important for the antimicrobial activity on microorganisms.
There is compelling evidence that many AMPs, including the defensins and the cathelicidin LL-37, interact and increase the permeability of the bacterial cytoplasmic membrane as part of their killing mechanism (5, 44). The initial binding is thought to depend on electrostatic interactions between the positively charged peptides and the negatively charged bacterial membrane. In a second step, the peptide induces membrane permeation either via pore formation or via membrane disintegration, which is accompanied by a loss of the bacterial membrane potential (5, 44). It has been shown that, in contrast to these membrane-active peptides, other AMPs, i.e., buforin, kill microorganisms by disruption of critical intracellular processes (44) such as the inhibition of macromolecular biosynthesis (16, 28, 41) or by interacting with specific vital components inside the microorganisms (14, 36, 43).
Dermcidin (DCD) is a recently discovered AMP with no homology to other known AMPs. In contrast to most other AMPs which are cationic, DCD has a net negative charge. DCD is constitutively expressed in eccrine sweat glands, secreted into sweat, and transported to the epidermal surface (37). DCD expression could not be observed in epidermal keratinocytes of healthy human skin (32). Recently, we described that patients with atopic dermatitis have a reduced amount of DCD peptides in sweat, which correlated with a diminished antimicrobial activity of eccrine sweat in vivo (34).
Analogous to other known AMPs a 110-amino-acid precursor protein of dermcidin is produced, which is proteolytically processed to antimicrobially active DCD peptides (10, 37). Antimicrobially active DCD peptides are derived from the C-terminal region of the precursor protein, DCD-1L (consisting of 48 amino acids) and DCD-1 (47 amino acids, lacking the last leucine). Both peptides show antimicrobial activity against a variety of pathogenic microorganisms, including Staphylococcus aureus, Escherichia coli, Enterococcus faecalis, and Candida albicans under in vitro conditions resembling human sweat (37). Further investigations revealed an extended antimicrobial spectrum, including Staphylococcus epidermidis (42), Pseudomonas putida, methicillin-resistant S. aureus, rifampin- and isoniazid-resistant Mycobacterium tuberculosis (20), and Listeria monocytogenes and Salmonella enterica serovar Typhimurium (8).
Recently, we identified in eccrine sweat 13 different DCD peptides derived from the C-terminal end and one N-terminal derived peptide named YDP-42, generated by postsecretory proteolytic processing (33). Recent evidence suggests that the sweat-derived aspartate protease cathepsin D and a 1,10-phenanthroline-sensitive carboxypeptidase (together with an as-yet-unidentified endopeptidase) are involved in the postsecretory processing of dermcidin (1). This indicated that several DCD-derived peptides are generated in human eccrine sweat by postsecretory processing. Interestingly, proteolytically processed DCD peptides possess net charges between −2 and +2; thus, initially, anionic DCD-1L and DCD-1 are the source of the generation of neutral or cationic shorter DCD-fragments due to subsequent processing in human sweat.
In the present study, we addressed the following questions. (i) Does the alteration from anionic to cationic peptides during proteolytic processing of DCD-1L lead to a different antimicrobial spectrum or result in loss of antimicrobial activity? (ii) Do dermcidin-derived peptides kill the microorganisms by permeabilization of the bacterial membrane? We conducted studies with a range of naturally processed DCD peptides which differ in length and charge and are ideally suited for comparative studies of the mechanism of antimicrobial activity.
MATERIALS AND METHODS
Peptide synthesis and purification.
Peptides were synthesized utilizing the Fmoc (9-fluorenylmethoxy carbonyl)/tBu chemistry using a multiple peptide synthesizer Syro II (MultiSynTech, Witten, Germany). After cleavage, the crude peptide was purified by HPLC on a reversed-phase C18 Nucleosil 100-5C column to a purity of >95% using a linear gradient of 5 to 80% acetonitrile in 0.05% trifluoroacetic acid for 45 min. All peptides were characterized by matrix-assisted laser desorption ionization-time of flight mass spectroscopy (MALDI-TOF-MS) and electrospray ionization and were in all cases in agreement with the calculated masses.
Antimicrobial assays.
Antimicrobial assays were performed using the CFU assay as previously described (34, 37). The antibacterial activity of DCD-derived peptides and LL-37 was tested against the following bacterial strains: Escherichia coli ATCC 25922, methicillin-susceptible Staphylococcus aureus ATCC 25923 (MSSA), methicillin-resistant Staphylococcus aureus (MRSA; clinical isolate), Staphylococcus epidermidis ATCC 12228, and Pseudomonas aeruginosa ATCC 27853. E. coli ML-35p was generously provided by Robert Lehrer (Department of Medicine, Center for Health Sciences, Los Angeles, CA) and cultivated on Luria-Bertani (LB) plates containing 50 μg of ampicillin/ml. This strain constitutively expresses cytoplasmic β-lactosidase but lacks lactose permease (22). The bacterial membrane mutants Staphylococcus aureus mprF and dltA and Staphylococcus epidermidis Δica and the corresponding wild-type strains S. aureus 113 and S. epidermidis 1457 were previously described (13, 30). Bacterial cultures were grown to mid-exponential growth phase and washed three times with 10 mM sodium phosphate buffer-10 mM NaCl (pH 7.0). The bacterial concentration was estimated photometrically at 600 nm. Absorbance of 1.0 corresponded to 8.56 × 108/ml for E. coli; 1 × 108/ml for E. coli ML-35p; 1.97 × 108/ml for S. aureus, MRSA, and the wild-type and membrane mutants of S. aureus and S. epidermidis; and 5.07 × 108/ml for P. aeruginosa.
After dilution to a concentration of 106 CFU/ml, 10-μl portions of the dilutions were incubated at 37°C for 2 to 4 h with the respective peptide diluted in water in a total volume of 30 μl in 10 mM sodium phosphate buffer-10 mM NaCl (pH 7.0). After incubation, the cells were diluted 1:100 in 10 mM sodium phosphate buffer-10 mM NaCl (pH 7.0), and 90 μl of the diluted bacterial suspension was plated in triplicates on blood agar. Bacterial colonies were counted after incubation for 18 to 24 h at 37°C. The antimicrobial activity was calculated by using [(cell survival after peptide incubation)/(cell survival in buffer without peptide) × 100]. The LC90 describes the lethal concentration of the current synthetic peptide in μg/ml or μM that leads to a 90% reduction of CFU compared to the buffer control.
Membrane permeabilization.
To examine the effects of DCD-derived peptides and LL-37 to permeabilize the inner membrane of gram-negative bacteria, we used a previously described method using the permease-deficient strain E. coli ML-35p, which constitutively expresses cytoplasmic β-galactosidase (21, 22). Bacteria were grown for 18 h at 37°C and washed three times with 10 mM sodium phosphate buffer (pH 7.0) and resuspended in this buffer to 108 CFU/ml. Then, 100 μl of the bacterial suspension was pipetted into a cuvette containing 70 μl of 20 mM sodium phosphate buffer (pH 7.0) and 30 μl of 30 mM β-galactosidase substrate ONPG (o-nitrophenyl-β-d-galactopyranoside) in 20 mM sodium phosphate and 100 μl of the test peptide solution. The production of o-nitrophenol (ONP) over time was monitored spectrophotometrically at a wavelength of 420 nm. An equivalent volume of water replaced the peptide solution in the control assay.
Outer membrane (OM) permeability was assessed by using E. coli ML-35p by 1-N-phenyl-naphtylamine (NPN; Sigma-Aldrich, Germany) uptake assay as already described (23). This hydrophobic probe fluoresces strongly in phospholipid environments but only weakly in an aqueous environment. Normally, intact OM excludes hydrophobic molecules, but through the action of permeabilizers, the phospholipids become accessible and allow NPN access into the phospholipid layer. Thus, increased fluorescence in NPN-containing bacterial suspensions indicates OM damage. E. coli ML-35p was incubated with DCD-derived peptides or LL-37. EDTA as a chelating agent was used as a control. NPN was prepared as a 0.5 mM acetone stock solution and diluted freshly in 5 mM HEPES buffer (pH 7.2) to a concentration of 10 μM. Cells were grown to the logarithmic phase in LB medium and were washed twice in 5 mM HEPES buffer (pH 7.2) and resuspended in the same buffer to an optical density (A600) of 0.5. Next, 50 μl of the bacterial suspension was added to a 96-microwell plate containing 5 mM HEPES buffer (23 μl), test peptide solution (25 μl), and 2 μl of 0.5 mM NPN. The permeabilization of outer membrane was monitored by measuring the increase in fluorescence intensity at 460 nm with excitation at 355 nm in a spectrofluorophotometer (Tecan SPECTRAFluor, Crailsheim, Germany). The results are expressed as NPN uptake factors, which was calculated as a quotient of background-corrected fluorescence and the highest fluorescence value.
Preparation of liposomes and CF release assay.
Large unilamellar vesicles made of 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) and 1,2-dioleoyl-sn-glycero-3-phosphoglycerol (DOPG)-DOPC at a 1:1 molar ratio were used in liposome lysis and CD measurements and prepared as already described (25). Phospholipids were purchased from Avanti-Polar Lipids (Alabaster, AL). Phospholipid stock solutions (2 μmol) were dissolved in CHCl3-MeOH (2:1) and then dried in an exsiccator by vacuum. Multilamellar liposomes were formed by hydrating the dry lipids at room temperature with 600 μl of 50 mM MES (morpholineethanesulfonic acid)-50 mM K2SO4 (pH 6.5) containing 50 mM carboxyfluorescein (CF; Sigma, Steinheim, Germany) to encapsulate it into liposomes for CF release assay and with 600 μl of 10 mM sodium phosphate buffer (pH 7.0) for CD measurement. Multilamellar liposomes were freeze-thawed 10 times to enhance encapsulation. In order to get unilamellar liposomes, multilamellar liposomes were extruded 10 times through two stacked 400-nm-pore-size polycarbonate membranes (Isopore membrane filters; Millipore). The CF-entrapped vesicles were separated from free carboxyfluorescein by gel filtration using Sephadex G-50 columns. The liposome concentration was determined by phosphorous analysis (35).The liposomal preparations were kept at 4°C and used within a few hours.
The release of CF from liposomes was measured by monitoring the increase in fluorescence intensity at 515 nm with excitation at 492 nm in a RF-5301 PC Spectrofluorophotometer (Schimadzu). CF release was initiated by addition of known concentrations of DCD peptide or LL-37 (1 to 10 μM) into a magnetically stirred cuvette containing the CF-loaded liposomes (25 μM phospholipid/ml) in 1.5 ml of assay buffer. CF release was expressed as the percentage of CF released relative to the fluorescence released after addition of 20 μl of a 20% Triton X-100 solution at the end of each experiment.
Preparation of unilamellar vesicles for carboxyfluorescein efflux experiments.
Large unilamellar vesicles were prepared for CF experiments by the extrusion technique (25). Fluorescence measurements were recorded using a RF-5301 spectrophotometer (Shimadzu).
CF-loaded vesicles were prepared with 50 mM CF and then diluted in 1.5 ml of K+ buffer (50 mM MES-KOH [pH 6.0], 100 mM K2SO4) in a final concentration of 25 μM phospholipid on a phosphorous base. After addition of the peptide, the increase of fluorescence intensity was measured at 520 nm (excitation at 492 nm) at room temperature. Peptide induced leakage was documented relative to the total amount of marker release after solubilization of the vesicles by the addition of 10 μl of 20% Triton X-100.
CD spectroscopy.
Circular dichroism (CD) measurements were performed with a Jasco J-720 CD spectrapolarimeter (Jasco, Tokyo, Japan) to determine the secondary structure of DCD-derived peptides. The data are reported as the average of three to four scans at 50 nm/min with a 1-nm step resolution. The CD spectra of the peptides (50 μM) in 10 mM sodium phosphate buffer (pH 7.0) with different amounts of NaCl (10, 100, and 150 mM) in 0 to 60% (vol/vol) trifluoroethanol (TFE) and in the presence of artificial phospholipid vesicles such as DOPC and DOPC-DOPG (1:1 molar ratio) were recorded at room temperature in the 180- to 260-nm wavelength range. The data are reported as the mean residue ellipticity in units of degrees cm2 dmol−1 and plotted versus wavelength. The secondary structural predictions for peptides were carried out by Jasco secondary structure software.
Aggregation assay.
Labeling of the N terminus of DCD-1L, SSL-23, and LL-37 with fluorescein isothiocyanate (FITC) was achieved by elongation of the N-terminal bound peptide with β-alanine and afterwards by FITC. The molecular mass was determined by MALDI-MS analysis and agreed with the calculated mass. Oligomerization of the peptides in solution was determined by fluorescence dequenching as previously described (26). Fluorescein-labeled peptides (stock solution 10 μM in 1× phosphate-buffered saline [PBS] pH 7.4) were added at different concentrations (0.0625 to 0.5 μM) to a white 96-well plate (final volume, 100 μl; Nunc, Wiesbaden, Germany). For the determination of the time-kinetics of oligomerization, peptides were incubated for different time points (0 to 220 min) at room temperature in PBS before proteinase K (50 μg/ml; Sigma) was added to each peptide. For the estimation of the correlation of the peptide concentration and the aggregation state, the peptides were preincubated 2 h in PBS before proteinase K treatment. Enzyme treatment resulted in an increase in the fluorescence emission as a result of the dequenching of the FITC fluorescence. Excitation was set at 485 nm, and emission was set at 535 nm on a spectrofluorophotometer (Tecan SPECTRAFluor; Crailsheim, Germany). Fluorescence measurements were performed in triplicates at room temperature.
Western blot analysis.
After physical exercise sweat of the forehead was collected and analyzed by Western blot as already described (33). Briefly, sweat was centrifuged 3 min at 13,000 rpm (16,000 × g) to remove particles and frozen. Then, 15-μl portions of the samples and 4 μg of the DCD peptides LEK-45 and DCD-1L dissolved in water were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (using 15% acrylamide with 0.05% bisacrylamide), transferred electrophoretically (1200 mA/h) onto polyvinylidene difluoride membrane sheets (Immobilon-PSQ; Millipore, Germany) with a tank type blotter and then blocked in 10% nonfat milk in PBS (pH 7.4) for 30 min at room temperature. After incubation with a rabbit polyclonal anti-DCD antibody (1:2,000) that detects the C terminus of DCD-1L overnight at 4°C, the membrane sheets were washed with phosphate buffer and incubated with an anti-rabbit secondary biotin-conjugated polyclonal antibody (1:1,500 in blocking solution). After washing in phosphate buffer the Streptavidin-AP-conjugate (Roche, Mannheim, Germany) was used for the detection of biotin-labeled secondary antibody. The membrane was immersed in CDP-Star solution (Western Lightning chemiluminescence reagents for AP; Roche) for 10 min and then exposed to X-ray film (Eastman Kodak, Rochester, NY).
Hemolytic assay.
The hemolytic activity of DCD peptides was determined on fresh human erythrocytes, which were separated from human EDTA blood using a Ficoll gradient. Isolated erythrocytes were washed three times in 5 ml of PBS (50 mM sodium phosphate buffer, 150 mM NaCl [pH 7.0]) and centrifuged at 1,000 × g for 6 min at room temperature. The pellets were resuspended in PBS and further diluted to a concentration of 109 human erythrocytes/ml. Hemolytic activity was determined as follows. To the human erythrocyte solution (50 μl), PBS (1 ml) was added, followed by different concentrations of the peptides. The suspension was incubated for 30 min at 37°C and then centrifuged at 1,000 × g for 6 min at 4°C. The supernatant was monitored at 415 nm by using a spectrophotometer (Bio-Rad, Munich, Germany). The hemolytic activity of each peptide was expressed as the percentage of the total hemoglobin release compared to the release after incubation with Millipore water.
TEM and IEM.
Liquid cultures of S. aureus (ATCC 25923) cells grown up to mid-logarithmic phase were washed twice in 10 mM sodium phosphate buffer (pH 7.4). Cells (108 CFU) were treated with DCD-1 or SSL-23 (100 μg/ml) for 4 h at 37°C. In parallel, antimicrobial assays with both peptides were performed which confirmed the antibacterial activity of both peptides. For immune electron microscopy (IEM), cells were fixed in PLP and centrifuged, and the resulting bacterial pellet was embedded in 3.5% agarose at 37°C and cooled on ice. Small parts of agarose blocks were embedded in Lowicryl (Polysciences, Eppelheim, Germany). Ultrathin sections (50 nm) were mounted on Formvar-coated nickel grids and incubated with rabbit anti-DCD, followed by 10-nm gold-conjugated goat anti-rabbit immunoglobulin G (Auroprobe EM; Amersham, Freiburg, Germany). In control samples the primary antibody was omitted. Samples were examined by using a Zeiss 109 transmission electron microscope (Zeiss, Oberkochen, Germany). For transmission EM (TEM) bacterial pellets were fixed in Karnovsky's fixative, postfixed in 1% osmium tetroxide, and embedded in Epon.
RESULTS
DCD-derived peptides have a diverse and overlapping spectrum of antimicrobial activity.
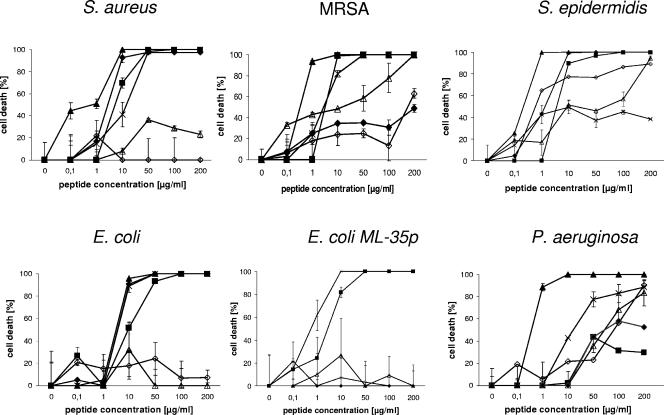
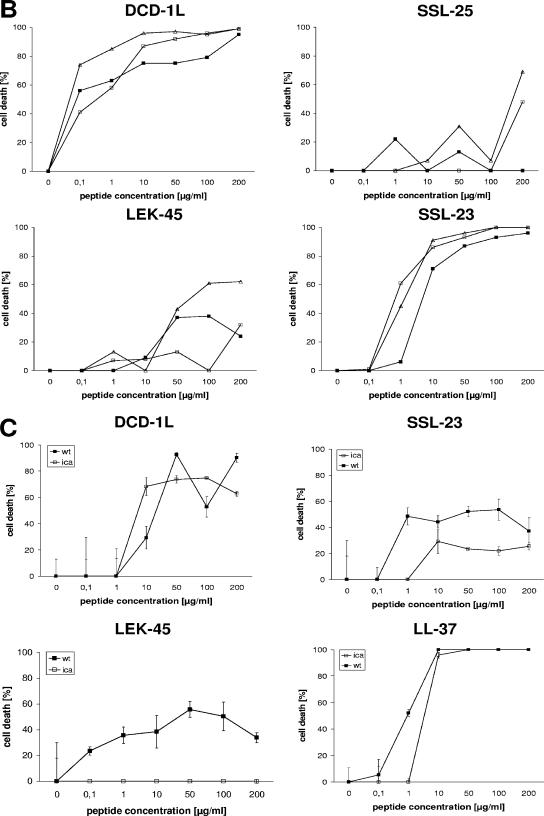
We previously showed (10) that several DCD-derived peptides are generated in human eccrine sweat by postsecretory proteolytic processing. First, we wanted to address the question whether the alteration from anionic to cationic peptides during proteolytic processing of DCD-1L leads to a different spectrum of antimicrobial activity. For this purpose, we performed antimicrobial assays with the most prominent DCD-derived peptides found in human eccrine sweat: DCD-1L and LEK-45 (both anionic), SSL-29 (neutral net charge), and SSL-25 and SSL-23 (both cationic). As a positive control peptide, we used the cathelicidin LL-37 and as a negative control peptide the unrelated scrambled peptide DPI (Table 1). The concentration-dependent antimicrobial activities of the DCD peptides are shown in Fig. 1, and the 90% inhibitory concentrations are summarized in Table 2. Interestingly, the different DCD peptides demonstrate different spectra of activity. Whereas the anionic peptide DCD-1L and the cationic derivatives SSL-25 and SSL-23 show antibiotic activity against most tested microorganisms (E. coli, S. aureus MSSA and MRSA, and S. epidermidis) (IC90 < 50 μg/ml), the DCD-derived peptides LEK-45 and SSL-29 show in most cases only minimal inhibitory activity against the tested microorganisms or only at higher peptide concentrations (IC90 > 180 μg/ml). Interestingly, SSL-25 and SSL-23 show similar or even better antibacterial activity than the parental peptide DCD-1L on the majority of the tested microorganisms. However, in contrast to DCD-1L, they possess only limited activity against S. epidermidis and MRSA, respectively (see Table 2 and Fig. 1). Furthermore, all tested DCD peptides show only a low level of antibacterial activity against P. aeruginosa. Our study demonstrates that the naturally processed DCD peptides have a different spectrum of activity and that DCD peptides exhibit antimicrobial activity irrespective of their charge. Since LEK-45 and further N-terminal-shortened derivatives (data not shown) have a significantly lower activity than the parental peptide DCD-1L, it seem that at least the first three amino acids SSL are important for the antimicrobial function. Furthermore, since the peptides SSL-23 and SSL-25 show antibacterial activity similar to that of DCD-1L, this indicates that the minimal region responsible for antimicrobial activity lies in the range of the first 23 amino acids of the DCD-1L peptide. Interestingly, a peptide with a few additional amino acids and a neutral net charge (SSL-29) lost the ability to kill several microorganisms (see Fig. 1 and Table 2).
TABLE 1.
Amino acid sequence and biochemical properties of DCD- derived peptides and control peptides used in this study
| Peptide | Amino acid sequence | Charge/pI |
|---|---|---|
| DCD peptides | ||
| DCD-1L | SSLLEKGLDGAKKAVGGLGKLGKDAVEDLESVGKGAVHDVKDVLDSVL | −2/5.07 |
| DCD-1 | SSLLEKGLDGAKKAVGGLGKLGKDAVEDLESVGKGAVHDVKDVLDSV | −2/5.07 |
| LEK-45 | LEKGLDGAKKAVGGLGKLGKDAVEDLESVGKGAVHDVKDVLDSVL | −2/5.08 |
| SSL-29 | SSLLEKGLDGAKKAVGGLGKLGKDAVEDL | 0/5.97 |
| SSL-25 | SSLLEKGLDGAKKAVGGLGKLGKDA | +2/9.4 |
| SSL-23 | SSLLEKGLDGAKKAVGGLGKLGK | +3/9.82 |
| Control peptides | ||
| DPI | DPYAEAASGPNPGSKSHESAQAENCGADPE | −5/4.08 |
| LL-37 | LLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES | +6/10.6 |
FIG. 1.
Antimicrobial activity of DCD-derived peptides against several bacterial strains. The concentration-dependent antimicrobial activity of the DCD-derived peptides DCD-1L (▪), LEK-45 (▵), SSL-29 (⋄), SSL-25 (+), SSL-23 (♦), and LL-37 (▴) on the bacterial strains S. aureus, MRSA, S. epidermidis, E. coli, E. coli ML-35p, and Pseudomonas aeruginosa after 2 to 3 h of incubation in 10 mM phosphate buffer-10 mM NaCl (pH 7.0) is presented. The number of bacterial colonies were counted, and the percentage of cell death calculated as described previously (37). The microbicidal activity was expressed as [1 − (cell survival after peptide incubation)/(cell survival after control peptide incubation)] × 100, which represents the percentage killing of the cells.
TABLE 2.
Antimicrobial activities of DCD-derived peptides and, as a control peptide, LL-37 on different microorganisms and bacterial mutants
| Microorganism | IC90 (μg/ml [μM])a |
|||||
|---|---|---|---|---|---|---|
| DCD-1L | LEK-45 | SSL-29 | SSL-25 | SSL-23 | LL-37 | |
| Gram-positive bacteria | ||||||
| Staphylococcus aureus (MSSA) | 45 (9.3) | >200 (>44) | >200 (>69) | 48 (19.9) | 10 (4.5) | 9 (2.0) |
| Staphylococcus aureus (MRSA) | 8 (1.7) | 180 (40) | >200 (>69) | 35 (14.5) | >200 (>89) | 0.9 (0.2) |
| Staphylococcus epidermidis | 10 (2.1) | 200 (44) | 200 (69) | >200 (>80) | 8 (3.6) | 0.9 (0.2) |
| Gram-negative bacteria | ||||||
| Escherichia coli | 45 (9.3) | >200 (>44) | >200 (>69) | 10 (4.1) | 10 (4.5) | 9 (2.0) |
| Escherichia coli ML-35p | 30 (6.2) | >200 (>44) | >200 (>69) | 9 (3.7) | ND | ND |
| Pseudomonas aeruginosa | >200 (>41) | >200 (>44) | >200 (>69) | 200 (83) | >200 (>89) | 1 (0.2) |
| Bacterial mutants | ||||||
| S. aureus 113 wild type | 180 (35) | >200 (>44) | >200 (>69) | >200 (>82) | >75 (>33) | 6 (1.3) |
| S. aureus mprF | 50 (10) | >200 (>44) | 180 (>62) | >200 (>82) | 50 (22) | 6 (1.3) |
| S. aureus dltA | 6 (1.2) | >200 (>44) | ND | >200 (>82) | 10 (4.5) | 6 (1.3) |
| S. epidermidis 1457 wild type | 50 (10) | >200 (>44) | >200 (>69) | 6 (2.5) | >200 (>89) | 9 (2.0) |
| S. epidermidis Δica | >200 (>41) | >200 (>44) | ND | ND | >200 (>89) | 9 (2.0) |
The 90% inhibitory concentration (IC90) is the concentration of peptide in μg/ml (or in μM [in parentheses]) able to kill 90% of the microorganisms in 10 mM NaP-10 mM NaCl in a CFU assay compared to the control incubated only in buffer without peptide. ND, not determined.
Conformational studies.
For several AMPs, a close correlation between the antimicrobial activity and an α-helical secondary structure of the peptide has been described (5). DCD-1L is able to arrange in an amphipathic α-helical conformation. To examine the secondary structure of the different DCD-derived peptides, we performed CD measurements of DCD-1L, LEK-45, SSL-29, SSL-25, and SSL-23 and the control peptides DPI and LL-37 in different solutions. As shown in Table 3, when the DCD peptides were dissolved in aqueous buffer such as water or sodium phosphate buffer, no α-helicity was observed and all peptides had predominantly a random coil secondary structure. This did not change when the peptides were incubated in a buffer with an NaCl concentration of up to 150 mM. In contrast, the human cathelicidin LL-37 presented a 50% α-helical content already in 10 mM sodium phosphate buffer. Furthermore, to simulate the contact of the peptides with a bacterial membrane, we incubated the DCD peptides with unilamellar liposomes that differ in charge due to the content of artificial phospholipids such as DOPC and DOPG-DOPC (1:1 molar ratio) in sodium phosphate buffer. Neither the cationic nor the anionic DCD peptides adopted an α-helix under these conditions, in contrast to LL-37, where the α-helical secondary structure increased from 50 to 80% upon contact with the negatively charged liposomes (Table 3). Similar results for the different DCD peptides were achieved after incubation with S. aureus (data not shown). Interestingly, all DCD-derived peptides can adopt an α-helix in the presence of increasing concentrations of TFE, although to different extents. Whereas DCD-1L and LEK-45 displayed in the presence of 60% TFE 34 and 28% α-helical contents, respectively, SSL-29, SSL-25, and SSL-23 demonstrated 12, 31, and 14% α-helical content, respectively (Table 3). Since, in contrast to DCD-1L, SSL-25, and SSL-23, the DCD-derived peptides LEK-45 and SSL-29 show a diminished antimicrobial activity on the microorganisms tested (see Table 2), there was no clear relationship between the α-helical content of N and C terminally truncated DCD-1L analogs with their antimicrobial activity.
TABLE 3.
Summary of the CD analysis of several DCD-derived and control peptides in different solutions: distilled water, 10 mM sodium phosphate (NaP), 10 mM NaP with 10 or 150 mM NaCl, TFE, or in the presence of unilamellar liposomes, such as DOPC and DOPC-DOPG (1:1 molar ratio)a
| Peptide | Solution | Secondary structure (%) |
|||
|---|---|---|---|---|---|
| α-Helix | β-Sheet | Turn | Random coil | ||
| DCD-1L | dH2O | 0.0 | 28.8 | 16.4 | 54.9 |
| 10 mM NaP | 0.0 | 17.5 | 20.5 | 62.0 | |
| 10 mM NaP-10 mM NaCl | 0.0 | 19.1 | 19.4 | 61.4 | |
| 10 mM NaP-150 mM NaCl | 0.0 | 32.1 | 0.0 | 67.9 | |
| DOPC | 0.0 | 35.2 | 9.8 | 55.0 | |
| DOPC-DOPG | 0.0 | 27.8 | 15.4 | 56.8 | |
| 60% TFE | 34.5 | 27.2 | 0.0 | 38.3 | |
| LEK-45 | dH2O | 0.0 | 30.3 | 16.1 | 53.6 |
| 10 mM NaP | 0.0 | 13.6 | 21.2 | 62.2 | |
| 10 mM NaP-10 mM NaCl | 0.0 | 17.0 | 19.5 | 63.5 | |
| 10 mM NaP-150 mM NaCl | 0.0 | 41.5 | 0.0 | 58.5 | |
| 60% TFE | 28.0 | 31.0 | 0.0 | 41.0 | |
| SSL-23 | dH2O | 0.0 | 38.2 | 14.8 | 47.0 |
| 10 mM NaP | 0.0 | 23.8 | 18.4 | 57.7 | |
| DOPC | 0.0 | 13.7 | 21.5 | 64.8 | |
| DOPC-DOPG | 2.2 | 49.2 | 3.8 | 44.8 | |
| 60% TFE | 14.1 | 40.8 | 4.9 | 49.2 | |
| 80% TFE | 32.9 | 27.6 | 0.0 | 39.5 | |
| SSL-25 | dH2O | 0.0 | 22.3 | 20.7 | 57.0 |
| 60% TFE | 31.1 | 27.1 | 0.0 | 41.8 | |
| SSL-29 | dH20 | 0.0 | 24.3 | 20.3 | 55.4 |
| 60% TFE | 12.2 | 46.2 | 0.0 | 41.7 | |
| LL-37 | 10 mM NaP | 50.9 | 6.0 | 16.6 | 26.5 |
| DOPC | 54.1 | 6.4 | 14.0 | 25.5 | |
| DOPC-DOPG | 80.2 | 0.0 | 8.7 | 11.0 | |
| DPI | dH2O | 0.0 | 34.3 | 15.4 | 50.3 |
| 10 mM NaP | 0.0 | 25.8 | 19.4 | 54.8 | |
| DOPC-DOPG | 0.0 | 29.7 | 19.1 | 51.2 | |
| 60% TFE | 9.6 | 42.0 | 6.0 | 42.5 | |
Data are based on Jasco secondary structure estimation software. dH2O, distilled water.
DCD-1L self-associates in solution.
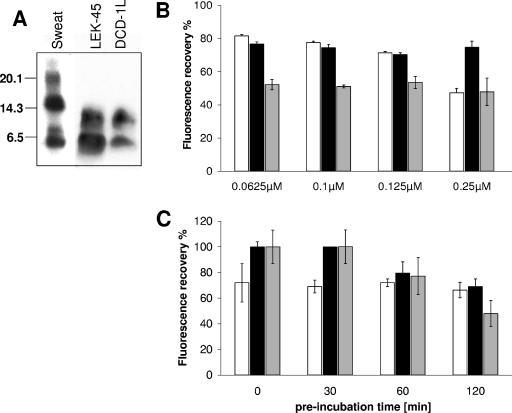
In human sweat several sodium dodecyl sulfate-stable oligomers of DCD peptides are found in a Western blot analysis with an anti-DCD antiserum (Fig. 2A). Interestingly, the synthetic peptides DCD-1L and LEK-45 (Fig. 2A) are also able to form SDS-stable dimers. These data suggest that DCD-1L and LEK-45 are able to self-associate.
FIG. 2.
Determination of the oligomerization of the peptides DCD-1L, LEK-45, SSL-23, and LL-37 in solution. (A) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot analysis of human eccrine sweat and 4 μg of the DCD peptides LEK-45 and DCD-1L dissolved in water using a polyclonal anti-DCD antibody, which detects the C terminus of DCD-1L. Seen are SDS-stable dimers for LEK-45 and DCD-1L and higher oligomers in sweat. (B) Percent fluorescence recovery of FITC-labeled peptides LL-37 (□), DCD1L (▪), and SSL-23 (░⃞) in 1× PBS (pH 7.4) at different concentrations (0.0625 to 0.25 μM). Peptides were preincubated 2 h in PBS before proteinase K (10 μg/ml) treatment. Oligomerization of the peptides in solution was determined by fluorescence dequenching. (C) Determination of the time-kinetics of oligomerization: peptides (0.25 μM) were incubated for different time points (0 to 120 min) at room temperature in PBS before proteinase K treatment, and the percentage of fluorescence recovery was determined.
Furthermore, to investigate the ability of DCD-1L and the shortened form SSL-23 to self-associate in solution, we used FITC-labeled peptides. As a control peptide we used FITC-labeled LL-37. We used the fluorescence quenching assay described by Oren et al. (26). This assay is based on the principle that the fluorescence is quenched when several molecules are in close proximity, i.e., when they self-associate or form oligomers. The fluorescence of the respective peptide at different concentrations is compared to the fluorescence of the peptide after treatment with proteinase K, which resulted in total degradation of the peptides (the 100% value). The percentage of fluorescence recovery of the peptide in solution is a measurement of the aggregation state. As seen in Fig. 2B and C, DCD-1L and SSL-23 self-associates in PBS in a time-dependent, but not concentration-dependent, manner. The fluorescence dropped successively to 70 and 50% of the fluorescence after protease treatment for DCD-1L and SSL-23, respectively, after 2 h. In contrast, LL-37 was able to self-associate concentration dependently up to 50%. In contrast to DCD-1L, however, a stable fluorescence level was achieved already from the earliest time point analyzed (Fig. 2C). This indicates that the DCD peptides DCD-1L and SSL-23 can self-associate but that it takes approximately 2 h to achieve this.
Kinetics of antimicrobial activity.
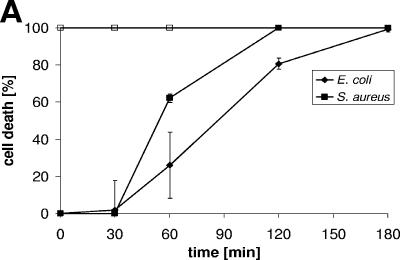
To analyze the time kinetics of antimicrobial activity, we incubated DCD-1L for 30, 60, 120, or 180 min with S. aureus or E. coli and analyzed the number of CFU after the respective time points. As shown in Fig. 3A, it takes approximately 2 h for DCD-1L to kill the majority of gram-positive and gram-negative bacteria (Fig. 3A) in a concentration-dependent manner (data not shown). After 1 h of incubation with DCD-1L, the bacterial cell number is reduced; however, killing was significantly increased after 2 and 3 h of incubation with the peptide. This is in contrast to incubation of the bacteria with LL-37, which results in an immediate decrease in bacterial cell numbers (Fig. 3A). This may point to different mechanisms of activity of LL-37 and DCD-1L.
FIG.3.
Time kinetics and antimicrobial activity of DCD-derived peptides against bacterial cell envelope mutants. (A) Time-dependent killing of S. aureus (▪) and E. coli (⧫) by DCD-1L using the CFU assay. Bacteria in the mid-logarithmic phase of growth were incubated with DCD-1L (200 μg/ml, black symbols) at different time intervals (0-180 min). The open squares indicate the antimicrobial activity of the control peptide LL-37 (100 μg/ml). (B) S. aureus cell envelope mutants mprF (□) and dltA (▵) and wild-type SA113 (▪) were incubated with various concentrations of peptides (0.1 to 200 μg/ml) in 10 mM phosphate buffer-10 mM NaCl (pH 7.0) for 2 to 3 h at 37°C. Aliquots of bacterial suspensions were diluted and plated in triplicate on blood agar. The percentage of cell death was determined as described above. (C) S. epidermdis Δica and wild-type S. epidermidis 1457 were incubated with various concentrations of peptides (0.1 to 200 μg/ml) in 10 mM phosphate buffer-10 mM NaCl (pH 7.0) for 2 to 3 h at 37°C, and the percentage of cell death was determined as described above.
Antimicrobial activity against bacterial cell envelope mutants.
To analyze whether bacterial membrane mutants lacking specific cell envelope modifications are more susceptible to cationic and anionic DCD-derived peptides, we performed antimicrobial assays with the S. aureus mutants mprF and dltA, the S. epidermidis mutant Δica, and the corresponding wild-type strains. As can be seen in Fig. 3B and Table 2, the S. aureus mutants mprF and dltA are more sensitive to DCD-derived peptides, especially the dltA mutant. Furthermore, whereas LEK-45 and SSL-29 did not kill wild-type bacteria up to a concentration of 200 μg/ml, both peptides are able to kill the mutants at high concentrations. In contrast, the ica mutant seemed to be less sensitive to DCD peptides than the wild-type S. epidermidis strain (Fig. 3C and Table 2). These data indicate that gram-positive bacterial mutants with specific modifications in the bacterial envelope exhibit altered susceptibilities to the DCD-derived peptides.
Permeabilization of the outer and inner membrane of gram-negative bacteria.
Since many antibacterial peptides exert their effect by perturbing the permeability properties of the inner or outer membrane in gram-negative bacteria, we examined the effect of DCD-derived peptides on the integrity of the cytoplasmic and outer membrane of E. coli.
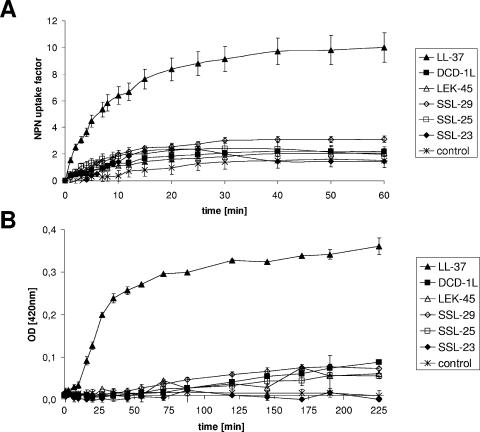
The activity of DCD peptides on the inner membrane of E. coli was investigated by monitoring the leakage of cytoplasmic β-galactosidase in the strain E. coli ML-35p as described by Lehrer et al. (21). Only when the cytoplasmic membrane is permeabilized by AMPs can the activity of cytoplasmic β-galactosidase be detected extracellularly by the hydrolysis of the substrate ONPG into ONP. The release of ONP was monitored spectrophotometrically at 420 nm. As shown in Fig. 4B, the DCD peptides DCD-1L, LEK-45, SSL-29, SSL-25, and SSL-23 at a concentration of 100 μg/ml did not permeabilize the inner membrane of E. coli ML-35p over a time period of more than 3 h, although DCD-1L and SSL-25 are highly active against this strain (see Table 2). In contrast, the cathelicidin LL-37 permeabilized the inner membrane of E. coli ML-35p after a few minutes at a concentration of 10 μg/ml.
FIG.4.
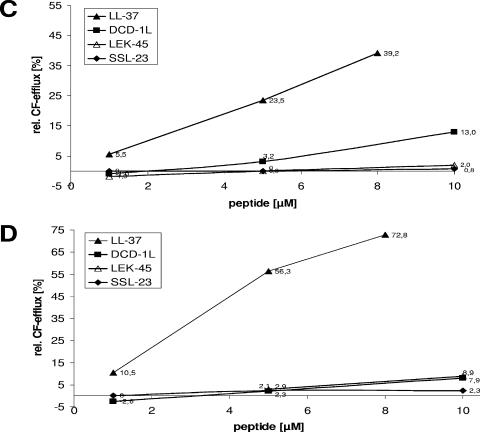
Effect of DCD-derived peptides on membrane permeability. (A) Outer membrane permeability measured by peptide-mediated NPN uptake in E. coli ML-35p. E. coli cells were incubated with 10 μM NPN in the presence of various concentrations of DCD peptides in 5 mM sodium HEPES buffer (pH 7.4). Enhanced uptake due to membrane permeability was measured by an increase in fluorescence intensity (Ex350 and Em460) caused by partition of NPN into the hydrophobic interior of the outer membrane. At time point 0 min, intact E. coli ML-35p cells were added to the peptides. The results are expressed as NPN uptake factor of fluorescence in arbitrary units. All analyses were performed in triplicates. (B) Inner membrane permeability measured as the influx of ONPG in E. coli ML-35p after the addition of DCD peptides. Stationary-growth-phase E. coli were incubated for 3 h at room temperature in 10 mM NaP (pH 7.0) with 1.67 mM ONPG. The release of ONP by cytoplasmic β-galactosidase was spectrophotometrically monitored at 420 nm. In the reference cuvette, peptides were placed in solvent. All samples were analyzed in triplicates. (C and D) Influence of the peptides DCD-1L (▪), LEK-45 (▵), and SSL-23 (⧫) on CF efflux of unilamellar liposomes made of DOPC (C) and DOPC-DOPG (1:1 molar ratio) (D). Release was determined 4 min after peptide addition at concentrations of 1 to 10 μM. At 4 min the amount of leakage reached a plateau when liposomes still contained a significant amount of CF. Reaction progress was expressed as the percentage of CF released relative to the total fluorescence released after the addition of Triton X-100 solution at the end of each experiment.
Next, we investigated whether DCD-derived peptides are able to permeabilize the outer membrane of E. coli ML-35p. For this purpose, we incubated the DCD peptides DCD-1L, LEK-45, SSL-29, SSL-25, and SSL-23 at a concentration of 200 μg/ml with E. coli ML-35p. Permeabilization of the outer membrane was measured by an increase in the fluorescence of NPN. The outer membrane normally excludes hydrophobic molecules such as NPN unless it is damaged. In this case, NPN can get access to the hydrophobic membrane interior, which results in an increase in NPN fluorescence. As shown in Fig. 4A, incubation of E. coli ML-35p with 200 μg/ml of the DCD peptides DCD-1L, LEK-45, SSL-29, SSL-25, and SSL-23 did not result in significant NPN uptake compared to a buffer control with the same amount of NPN. In contrast, LL-37 permeabilized the outer membrane of E. coli ML-35p in a few minutes (Fig. 4A). These data indicate that—in contrast to LL-37—cationic and anionic DCD peptides did not permeabilize the inner and outer membranes of E. coli ML-35p.
Interaction with phospholipid bilayers.
By using a model membrane system such as CF-loaded liposomes, we wanted to assess the mechanisms of natural peptide activity. The DCD peptides DCD-1L, LEK-45, and SSL-23 and, as a control, the irrelevant peptide DPI were tested for their ability to interact with phospholipid bilayers. We used unilamellar liposomes of different lipid compositions and charges: DOPC (neutral) and DOPC-DOPG (1:1 molar ratio) (50% negatively charged phospholipids). An increase in fluorescence intensity corresponds to CF release and is indicative of liposome leakage or lysis (4). In Fig. 4 it is shown that DCD-derived peptides caused only weak leakage (<13%) from liposomes made of DOPC (Fig. 4C) and DOPC-DOPG (Fig. 4D) even after the addition of 10 μM peptide. In contrast, LL-37 caused a rapid release of CF in the first minute (data not shown). LL-37 induced CF efflux from the DOPC liposomes was lower (∼39%) (Fig. 4C) than from DOPC-DOPG liposomes with a negative charged surface (∼73%) (Fig. 4D). Incubation of the liposomes with the irrelevant peptide DPI resulted in very low CF release (≤2%) (data not shown). These data demonstrate that DCD-derived peptides do not permeabilize unilamellar liposomes made of different lipid compositions as bacterial model membranes. This suggests that the antimicrobial activity of DCD peptides is not due to pore formation or a destabilization of the bacterial membrane.
Morphological changes.
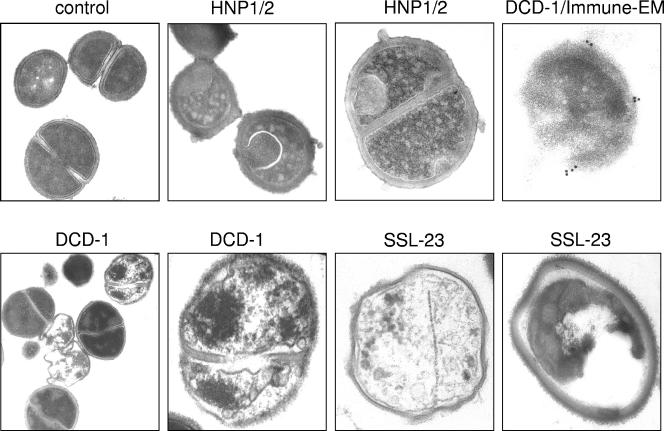
The experiments described above indicated that DCD peptides did not kill gram-negative bacteria by permeabilization of the bacterial membranes. To investigate how DCD peptides kill gram-positive bacteria, we examined the morphological changes of S. aureus by using transmission EM after incubation with 100 μg of the antimicrobially active DCD peptides DCD-1 or SSL-23/ml for 4 h. As shown in Fig. 5, we found no signs of cell wall damage. Instead, we observed cellular disintegration, suggesting that the anionic DCD-1 and cationic SSL-23 kill these bacteria by some unknown mechanism that does not initially disrupt their cytoplasmic membrane. In contrast, the alpha-defensins HNP-1/2 induced membrane blebbing of S. aureus as already described (21, 38) (Fig. 5). Bacteria from the control culture (Fig. 5) did not show any detectable deformation or alteration of the membrane or the cytoplasm. Furthermore, to investigate whether DCD-1 binds to the bacterial surface or is found in the cytoplasm, we performed immune-EM using an antiserum to DCD-1. As shown in Fig. 5, DCD-1 is able to bind to the cell surface but is not found in the bacterial cytoplasm.
FIG. 5.
Morphology of peptide-treated S. aureus. Transmission EM and immune-EM of S. aureus (ATCC 25923) treated with either DCD-1, SSL-23, or the alpha-defensins HNP-1 and -2 as a positive control for pore formation in 10 mM sodium phosphate buffer. Bacteria were incubated with 100 μg of DCD-1, SSL-23, and HNP1/2 per ml for 4 h. As a negative control cells were incubated in buffer without peptide. For the immune-EM, bacteria were incubated with a polyclonal antiserum to DCD-1, and the reactivity was detected by immunogold labeling. Seen is the binding of DCD to the bacterial surface.
Hemolytic activity.
Since several cationic AMPs have been described to have cytotoxic activity against eukaryotic cells, we analyzed whether DCD peptides are able to lyse eukaryotic cells. Therefore, we incubated the DCD peptides DCD-1L, LEK-45, SSL-29, SSL-25, and SSL-23 with human erythrocytes and analyzed photometrically the hemolytic activity. The DCD peptides did not exhibit hemolytic activity up to a concentration of 100 μg/ml. Similar results were obtained for LL-37: little hemolytic activity (up to 2.5%) was seen at a high LL-37 concentration (100 μg/ml). These data indicate that DCD-derived peptides do not damage the membranes of either prokaryotic or eukaryotic cells.
DISCUSSION
Until now, more than 700 AMPs have been isolated from diverse species such as plants, amphibians, insects, and mammals (2). Despite diverse structural motifs, a common feature of most of these peptides is that they are cationic and form amphipathic structures (15). Cationic AMPs display a net positive charge ranging from +2 to +9. It is believed that the charge is important for the initial electrostatic attraction of AMPs to negatively charged phospholipid membranes of bacteria or other microorganisms (5, 44). Cell death due to cationic AMPs may begin as quickly as 2 to 3 min after initial exposure (3, 14, 21, 40). It has been primarily attributed to membrane perturbation due to pore formation, membrane permeabilization, or depolarization of the bacterial membrane that leads to the loss of ions and metabolites, the cessation of essential vital functions, and ultimately to cell death (12, 17, 24).
We previously showed that by postsecretory proteolytic processing in sweat the dermcidin gene product gives rise to a whole group of truncated DCD peptides (10). Interestingly, proteolytically processed DCD peptides possess net charges between −2 and +2. DCD-1L and DCD-1 exhibit antimicrobial activity against gram-positive organisms, including S. aureus, E. faecalis, and gram-negative organisms including E. coli (37), as well as against S. epidermidis (42), Pseudomonas putida, MRSA, and rifampin- and isoniazid-resistant M. tuberculosis (20). In the present study we show that the cationic truncated DCD peptides SSL-25 and SSL-23 additionally exhibit antimicrobial activity against several gram-positive and gram-negative bacteria, including MRSA, with a similar spectrum of activity than the parental peptide DCD-1L. This indicates that the net charge of the DCD peptides is not essential for the antimicrobial function. However, the DCD peptide SSL-29 with four additional amino acids compared to SSL-25 and which has a neutral charge did not kill any of the microorganisms analyzed up to a concentration of 200 μg/ml. This suggests that the charge of the DCD peptide, irrespective of a positive or negative net charge, is essential either for binding to the bacterial membrane or for structure formation. Furthermore, a peptide lacking the first N-terminal three amino acids (SSL) from the DCD-1L sequence is not able to kill the microorganisms analyzed. This suggests that these N-terminal amino acids are essential for the activity. Our comprehensive analysis furthermore suggested that the active part of the DCD-1L sequence resides in the first 23 amino acids. It might be that this cationic part of DCD-1L is crucial for binding of the peptide to the bacterial membrane or for oligomerization.
Membrane permeability studies with gram-negative bacteria or liposomes as model bacterial membranes indicated that all DCD peptides did not permeabilize the bacterial membrane in contrast to the cathelicidin LL-37. The same seems to be the case for eukaryotic cells such as red blood cells since we see no hemolysis up to a concentration of 100 μg/ml, which is in agreement with a previous publication (20). Time kinetics indicated that the killing of bacteria by DCD peptides is a rather slow process, taking at least 2 h in vitro. This is in contrast to LL-37, which is able to kill the bacteria by membrane permeabilization after the first few minutes. Interestingly, analysis of the ability of DCD-1L and SSL-23 to aggregate indicated that it also takes approximately 2 h to self-associate to a stable plateau. The ability to form oligomers was also seen in vivo in human sweat. These data suggest that DCD-1L has to form stable complexes in order to kill microorganisms. Aggregation occurs most likely in solution or on the bacterial membrane. Indeed, our studies suggest that the bacterial membrane interacts with DCD peptides. Immune-EM studies showed that DCD-1 binds to the bacterial surface in clusters. Furthermore, the bacterial envelope mprF and dltA mutants lacking cell envelope modifications are more sensitive to the activity of DCD peptides irrespective of the charge of the latter. Therefore, we suggest a model in which negatively or positively charged DCD peptides form oligomers, which in turn bind to the bacterial membrane without causing massive permeabilization. Binding to and possibly insertion into the membrane may impair vital functions to such an extent that the system gets highly stressed and eventually out of balance. Interaction with defined targets outside and possibly also inside the bacteria may enforce the stress and finally result in cell death. It may be that DCD peptides act similar to the non-membrane-permeabilizing AMPs already described (5, 27, 44).
Although DCD is able to form multimers in solution, it is unclear which structure is necessary for the antimicrobial activity and whether DCD peptides have to multimerize in lipid membranes to achieve the toxic activity on microorganisms. We could show that all analyzed DCD peptides, irrespective of charge or activity, can adopt an alpha-helical conformation in helix-inducing solvents. These data are in agreement with the determination of the secondary structure of recombinant DCD-1L (20). Whereas in buffer or after incubation with artificial phospholipid membranes for more than 2 h DCD peptides have mainly a random structure, the alpha-helical content of LL-37 rapidly increased after incubation with negatively charged liposomes. These data indicate that in contrast to LL-37 the secondary structure does not correlate with the antimicrobial activity of DCD peptides. A number of models for membrane permeation by amphipathic alpha-helical peptides have been described, in some of them aggregation or oligomerization of the peptides seems to be important for disrupting the membranes of the target cells (19, 26). Many AMPs exist in relatively unstructured conformations prior to interaction with the target cell. Upon binding to bacterial membranes, peptides may undergo significant conformational dynamics to helical or other structures that affect antimicrobial activity (5).
Gram-positive bacteria, such as staphylococci, are distinguished by the presence of a thick cell wall composed of peptidoglycan and teichoic acid polymers and the absence of an outer membrane (29). S. aureus mutants lacking specific modifications in the bacterial membrane are highly susceptible to a variety of cationic AMPs. For example, incorporation of d-alanine into S. aureus teichoic acids by the dltA enzymes (31) or the lysylation of phosphatidylglycerol by mprF (39) confers resistance to defensins, protegrins, and other AMPs by repulsion of the cationic peptides. Disruption of the dltA or mprF gene in S. aureus increases the susceptibility to several cationic AMPs such as defensins, protegrins, or amphibian magainin (30, 31). Furthermore, mutations in the ica operon in S. epidermidis, the genes responsible for the biosynthesis of the slime polymer polysaccharide intercellular adhesin, reduces the formation of a biofilm, and increases the sensitivity of S. epidermidis to cationic AMPs (13, 42). Interestingly, we could show that the dltA and mprF mutants are susceptible to cationic and anionic DCD peptides, i.e., irrespective of charge of the peptide. In contrast, the ica mutant seemed to be less sensitive to DCD peptides than the wild-type S. epidermidis strain. It has been reported that by increasing the salt concentration of the incubation buffer, the efficacy of DCD-1L against the ica mutant is increased (42). This could indicate that a complex is formed between DCD peptides and salt ions which increases the ability to kill bacteria. We could show that DCD-1L and DCD-1 are also active under high-salt conditions and in a buffer resembling human sweat (37). Futhermore, our own experiments indicated that DCD-1L has a higher killing activity in a buffer with 10 mM NaCl compared to phosphate buffer alone. Increasing the salt concentration up to 150 mM did not alter the activity (data not shown). In a recently published study it was shown that the ionic environment dictates microbial susceptibility to AMPs (9). Therefore, it is possible that under the complex conditions in human sweat the antimicrobial activity of DCD peptides is much higher than under the in vitro conditions used in this study.
Whereas several AMPs are suggested to target bacterial components other than membranes, some studies indicated that receptor-type interactions may be important for some peptides in targeting specific epitopes on the microbial surface (5). Nisin exhibits antimicrobial activity in the nanomolar range and specifically binds to bacterial lipid II, a membrane-bound component involved in peptidoglycan synthesis (7). Similarly, the lantibiotic mersacidin interferes with transglycosylation and peptidoglycan synthesis in gram-positive bacteria by direct the targeting of lipid II (6). Moreover, tachyplesin has an affinity for lipopolysaccharide (18). Similarly, our own immune-EM studies showed that DCD-1 binds to the bacterial membrane in the form of clusters, which could indicate that there is a receptor-type interaction with a yet-unknown target.
In conclusion, in human eccrine sweat several dermcidin-derived peptides are generated by postsecretory proteolytic processing. Some of the dominant peptides such as DCD-1L, DCD-1, SSL-46 (34), SSL-25, and SSL-23 have a diverse and overlapping spectrum of antimicrobial activity, whereas the other dominant peptides in sweat such as LEK-45 and SSL-29 found also in the majority of sweat samples (33) are inactive. Thus, by postsecretory proteolytic processing the immune response against skin pathogens is modulated. Further studies will reveal whether there is a synergistic antimicrobial activity of the naturally occurring DCD peptides in human sweat and how bacteria are killed by these peptides.
Acknowledgments
We gratefully acknowledge the gift of E. coli ML-35p strain from Robert Lehrer (Department of Medicine, Center for Health Sciences, Los Angeles). We thank Alida Theil and Birgit Fehrenbacher for expert technical assistance.
This study was supported by Deutsche Forschungsgemeinschaft (DFG SCHI 510/3-1 and 3-2).
REFERENCES
- 1.Baechle, D., T. Flad, A. Cansier, H. Steffen, B. Schittek, J. Tolson, T. Herrmann, H. Dihazi, A. Beck, G. A. Mueller, M. Mueller, S. Stevanovic, C. Garbe, C. A. Mueller, and H. Kalbacher. 2006. Cathepsin D is present in human eccrine sweat and involved in the postsecretory processing of DCD-1L. J. Biol. Chem. 281:5406-5415. [DOI] [PubMed] [Google Scholar]
- 2.Beisswenger, C., and R. Bals. 2005. Functions of antimicrobial peptides in host defense and immunity. Curr. Protein Peptide Sci. 6:255-264. [DOI] [PubMed] [Google Scholar]
- 3.Blondelle, S. E., K. Lohner, and M. Aguilar. 1999. Lipid-induced conformation and lipid-binding properties of cytolytic and antimicrobial peptides: determination and biological specificity. Biochim. Biophys. Acta 1462:89-108. [DOI] [PubMed] [Google Scholar]
- 4.Breukink, E., C. van Kraaij, R. A. Demel, R. J. Siezen, O. P. Kuipers, and B. de Kruijff. 1997. The C-terminal region of nisin is responsible for the initial interaction of nisin with the target membrane. Biochemistry 36:6968-6976. [DOI] [PubMed] [Google Scholar]
- 5.Brogden, K. A. 2005. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3:238-250. [DOI] [PubMed] [Google Scholar]
- 6.Brotz, H., G. Bierbaum, K. Leopold, P. E. Reynolds, and H. G. Sahl. 1998. The lantibiotic mersacidin inhibits peptidoglycan synthesis by targeting lipid II. Antimicrob. Agents Chemother. 42:154-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brotz, H., M. Josten, I. Wiedemann, U. Schneider, F. Gotz, G. Bierbaum, and H. G. Sahl. 1998. Role of lipid-bound peptidoglycan precursors in the formation of pores by nisin, epidermin, and other lantibiotics. Mol. Microbiol. 30:317-327. [DOI] [PubMed] [Google Scholar]
- 8.Cipakova, I., J. Gasperik, and E. Hostinova. 2006. Expression and purification of human antimicrobial peptide, dermcidin, in Escherichia coli. Protein Expr. Purif. 45:269-274. [DOI] [PubMed] [Google Scholar]
- 9.Dorschner, R. A., B. Lopez-Garcia, A. Peschel, D. Kraus, K. Morikawa, V. Nizet, and R. L. Gallo. 2006. The mammalian ionic environment dictates microbial susceptibility to antimicrobial defense peptides. FASEB J. 20:35-42. [DOI] [PubMed] [Google Scholar]
- 10.Flad, T., R. Bogumil, J. Tolson, B. Schittek, C. Garbe, M. Deeg, C. A. Mueller, and H. Kalbacher. 2002. Detection of dermcidin-derived peptides in sweat by ProteinChip technology. J. Immunol. Methods 270:53-62. [DOI] [PubMed] [Google Scholar]
- 11.Ganz, T. 2003. Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 3:710-720. [DOI] [PubMed] [Google Scholar]
- 12.Gazit, E., A. Boman, H. G. Boman, and Y. Shai. 1995. Interaction of the mammalian antibacterial peptide cecropin P1 with phospholipid vesicles. Biochemistry 34:11479-11488. [DOI] [PubMed] [Google Scholar]
- 13.Gotz, F. 2002. Staphylococcus and biofilms. Mol. Microbiol. 43:1367-1378. [DOI] [PubMed] [Google Scholar]
- 14.Hancock, R. E., and D. S. Chapple. 1999. Peptide antibiotics. Antimicrob. Agents Chemother. 43:1317-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hancock, R. E., and R. Lehrer. 1998. Cationic peptides: a new source of antibiotics. Trends Biotechnol. 16:82-88. [DOI] [PubMed] [Google Scholar]
- 16.Haukland, H. H., H. Ulvatne, K. Sandvik, and L. H. Vorland. 2001. The antimicrobial peptides lactoferricin B and magainin 2 cross over the bacterial cytoplasmic membrane and reside in the cytoplasm. FEBS Lett. 508:389-393. [DOI] [PubMed] [Google Scholar]
- 17.Heller, W. T., A. J. Waring, R. I. Lehrer, T. A. Harroun, T. M. Weiss, L. Yang, and H. W. Huang. 2000. Membrane thinning effect of the beta-sheet antimicrobial protegrin. Biochemistry 39:139-145. [DOI] [PubMed] [Google Scholar]
- 18.Hirakura, Y., S. Kobayashi, and K. Matsuzaki. 2002. Specific interactions of the antimicrobial peptide cyclic beta-sheet tachyplesin I with lipopolysaccharides. Biochim. Biophys. Acta 1562:32-36. [DOI] [PubMed] [Google Scholar]
- 19.Johansson, J., G. H. Gudmundsson, M. E. Rottenberg, K. D. Berndt, and B. Agerberth. 1998. Conformation-dependent antibacterial activity of the naturally occurring human peptide LL-37. J. Biol. Chem. 273:3718-3724. [DOI] [PubMed] [Google Scholar]
- 20.Lai, Y. P., Y. F. Peng, Y. Zuo, J. Li, J. Huang, L. F. Wang, and Z. R. Wu. 2005. Functional and structural characterization of recombinant dermcidin-1L, a human antimicrobial peptide. Biochem. Biophys. Res. Commun. 328:243-250. [DOI] [PubMed] [Google Scholar]
- 21.Lehrer, R. I., A. Barton, K. A. Daher, S. S. Harwig, T. Ganz, and M. E. Selsted. 1989. Interaction of human defensins with Escherichia coli. Mechanism of bactericidal activity. J. Clin. Investig. 84:553-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lehrer, R. I., A. Barton, and T. Ganz. 1988. Concurrent assessment of inner and outer membrane permeabilization and bacteriolysis in E. coli by multiple-wavelength spectrophotometry. J. Immunol. Methods 108:153-158. [DOI] [PubMed] [Google Scholar]
- 23.Loh, B., C. Grant, and R. E. Hancock. 1984. Use of the fluorescent probe 1-N-phenylnaphthylamine to study the interactions of aminoglycoside antibiotics with the outer membrane of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 26:546-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuzaki, K. 1998. Magainins as paradigm for the mode of action of pore forming polypeptides. Biochim. Biophys. Acta 1376:391-400. [DOI] [PubMed] [Google Scholar]
- 25.Mayer, L. D., M. J. Hope, and P. R. Cullis. 1986. Vesicles of variable sizes produced by a rapid extrusion procedure. Biochim. Biophys. Acta 858:161-168. [DOI] [PubMed] [Google Scholar]
- 26.Oren, Z., J. C. Lerman, G. H. Gudmundsson, B. Agerberth, and Y. Shai. 1999. Structure and organization of the human antimicrobial peptide LL-37 in phospholipid membranes: relevance to the molecular basis for its non-cell-selective activity. Biochem. J. 341(Pt. 3):501-513. [PMC free article] [PubMed] [Google Scholar]
- 27.Otvos, L., Jr. 2005. Antibacterial peptides and proteins with multiple cellular targets. J. Peptide Sci. 11:697-706. [DOI] [PubMed] [Google Scholar]
- 28.Park, C. B., H. S. Kim, and S. C. Kim. 1998. Mechanism of action of the antimicrobial peptide buforin II: buforin II kills microorganisms by penetrating the cell membrane and inhibiting cellular functions. Biochem. Biophys. Res. Commun. 244:253-257. [DOI] [PubMed] [Google Scholar]
- 29.Peschel, A. 2002. How do bacteria resist human antimicrobial peptides? Trends Microbiol. 10:179-186. [DOI] [PubMed] [Google Scholar]
- 30.Peschel, A., R. W. Jack, M. Otto, L. V. Collins, P. Staubitz, G. Nicholson, H. Kalbacher, W. F. Nieuwenhuizen, G. Jung, A. Tarkowski, K. P. van Kessel, and J. A. van Strijp. 2001. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. J. Exp. Med. 193:1067-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peschel, A., M. Otto, R. W. Jack, H. Kalbacher, G. Jung, and F. Gotz. 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274:8405-8410. [DOI] [PubMed] [Google Scholar]
- 32.Rieg, S., C. Garbe, B. Sauer, H. Kalbacher, and B. Schittek. 2004. Dermcidin is constitutively produced by eccrine sweat glands and is not induced in epidermal cells under inflammatory skin conditions. Br. J. Dermatol. 151:534-539. [DOI] [PubMed] [Google Scholar]
- 33.Rieg, S., S. Seeber, H. Steffen, A. Humeny, H. Kalbacher, S. Stevanovic, A. Kimura, C. Garbe, and B. Schittek. 2006. Generation of multiple stable dermcidin-derived antimicrobial peptides in sweat of different body sites. J. Investig. Dermatol. 126:354-365. [DOI] [PubMed] [Google Scholar]
- 34.Rieg, S., H. Steffen, S. Seeber, A. Humeny, H. Kalbacher, K. Dietz, C. Garbe, and B. Schittek. 2005. Deficiency of dermcidin-derived antimicrobial peptides in sweat of patients with atopic dermatitis correlates with an impaired innate defense of human skin in vivo. J. Immunol. 174:8003-8010. [DOI] [PubMed] [Google Scholar]
- 35.Rouser, G., S. Fkeischer, and A. Yamamoto. 1970. Two dimensional then layer chromatographic separation of polar lipids and determination of phospholipids by phosphorus analysis of spots. Lipids 5:494-496. [DOI] [PubMed] [Google Scholar]
- 36.Ruissen, A. L., J. Groenink, E. J. Helmerhorst, E. Walgreen-Weterings, W. Van't Hof, E. C. Veerman, and A. V. Nieuw Amerongen. 2001. Effects of histatin 5 and derived peptides on Candida albicans. Biochem. J. 356:361-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schittek, B., R. Hipfel, B. Sauer, J. Bauer, H. Kalbacher, S. Stevanovic, M. Schirle, K. Schroeder, N. Blin, F. Meier, G. Rassner, and C. Garbe. 2001. Dermcidin: a novel human antibiotic peptide secreted by sweat glands. Nat. Immunol. 2:1133-1137. [DOI] [PubMed] [Google Scholar]
- 38.Selsted, M. E., and A. J. Ouellette. 2005. Mammalian defensins in the antimicrobial immune response. Nat. Immunol. 6:551-557. [DOI] [PubMed] [Google Scholar]
- 39.Staubitz, P., H. Neumann, T. Schneider, I. Wiedemann, and A. Peschel. 2004. MprF-mediated biosynthesis of lysylphosphatidylglycerol, an important determinant in staphylococcal defensin resistance. FEMS Microbiol. Lett. 231:67-71. [DOI] [PubMed] [Google Scholar]
- 40.Tossi, A., C. Tarantino, and D. Romeo. 1997. Design of synthetic antimicrobial peptides based on sequence analogy and amphipathicity. Eur. J. Biochem. 250:549-558. [DOI] [PubMed] [Google Scholar]
- 41.Ulvatne, H., O. Samuelsen, H. H. Haukland, M. Kramer, and L. H. Vorland. 2004. Lactoferricin B inhibits bacterial macromolecular synthesis in Escherichia coli and Bacillus subtilis. FEMS Microbiol. Lett. 237:377-384. [DOI] [PubMed] [Google Scholar]
- 42.Vuong, C., J. M. Voyich, E. R. Fischer, K. R. Braughton, A. R. Whitney, F. R. DeLeo, and M. Otto. 2004. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell Microbiol. 6:269-275. [DOI] [PubMed] [Google Scholar]
- 43.Wu, M., E. Maier, R. Benz, and R. E. Hancock. 1999. Mechanism of interaction of different classes of cationic antimicrobial peptides with planar bilayers and with the cytoplasmic membrane of Escherichia coli. Biochemistry 38:7235-7242. [DOI] [PubMed] [Google Scholar]
- 44.Yeaman, M. R., and N. Y. Yount. 2003. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 55:27-55. [DOI] [PubMed] [Google Scholar]
- 45.Zasloff, M. 2002. Antimicrobial peptides in health and disease. N. Engl. J. Med. 347:1199-1200. [DOI] [PubMed] [Google Scholar]
- 46.Zasloff, M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389-395. [DOI] [PubMed] [Google Scholar]