Abstract
A nationwide survey of extended-spectrum β-lactamase (ESBL) production among Enterobacteriaceae, carried out in 2003, showed that CTX-M-type enzymes have achieved a sizeable prevalence among ESBL producers in Italy, mostly in Escherichia coli and, to a lesser extent, in Klebsiella pneumoniae. In this work, we report on the molecular epidemiology of the CTX-M-producing isolates from that survey and on the mechanisms of dissemination of these emerging resistance determinants. The CTX-M-producing isolates were detected in 10 of the 11 participating centers distributed across the Italian national territory, although at remarkably variable rates in different centers (1.2 to 49.5% of the ESBL producers). All CTX-M determinants were of group 1, with CTX-M-15 and CTX-M-1 being the most prevalent variants (60% and 35%, respectively) and CTX-M-32 carried by a minority (5%) of isolates. Each variant was detected both in E. coli and in K. pneumoniae. Genotyping of the CTX-M-producing isolates by random amplification of polymorphic DNA revealed a notable diversity, especially among those producing CTX-M-1, while clonal expansion was evident with some CTX-M-15-producing strains. Mating experiments revealed a higher overall transferability of blaCTX-M-1 and blaCTX-M-32 than of blaCTX-M-15. Coresistance to quinolones and aminoglycosides was overall higher with the CTX-M-15-producing isolates. The present results indicate that CTX-M-producing strains are now widespread across the Italian territory and underscore the emerging role of these ESBL determinants in the European setting. They also reveal notable differences in the dissemination mechanisms of genes encoding different CTX-M variants of the same lineage.
Plasmid-mediated extended-spectrum β-lactamases (ESBLs) capable of degrading the expanded-spectrum cephalosporins and monobactams are among the most important resistance determinants emerging worldwide in Enterobacteriaceae (6, 18, 30). Strains producing ESBLs are resistant to the above-mentioned compounds and often exhibit a multidrug-resistant phenotype, including resistance to aminoglycosides and fluoroquinolones (13, 31), leaving only a few reliable therapeutic options (30, 32). Infections caused by ESBL producers are associated with increased morbidity, mortality, and health care-associated costs (14, 22, 41).
The CTX-M-type β-lactamases, encoded by genes that have been captured on transferable plasmids from the chromosomes of Kluyvera spp., are among the most common and widespread ESBLs encountered in Enterobacteriaceae (4, 30). Although discovered later than the TEM- and SHV-type ESBLs (2, 3), it is now clear that the CTX-M-type β-lactamases are playing a major role as emerging resistance determinants in Enterobacteriaceae (4, 30). A worldwide distribution of these enzymes has been reported (4), and in some settings (e.g., Argentina, Greece, Japan, Spain, and Taiwan), the CTX-M-type enzymes are more prevalent than TEM- and SHV-type ESBLs (35, 36, 43, 45, 46). In Europe, where the TEM- and SHV-type ESBLs were first reported (20, 40) and are widespread overall (12, 26, 30, 33, 37), a rapid and massive dissemination of isolates producing CTX-M-type ESBLs has recently been reported in some countries (1, 16, 17, 21, 25, 44) and is a matter of major concern.
At least five different lineages of CTX-M-type enzymes have been identified, indicated as CTX-M-1, CTX-M-2, CTX-M-8, CTX-M-9, and CTX-M-25 groups after the representative enzymes of each lineage (http://www.lahey.org/studies/webt.htm) (4).
In Italy, the presence of CTX-M-type ESBLs was previously reported in clinical isolates of Enterobacteriaceae from some hospitals (7, 29, 39), as well as from companion animals (8). In 2003, the second Italian nationwide survey on ESBL production among Enterobacteriaceae was carried out, and the results showed that CTX-M-type enzymes were common overall (around 20%) among ESBL producers (24). In this work, we report on the molecular epidemiology of the CTX-M-producing isolates from that survey and on the mechanisms of dissemination of these emerging resistance determinants.
(These results were presented in part at the 44th Interscience Conference on Antimicrobial Agents and Chemotherapy, Washington, D.C., 2004.)
MATERIALS AND METHODS
Clinical isolates.
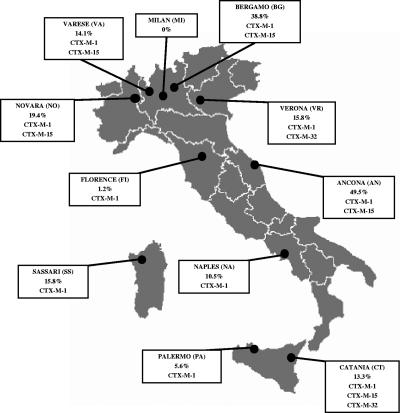
The clinical isolates investigated in this work were collected during the second Italian nationwide survey of ESBL production in Enterobacteriaceae (24). In that survey, nonreplicate clinical isolates of Enterobacteriaceae suspect for ESBL production (showing cefotaxime, ceftazidime, ceftriaxone, and/or aztreonam MICs of >1 μg/ml) were consecutively collected at the clinical microbiology laboratories of 11 teaching hospitals located across the Italian national territory (Fig. 1). In each center, the collection of isolates was carried out during the period from September to December 2003 and went on until the end of the sampling period or until a maximum of 750 isolates from inpatients and 250 from outpatients had been collected (whichever occurred first). Production of ESBL activity was confirmed in all isolates by a double-disk synergy test, and the presence of major lineages of ESBL genes (blaTEM, blaSHV, blaCTX-M, and blaPER) was investigated by colony blot hybridization (24). All the ESBL-producing isolates recognized by the blaCTX-M probes (hybridization was performed under low-stringency conditions using a probe mix that was capable of recognizing members of all major blaCTX-M lineages) were further investigated in this study.
FIG. 1.
Map of the Italian territory showing the locations of the centers participating in the study, the prevalence of CTX-M producers observed among the ESBL-positive isolates from each center, and the CTX-M variants detected in each center.
In vitro susceptibility testing.
Susceptibility testing of ESBL producers was carried out by disk diffusion (19), and results were interpreted according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) (10). With some antibiotics, MICs were also determined using Etest (AB Biodisk, Solna, Sweden). Escherichia coli ATCC 25922 and Klebsiella pneumoniae ATCC 700603 were used for quality control of susceptibility testing.
Molecular characterization of β-lactamase determinants.
The blaCTX-M genes were initially amplified from all isolates using primers CTX-MU1 and CTX-MU2 (Table 1), designed on conserved regions and capable of amplification of an internal fragment of blaCTX-M genes of all major lineages, as described previously (29). E. coli Eco02SI (a CTX-M-1-producing isolate from our collection), Proteus vulgaris PV1SM01 (a CTX-M-2-producing isolate) (29), Enterobacter aerogenes Rio-3 (producing CTX-M-8) (5), and E. coli 785-D (producing CTX-M-9) (38) were used as positive controls in PCR experiments. Direct sequencing of the amplification products allowed group assignment. Complete nucleotide sequences of group 1 blaCTX-M genes (the only ones detected following the above-mentioned screening) were determined on both strands by direct sequencing of PCR products obtained with primers CTX-M3G-F and CTX-M3GE-R (Table 1), external to the coding sequence, as described previously (28). The presence of blaTEM and blaSHV genes in CTX-M producers was determined by PCR using primers TEM/F and TEM/R or SHV/F and SHV/R (Table 1), as described previously (33). The natures of blaTEM alleles were investigated by sequencing both strands of the amplification products as described previously (33).
TABLE 1.
Oligonucleotide primers used in this work
| Primer | Target | Sequence (5′-3′) | Reference |
|---|---|---|---|
| CTX-MU1 | blaCTX-M-like | ATGTGCAGYACCAGTAARGT | 29 |
| CTX-MU2 | blaCTX-M-like | TGGGTRAARTARGTSACCAGA | 29 |
| CTX-M3G-F | blaCTX-M (group 1) | GTTACAATGTGTGAGAAGCAG | 28 |
| CTX-M3GE-R | blaCTX-M (group 1) | AACGGAATGAGTTTCCCCCATT | 28 |
| TEM/F | blaTEM-like | ATGAGTATTCAACATTTCCG | 33 |
| TEM/R | blaTEM-like | TTACCAATGCTTAATCAGTGAG | 33 |
| SHV/F | blaSHV-like | GCCCGGGTTATTCTTATTTGTCGC | 33 |
| SHV/R | blaSHV-like | TCTTTCCGATGCCGCCGCCAGTCA | 33 |
| 1254 | —a | CCGCAGCCAA | 27 |
| AP12h | — | CGGCCCCTGT | 11 |
—, not applicable.
Genotyping of isolates.
Random amplification of polymorphic DNA (RAPD) was carried out using primer 1254 or AP12h (Table 1) for the E. coli and the K. pneumoniae isolates, respectively. Reactions were carried out in a 25-μl volume using 1 U of the Taq DNA polymerase enzyme (Promega, Madison, Wis.) in the reaction buffer provided by the manufacturer, containing 1.5 mM MgCl2, 150 μM of each deoxynucleoside triphosphate, 40 pmol of the selected primer, and 2 μl of a crude cell extract obtained by boiling a bacterial suspension (A600, 0.15) for 10 min in sterile distilled water. The cycling parameters were as follows: 1 cycle each of 5 min at 94, 36, and 72°C; 10 cycles of 1 min at 94°C, 1 min at 36°C, and 2 min at 72°C; 20 cycles of 1 min at 94°C, 1 min at 50°C, and 2 min at 72°C; and a final extension of 10 min at 72°C. The RAPD profiles were resolved by electrophoresis in 2% agarose gels in Tris-acetate-EDTA buffer; recorded as digital images after ethidium bromide staining; and analyzed using the Diversity Database Fingerprinting software (Bio-Rad, Richmond, Calif.). Under the above-mentioned experimental conditions reproducible profiles were consistently obtained in replicate experiments. Clustering of isolates according to the RAPD profiles was done according to Dice's coefficient in combination with the unweighted-pair group method using average linkages clustering method (the band intensity was not considered for this analysis). Isolates were considered to belong in the same lineage when the similarity score was ≥0.90.
Gene transfer assays.
Transfer of resistance genes by conjugation was assayed by mating experiments in Mueller-Hinton broth (Difco Laboratories, Detroit, Mich.) using E. coli J-53 (pro met Rifr Nalr) as a recipient and an initial donor/recipient ratio of 0.1. Mating tubes were incubated at 30°C for 20 h. Transconjugants were selected on Mueller-Hinton agar containing rifampin (300 μg/ml) plus cefotaxime (2 μg/ml), and their identities were always confirmed by testing for the recipient's genetic markers (pro met). The presence of blaCTX-M genes in transconjugants was always confirmed by PCR as described for the clinical isolates.
Statistical analysis.
The chi-squared test with Yates' correction was used for statistical evaluation of comparisons between frequencies.
RESULTS
CTX-M-type β-lactamases in Enterobacteriaceae.
During the second Italian nationwide survey of ESBL production in Enterobacteriaceae, carried out in 2003, CTX-M-type β-lactamase genes were detected in 115 of 583 (19.7%) ESBL producers overall by a colony blot hybridization assay that could detect all major lineages of blaCTX-M genes but could not discriminate among the different variants. The CTX-M-encoding genes were mostly detected in E. coli and K. pneumoniae, but also in single isolates of Citrobacter amalonaticus and Morganella morganii (Table 2). The last two isolates have been described previously (28) and will not be discussed further here.
TABLE 2.
Prevalences of CTX-M producers among ESBL-positive isolates of Enterobacteriaceae from the Italian nationwide survey carried out in 2003
| Species (no. of ESBL producers) | No. of CTX-M producers (%) | CTX-M variant (no.) | Centers with CTX-M producersa |
|---|---|---|---|
| Escherichia coli (188) | 103 (54.8)b | CTX-M-1 (35) | AN, BG, CT, FI, NA, NO, PA, SS, VA, VR |
| CTX-M-15 (64) | AN, BG, CT, NO, VA | ||
| CTX-M-32 (4) | VR | ||
| Klebsiella pneumoniae (81) | 10 (12.3)b | CTX-M-1 (3) | BG, CT, NO |
| CTX-M-15 (5) | BG | ||
| CTX-M-32 (2) | CT | ||
| Klebsiella oxytoca (18) | 0 | —c | — |
| Enterobacter spp. (57)d | 0 | — | — |
| Citrobacter amalonaticus (1) | 1 | CTX-M-1 (1) | VA |
| Citrobacter spp. (17)e | 0 | — | — |
| Proteus mirabilis (163) | 0 | — | — |
| Morganella morganii (3) | 1 | CTX-M-1 (1) | VA |
| Providencia spp.(45)f | 0 | — | — |
| Serratia marcescens (10) | 0 | — | — |
| Total (583) | 115 (19.7) | — | AN, BG, CT, FI, NA, NO, PA, SS, VA, VR |
AN, Ancona; BG, Bergamo; CT, Catania; FI, Florence; NA, Naples; NO, Novara; PA, Palermo; SS, Sassari; VA, Varese; VR, Verona.
Percentage of the total number of ESBL producers (of the corresponding species).
—, not applicable.
Including E. aerogenes and E. cloacae.
Including C. freundii and C. koseri.
Including P. stuartii and P. rettgeri.
Sequence analysis of PCR products obtained with the CTX-MU1 and CTX-MU2 primers revealed that all the blaCTX-M genes belonged in group 1. Sequencing of the complete coding regions identified the genes as blaCTX-M-1, blaCTX-M-15, or blaCTX-M-32. Each variant was detected both in E. coli and in K. pneumoniae (Table 2). CTX-M-15 was the most prevalent variant (60% of CTX-M producers) but was detected in only 5 of the 11 centers. CTX-M-1 was the second most prevalent variant (35%) and the most widespread, being detected in 10 of the 11 centers. CTX-M-32 was the less common variant (5%) and showed a more restricted distribution (Fig. 1 and Table 2). The rates of ESBL-positive isolates producing CTX-M-type enzymes were highly variable (range, 1.2 to 49.5%) in different centers (Fig. 1).
Association of blaCTX-M with other β-lactamase determinants.
In the CTX-M-positive E. coli and K. pneumoniae isolates, the presence of additional β-lactamase determinants of the blaTEM and blaSHV types was investigated by PCR. Of the 103 CTX-M-positive E. coli isolates, 92 (89%) also carried a blaTEM determinant, while none carried a blaSHV determinant. Of the 10 CTX-M-positive K. pneumoniae isolates, all carried a blaSHV determinant (as expected, given the presence of a chromosomal blaSHV gene in the species) (42), while 5 (50%) also carried a blaTEM determinant. The blaTEM genes always encoded TEM-1. Carriage of a blaTEM-1 gene was found to be more frequent among CTX-M-15 producers (96%) than among CTX-M-1 producers (71%) (P < 0.005). The nature of the SHV determinants present in the K. pneumoniae isolates was not further investigated in this work.
Genotyping of the CTX-M-positive isolates.
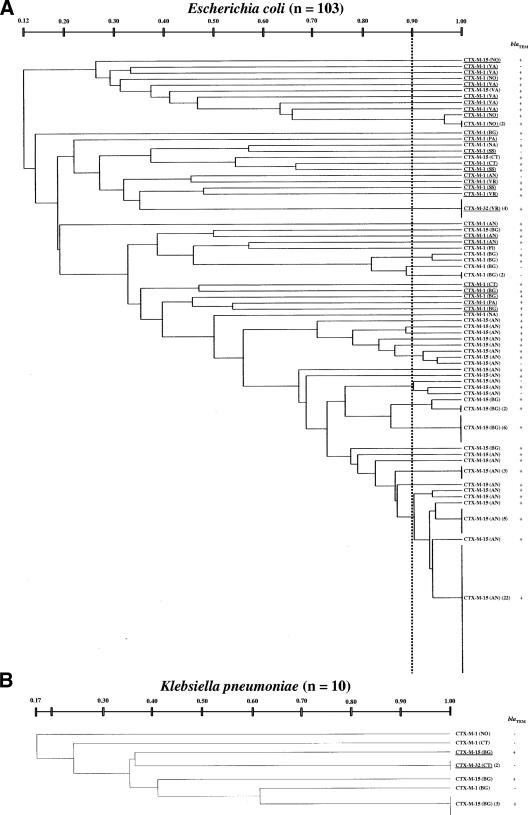
Genomic relatedness among the CTX-M-producing E. coli or K. pneumoniae isolates was investigated by RAPD profiling. The results revealed the presence of multiple lineages among the CTX-M producers of either species, even within the same center (Fig. 2). Isolates of the same lineage (i.e., sharing a similarity of ≥0.90) always carried the same blaCTX-M variant and were never detected in different centers (Fig. 2).
FIG. 2.
Dendrograms based on RAPD typing showing genetic relatedness among CTX-M-producing E. coli isolates (A) and among CTX-M-producing K. pneumoniae isolates (B) from various centers. The vertical dotted line indicates the 0.90 similarity score adopted to assign isolates to the same lineage. The origins of isolates are also indicated (abbreviations are the same as in Fig. 1). Lineages for which conjugative transfer of the CTX-M determinant was demonstrated are underlined.
In E. coli, the genotypic diversity appeared to be higher among isolates carrying blaCTX-M-1 (35 isolates distributed in 31 lineages; lineage/isolate ratio, 0.89) than among those carrying blaCTX-M-15 (64 isolates distributed in 21 lineages; lineage/isolate ratio, 0.33). The four blaCTX-M-32-positive isolates belonged to a single lineage (Fig. 2A).
The 10 K. pneumoniae isolates producing CTX-M enzymes were distributed in seven different lineages, three producing CTX-M-1, three producing CTX-M-15, and one producing CTX-M-32 (Fig. 2B).
Transferability of the blaCTX-M genes.
Transferability of the blaCTX-M genes was assayed with isolates representative of all of the different lineages (for those lineages including ≥3 isolates, a random sample of 2 to 5 isolates were selected for this analysis). Transferability of blaCTX-M-1 was observed for 25 (81%) of the 31 E. coli lineages and for none of the 3 K. pneumoniae lineages carrying that gene. Transferability of blaCTX-M-15 was observed in 1 (5%) of the 21 E. coli lineages and in 1 of the 3 K. pneumoniae lineages carrying that gene. Transferability of blaCTX-M-32 was observed in both the E. coli and the K. pneumoniae strains carrying that gene (Fig. 2).
Resistance phenotypes of the CTX-M-positive isolates.
All of the CTX-M-positive isolates were susceptible to imipenem, and most of them were also susceptible to amikacin and piperacillin-tazobactam. Lower susceptibility rates were observed for gentamicin, ciprofloxacin, and amoxicillin-clavulanate (Table 3). Coresistance to ciprofloxacin (in E. coli) and to aminoglycosides (in both E. coli and K. pneumoniae) was overall more frequent among the CTX-M-15-producing isolates (Table 3).
TABLE 3.
Resistance phenotypes of E. coli and K. pneumoniae isolates producing CTX-M-type ESBLs
| Species | Enzyme (no. of isolates) | % Susceptible toa,b:
|
|||||
|---|---|---|---|---|---|---|---|
| AMC | PTZ | IMI | GEN | AMK | CIP | ||
| E. colic | CTX-M-1 (35) | 77 | 100 | 100 | 89 | 97 | 54 |
| CTX-M-15 (64) | 22 | 89 | 100 | 46 | 98 | 2 | |
| CTX-M-32 (4) | 0 | 100 | 100 | 100 | 100 | 0 | |
| K. pneumoniae | CTX-M-1 (3) | 67 | 100 | 100 | 100 | 100 | 100 |
| CTX-M-15 (5) | 20 | 80 | 100 | 0 | 80 | 100 | |
| CTX-M-32 (2) | 0 | 100 | 100 | 100 | 100 | 100 | |
Based on disk diffusion testing.
AMC, amoxicillin-clavulanate; PTZ, piperacillin-tazobactam; IMI, imipenem; GEN, gentamicin; AMK, amikacin; CIP, ciprofloxacin.
Chi-squared test (CTX-M-1 and CTX-M-15): AMC, P < 0.001; PTZ, P < 0.05; GEN, P < 0.001; AMK, not significant; CIP, P < 0.001.
With all the CTX-M-producing isolates, cefotaxime and ceftazidime MICs were equal to or higher than the breakpoints (2 μg/ml) recommended by CLSI for suspicion of ESBL production (10), although the cefotaxime MICs were higher overall than those of ceftazidime (Table 4). Isolates producing CTX-M-15 or CTX-M-32 showed higher ceftazidime MICs than those producing CTX-M-1 (Table 4), in agreement with the enhanced ceftazidimase activities of the former enzymes (9, 34).
TABLE 4.
MICs of cefotaxime and ceftazidime and inhibitory-zone diameters of expanded-spectrum cephalosporins and aztreonam for CTX-M-producing isolates
| Species | Enzyme (no. of isolates) | MIC (μg/ml)a
|
Inhibitory zone diam (mm)a,b
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTX
|
CAZ
|
CTX
|
CAZ
|
ATM
|
FEP range | |||||||||
| Range | Median | MIC90 | Range | Median | MIC90 | Range | % Idd | Range | % Id | Range | % Id | |||
| E. coli | CTX-M-1 (35) | 32->256 | 64 | > 256 | 2-8 | 2 | 4 | 6-14 | 100 | 16-24 | 82 | 10-18 | 100 | 14-22 |
| CTX-M-15 (64) | 64->256 | 256 | > 256 | 4->256 | 32 | 128 | 6-14 | 100 | 6-20 | 100 | 6-18 | 100 | 10-22 | |
| CTX-M-32 (4) | 128->256 | 256 | 16-48 | 32 | 6-10 | 100 | 14-16 | 100 | 6-12 | 100 | 16 | |||
| K. pneumoniae | CTX-M-1 (3) | 24->256 | 32-64 | 6-10 | 100 | 14-20 | 100 | 6-14 | 100 | 10-18 | ||||
| CTX-M-15 (5) | 64->256 | 128 | 24-64 | 32 | 6-10 | 100 | 10-14 | 100 | 6-10 | 100 | 10-20 | |||
| CTX-M-32 (2) | >256 | >256 | 6 | 100 | 6 | 100 | 6 | 100 | 14-16 | |||||
CTX, cefotaxime; CAZ, ceftaxidime.
ATM, aztreonam; FEP, cefepime.
A 6-mm value means no zone of inhibition. The breakpoints for suspicion of ESBL production were as follows: CTX, ≤27 mm; CAZ, ≤22 mm; ATM, ≤27 mm (10).
% Id, percentage of isolates categorized as putative ESBL producers by disk diffusion testing, based on the CLSI breakpoint for suspicion of ESBL production, with the corresponding drug (10).
In disk diffusion testing, all the CTX-M-producing isolates yielded inhibitory-zone diameters lower than the breakpoints recommended by CLSI for suspicion of ESBL production with cefotaxime (≤27 mm) and with aztreonam (≤27 mm) (10), while 18% of the CTX-M-1-producing E. coli isolates yielded an inhibitory zone diameter of >22 mm with ceftazidime (Table 4).
DISCUSSION
ESBL production is the major emerging mechanism of resistance to expanded-spectrum cephalosporins and monobactams among Enterobacteriaceae and is a matter of major concern in the field of microbial drug resistance. In the European scenario, where the TEM- and SHV-type ESBLs were first detected (20, 40) and are widespread (12, 26, 30, 33, 37), recent reports have shown a rapid and alarming dissemination of Enterobacteriaceae producing ESBLs of the CTX-M type in some countries, with notable changes in the epidemiologies of these resistance determinants (15, 23, 44). The second Italian nationwide survey on ESBL production in Enterobacteriaceae, carried out in 2003, revealed that CTX-M-type ESBLs are now also widespread in Italy, where they are present in approximately 20% of ESBL-producing Enterobacteriaceae and in more than 60% of ESBL-producing E. coli isolates (24).
The results of this work provided some insights into the molecular epidemiology of this emerging problem in Italy. Isolates producing CTX-M-type enzymes were detected in 10 of the 11 centers distributed across Italian territory, showing that these enzymes have achieved a countrywide distribution. However, their prevalences in different areas appeared to be highly variable, which could reflect the scenario of a relatively early stage of dissemination of these resistance determinants in the clinical setting. Continuing surveillance will be necessary to monitor the evolution of this phenomenon and to verify whether the CTX-M-type ESBLs will eventually prevail over the TEM- and SHV-type ESBLs, which are still widespread, especially in some areas. It will also be interesting to investigate if these epidemiological differences could reflect differences in regional antimicrobial policies (the Italian Public Health System is organized on a strictly regional basis).
Unlike in other countries (e.g., Spain, France), where members of multiple CTX-M lineages have been reported (15, 43), a virtually absolute prevalence of members of the CTX-M-1 lineage was observed in Italy. The reasons for this finding, which is similar to that reported in Poland (1), remain to be clarified, but it might also be consistent with a stage of early dissemination of these ESBL determinants.
Molecular characterization of the CTX-M-producing isolates and investigation of transferability of the CTX-M-encoding determinants revealed significant differences between the two closely related allelic variants blaCTX-M-1 and blaCTX-M-15. In particular, the notable genotypic diversity among the E. coli isolates producing CTX-M-1 and the high frequency at which conjugative transfer of the blaCTX-M-1 gene could be detected suggest that plasmid-mediated horizontal transfer played a major role in the dissemination of this ESBL determinant in E. coli. On the other hand, the lower genotypic diversity among the E. coli isolates producing CTX-M-15 and the lower propensity of blaCTX-M-15 to be transferred by conjugation suggest that dissemination of blaCTX-M-15 was more heavily dependent on clonal expansion. These different behaviors, which likely reflect different natures of the genetic elements carrying the two CTX-M determinants, could account for the more restricted geographical dissemination of CTX-M-15 producers than of CTX-M-1 producers, despite their higher overall prevalence. Investigation of the genetic support of the CTX-M determinants is ongoing, to clarify the nature of the conjugative plasmids and to assess the locations of blaCTX-M genes that were apparently not transferable by conjugation.
Concerning antimicrobial susceptibility, all the CTX-M-producing isolates investigated in this study retained susceptibility to carbapenems and most of them also to amikacin and piperacillin-tazobactam. High resistance rates were observed with gentamicin, ciprofloxacin, and amoxicillin-clavulanate. The notable discrepancy in the behaviors observed with the two β-lactam/β-lactamase inhibitor combinations is likely due, at least in part, to the higher susceptibility to tazobactam than to clavulanate of the CTX-M-type enzymes (6). The higher rates of resistance to gentamicin and ciprofloxacin observed with the CTX-M-15 producers could be due to genetic linkage of blaCTX-M-15 with other resistance determinants on the same genetic element and/or to the expansion of clones carrying these resistance determinants.
Although ceftazidime MICs were overall lower than those of cefotaxime, especially for CTX-M-1-producing isolates, the MICs of both compounds for all the CTX-M-producing isolates were equal to or higher than the 2-μg/ml breakpoint recommended by CLSI for suspicion of ESBL production. In disk diffusion testing, however, 18% of the CTX-M-1 producers would have been missed as potential ESBL producers if ceftazidime alone were used as an expanded-spectrum cephalosporin for screening purposes. Thus, when disk diffusion is used for susceptibility testing, the use of ceftazidime alone is not advisable for screening of ESBL production. This is an important point that clinical laboratories should consider as the CTX-M-type β-lactamases continue to expand.
Acknowledgments
This study was supported in part by a research grant from Wyeth Pharmaceuticals. We are gratefully to Richard Bonnet and Elisenda Miro for providing us with the E. aerogenes Rio-3 and E. coli 758-D control strains, respectively.
REFERENCES
- 1.Baraniak, A., J. Fiett, A. Sulikowska, W. Hryniewicz, and M. Gniadkowski. 2002. Countrywide spread of CTX-M-3 extended-spectrum β-lactamase-producing microorganisms of the family Enterobacteriaceae in Poland. Antimicrob. Agents Chemother. 46:151-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barthélémy, M., J. Peduzzi, H. Bernard, C. Tancrede, and R. Labia. 1992. Close amino acid sequence relationship between the new plasmid-mediated extended-spectrum β-lactamase MEN-1 and chromosomally encoded enzymes of Klebsiella oxytoca. Biochim. Biophys. Acta 1122:15-22. [DOI] [PubMed] [Google Scholar]
- 3.Bauernfeind, A., H. Grimm, and S. Schweighart. 1990. A new plasmidic cefotaximase in a clinical isolate of Escherichia coli. Infection 18:294-298. [DOI] [PubMed] [Google Scholar]
- 4.Bonnet, R. 2004. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonnet, R., J. L. Sampaio, R. Labia, C. de Champs, D. Sirot, C. Chanal, and J. Sirot. 2000. A novel CTX-M β-lactamase (CTX-M-8) in cefotaxime-resistant Enterobacteriaceae isolated in Brazil. Antimicrob. Agents Chemother. 44:1936-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradford, P. A. 2001. Extended-spectrum β-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14:933-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brigante, G., F. Luzzaro, M. Perilli, G. Lombardi, A. Colì, G. M. Rossolini, G. Amicosante, and A. Toniolo. 2005. Evolution of CTX-M-type β-lactamases in isolates of Escherichia coli infecting hospital and community patients. Int. J. Antimicrob. Agents 25:157-162. [DOI] [PubMed] [Google Scholar]
- 8.Carattoli, A., S. Lovari, A. Franco, G. Cordaro, M. P. Di, and A. Battisti. 2005. Extended-spectrum β-lactamases in Escherichia coli isolated from dogs and cats in Rome, Italy, from 2001 to 2003. Antimicrob. Agents Chemother. 49:833-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cartelle, M., T. M. Del Mar, F. Molina, R. Moure, R. Villanueva, and G. Bou. 2004. High-level resistance to ceftazidime conferred by a novel enzyme, CTX-M-32, derived from CTX-M-1 through a single Asp240-Gly substitution. Antimicrob. Agents Chemother. 48:2308-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing; 16th informational supplement. Clinical Laboratory Standards Institute, Wayne, Pa.
- 11.Davin-Regli, A., D. Monnet, P. Saux, C. Bosi, R. Charrel, A. Barthelemy, and C. Bollet. 1996. Molecular epidemiology of Enterobacter aerogenes acquisition: one-year prospective study in two intensive care units. J. Clin. Microbiol. 34:1474-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Champs, C., C. Chanal, D. Sirot, R. Baraduc, J. P. Romaszko, R. Bonnet, A. Plaidy, M. Boyer, E. Carroy, M. C. Gbadamassi, S. Laluque, O. Oules, M. C. Poupart, M. Villemain, and J. Sirot. 2004. Frequency and diversity of class A extended-spectrum β-lactamases in hospitals of the Auvergne, France: a 2 year prospective study. J. Antimicrob. Chemother. 54:634-639. [DOI] [PubMed] [Google Scholar]
- 13.Diaz, P. Q., H. T. Bello, M. Y. Dominguez, N. F. Trabal, S. M. Mella, R. Z. Zemelman, and G. R. Gonzalez. 2004. Resistance to gentamicin, amikacin and ciprofloxacin among nosocomial isolates of Klebsiella pneumoniae subspecies pneumoniae producing extended spectrum β-lactamases. Rev. Med. Chil. 132:1173-1178. [DOI] [PubMed] [Google Scholar]
- 14.Du, B., Y. Long, H. Liu, D. Chen, D. Liu, Y. Xu, and X. Xie. 2002. Extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae bloodstream infection: risk factors and clinical outcome. Intensive Care Med. 28:1718-1723. [DOI] [PubMed] [Google Scholar]
- 15.Eckert, C., V. Gautier, M. Saladin-Allard, N. Hidri, C. Verdet, Z. Ould-Hocine, G. Barnaud, F. Delisle, A. Rossier, T. Lambert, A. Philippon, and G. Arlet. 2004. Dissemination of CTX-M-type β-lactamases among clinical isolates of Enterobacteriaceae in Paris, France. Antimicrob. Agents Chemother. 48:1249-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edelstein, M., M. Pimkin, I. Palagin, I. Edelstein, and L. Stratchounski. 2003. Prevalence and molecular epidemiology of CTX-M extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in Russian hospitals. Antimicrob. Agents Chemother. 47:3724-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang, H., C. Lundberg, B. Olsson-Liljequist, G. Hedin, E. Lindback, A. Rosenberg, and J. Struwe. 2004. Molecular epidemiological analysis of Escherichia coli isolates producing extended-spectrum β-lactamases for identification of nosocomial outbreaks in Stockholm, Sweden. J. Clin. Microbiol. 42:5917-5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacoby, G. A., and L. S. Munoz-Price. 2005. The new β-lactamases. N. Engl. J. Med. 352:380-391. [DOI] [PubMed] [Google Scholar]
- 19.Jorgensen, J. H., J. D. Turnidge, and J. A. Washington. 1999. Antibacterial susceptibility tests: dilution and disk diffusion methods, p. 1526-1577. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. American Society for Microbiology, Washington, D.C.
- 20.Knothe, H., P. Shah, V. Krcmery, M. Antal, and S. Mitsuhashi. 1983. Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infection 11:315-317. [DOI] [PubMed] [Google Scholar]
- 21.Lartigue, M. F., N. Fortineau, and P. Nordmann. 2005. Spread of novel expanded-spectrum β-lactamases in Enterobacteriaceae in a university hospital in the Paris area, France. Clin. Microbiol. Infect. 11:588-591. [DOI] [PubMed] [Google Scholar]
- 22.Lautenbach, E., J. B. Patel, W. B. Bilker, P. H. Edelstein, and N. O. Fishman. 2001. Extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae: risk factors for infection and impact of resistance on outcomes. Clin. Infect. Dis. 32:1162-1171. [DOI] [PubMed] [Google Scholar]
- 23.Livermore, D. M., and P. M. Hawkey. 2005. CTX-M: changing the face of ESBLs in the UK. J. Antimicrob. Chemother. 56:451-454. [DOI] [PubMed] [Google Scholar]
- 24.Luzzaro, F., M. Mezzatesta, C. Mugnaioli, M. Perilli, S. Stefani, G. Amicosante, G. M. Rossolini, and A. Toniolo. 2006. Trends in the production of extended-spectrum β-lactamases among enterobacteria of medical interest. Report of the second Italian survey. J. Clin. Microbiol. 44:1659-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Markovska, R., I. Schneider, E. Keuleyan, and A. Bauernfeind. 2004. Extended-spectrum β-lactamase (ESBL) CTX-M-15-producing Escherichia coli and Klebsiella pneumoniae in Sofia, Bulgaria. Clin. Microbiol. Infect. 10:752-755. [DOI] [PubMed] [Google Scholar]
- 26.Morris, D., C. O'Hare, M. Glennon, M. Maher, G. Corbett-Feeney, and M. Cormican. 2003. Extended-spectrum β-lactamases in Ireland, including a novel enzyme, TEM-102. Antimicrob. Agents Chemother. 47:2572-2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mueen, A., F. Nattress, G. Greer, C. Yost, C. Gill, and L. McMullen. 2003. Origin of contamination and genetic diversity of Escherichia coli in beef cattle. Appl. Environ. Microbiol. 69:2794-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mugnaioli, C., F. Luzzaro, F. De Luca, G. Brigante, G. Amicosante, and G. M. Rossolini. 2005. Dissemination of CTX-M-type extended-spectrum β-lactamase genes to unusual hosts. J. Clin. Microbiol. 43:4183-4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pagani, L., E. Dell'Amico, R. Migliavacca, M. M. D'Andrea, E. Giacobone, G. Amicosante, E. Romero, and G. M. Rossolini. 2003. Multiple CTX-M-type extended-spectrum β-lactamases in nosocomial isolates of Enterobacteriaceae from a hospital in northern Italy. J. Clin. Microbiol. 41:4264-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paterson, D. L., and R. A. Bonomo. 2005. Extended-spectrum β-lactamases: a clinical update. Clin. Microbiol. Rev. 18:657-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paterson, D. L., L. Mulazimoglu, J. M. Casellas, W. C. Ko, H. Goossens, G. A. Von, S. Mohapatra, G. M. Trenholme, K. P. Klugman, J. G. McCormack, and V. L. Yu. 2000. Epidemiology of ciprofloxacin resistance and its relationship to extended-spectrum β-lactamase production in Klebsiella pneumoniae isolates causing bacteremia. Clin. Infect. Dis. 30:473-478. [DOI] [PubMed] [Google Scholar]
- 32.Paterson, D. L., and L. B. Rice. 2003. Empirical antibiotic choice for the seriously ill patient: are minimization of selection of resistant organisms and maximization of individual outcome mutually exclusive? Clin. Infect. Dis. 36:1006-1012. [DOI] [PubMed] [Google Scholar]
- 33.Perilli, M., E. Dell'Amico, B. Segatore, M. R. De Massis, C. Bianchi, F. Luzzaro, G. M. Rossolini, A. Toniolo, G. Nicoletti, and G. Amicosante. 2002. Molecular characterization of extended-spectrum β-lactamases produced by nosocomial isolates of Enterobacteriaceae from an Italian nationwide survey. J. Clin. Microbiol. 40:611-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poirel, L., M. Gniadkowski, and P. Nordmann. 2002. Biochemical analysis of the ceftazidime-hydrolysing extended-spectrum β-lactamase CTX-M-15 and of its structurally related beta-lactamase CTX-M-3. J. Antimicrob. Chemother. 50:1031-1034. [DOI] [PubMed] [Google Scholar]
- 35.Pournaras, S., A. Ikonomidis, I. Kristo, A. Tsakris, and A. N. Maniatis. 2004. CTX-M enzymes are the most common extended-spectrum β-lactamases among Escherichia coli in a tertiary Greek hospital. J. Antimicrob. Chemother. 54:574-575. [DOI] [PubMed] [Google Scholar]
- 36.Quinteros, M., M. Radice, N. Gardella, M. M. Rodriguez, N. Costa, D. Korbenfeld, E. Couto, and G. Gutkind. 2003. Extended-spectrum β-lactamases in Enterobacteriaceae in Buenos Aires, Argentina, public hospitals. Antimicrob. Agents Chemother. 47:2864-2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodriguez-Bano, J., M. D. Navarro, L. Romero, M. A. Muniain, E. J. Perea, R. Perez-Cano, J. R. Hernandez, and A. Pascual. 2006. Clinical and molecular epidemiology of extended-spectrum β-lactamase-producing Escherichia coli as a cause of nosocomial infection or colonization: implications for control. Clin. Infect. Dis. 42:37-45. [DOI] [PubMed] [Google Scholar]
- 38.Sabate, M., R. Tarrago, F. Navarro, E. Miro, C. Verges, J. Barbe, and G. Prats. 2000. Cloning and sequence of the gene encoding a novel cefotaxime-hydrolyzing β-lactamase (CTX-M-9) from Escherichia coli in Spain. Antimicrob. Agents Chemother. 44:1970-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanguinetti, M., B. Posteraro, T. Spanu, D. Ciccaglione, L. Romano, B. Fiori, G. Nicoletti, S. Zanetti, and G. Fadda. 2003. Characterization of clinical isolates of Enterobacteriaceae from Italy by the BD Phoenix extended-spectrum β-lactamase detection method. J. Clin. Microbiol. 41:1463-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sougakoff, W., S. Goussard, G. Gerbaud, and P. Courvalin. 1988. Plasmid-mediated resistance to third-generation cephalosporins caused by point mutations in TEM-type penicillinase genes. Rev. Infect. Dis. 10:879-884. [DOI] [PubMed] [Google Scholar]
- 41.Stone, P. W., A. Gupta, M. Loughrey, P. Della-Latta, J. Cimiotti, E. Larson, D. Rubenstein, and L. Saiman. 2003. Attributable costs and length of stay of an extended-spectrum β-lactamase-producing Klebsiella pneumoniae outbreak in a neonatal intensive care unit. Infect. Control Hosp. Epidemiol. 24:601-606. [DOI] [PubMed] [Google Scholar]
- 42.Tzouvelekis, L. S., and R. A. Bonomo. 1999. SHV-type β-lactamases. Curr. Pharm. Des. 5:847-864. [PubMed] [Google Scholar]
- 43.Valverde, A., T. M. Coque, M. P. Sanchez-Moreno, A. Rollan, F. Baquero, and R. Canton. 2004. Dramatic increase in prevalence of fecal carriage of extended-spectrum β-lactamase-producing Enterobacteriaceae during nonoutbreak situations in Spain. J. Clin. Microbiol. 42:4769-4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woodford, N., M. E. Ward, M. E. Kaufmann, J. Turton, E. J. Fagan, D. James, A. P. Johnson, R. Pike, M. Warner, T. Cheasty, A. Pearson, S. Harry, J. B. Leach, A. Loughrey, J. A. Lowes, R. E. Warren, and D. M. Livermore. 2004. Community and hospital spread of Escherichia coli producing CTX-M extended-spectrum β-lactamases in the UK. J. Antimicrob. Chemother. 54:735-743. [DOI] [PubMed] [Google Scholar]
- 45.Wu, T. L., J. H. Chia, L. H. Su, A. J. Kuo, C. Chu, and C. H. Chiu. 2003. Dissemination of extended-spectrum β-lactamase-producing Enterobacteriaceae in pediatric intensive care units. J. Clin. Microbiol. 41:4836-4838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamasaki, K., M. Komatsu, T. Yamashita, K. Shimakawa, T. Ura, H. Nishio, K. Satoh, R. Washidu, S. Kinoshita, and M. Aihara. 2003. Production of CTX-M-3 extended-spectrum β-lactamase and IMP-1 metallo β-lactamase by five Gram-negative bacilli: survey of clinical isolates from seven laboratories collected in 1998 and 2000, in the Kinki region of Japan. J. Antimicrob. Chemother. 51:631-638. [DOI] [PubMed] [Google Scholar]