Abstract
tet(W) is one of the most abundant tetracycline resistance genes found in bacteria from the mammalian gut and was first identified in the rumen anaerobe Butyrivibrio fibrisolvens 1.230, where it is highly mobile and its transfer is associated with the transposable chromosomal element TnB1230. In order to compare the genetic basis for tet(W) carriage in different bacteria, we studied sequences flanking tet(W) in representatives of seven bacterial genera originating in diverse gut environments. The sequences 657 bp upstream and 43 bp downstream of tet(W) were 96 to 100% similar in all strains examined. A common open reading frame (ORF) was identified downstream of tet(W) in five different bacteria, while another conserved ORF that flanked tet(W) in B. fibrisolvens 1.230 was also present upstream of tet(W) in a human colonic Roseburia isolate and in another rumen B. fibrisolvens isolate. In one species, Bifidobacterium longum (strain F8), a novel transposase was located within the conserved 657-bp region upstream of tet(W) and was flanked by imperfect direct repeats. Additional direct repeats 6 bp long were identified on each end of a chromosomal ORF interrupted by the insertion of the putative transposase and the tet(W) gene. This tet(W) gene was transferable at low frequencies between Bifidobacterium strains. A putative minielement carrying a copy of tet(W) was identified in B. fibrisolvens transconjugants that had acquired the tet(W) gene on TnB1230. Several different mechanisms, including mechanisms involving plasmids and conjugative transposons, appear to be involved in the horizontal transfer of tet(W) genes, but small core regions that may function as minielements are conserved.
Tetracyclines are broad-spectrum antimicrobial agents that are active not only against a wide range of gram-positive and gram-negative bacteria but also against chlamydiae, mycoplasmas, rikettsiae, and protozoan parasites (8, 27, 28). Tetracyclines are widely used in both human and veterinary therapy, as prophylactics, and in many countries in animal feed as growth promoters. Resistance to tetracyclines is the most common bacterial antibiotic resistance found in nature, and tetracycline resistance (Tcr) genes are present in a variety of bacteria isolated from animal and human feces and from the environment. The majority of tetracycline resistance genes are located on mobilizable or conjugative elements, which may partially explain their wide distribution among bacterial species (8, 27).
Tetracyclines bind to the ribosome and inhibit the elongation phase of protein synthesis by interfering with the binding of the aminoacyl-tRNA to the ribosomal A site (8, 9, 27). Resistance to this antibiotic is commonly associated with tetracycline efflux proteins and ribosomal protection proteins (8, 27). In rare situations, resistance is mediated through direct inactivation of the antibiotic (34) or by 16S rRNA mutation (29). Most studies on bacterial resistance have concerned clinical pathogens, opportunistic pathogens, or antibiotic-producing bacteria. In recent years, however, there has been interest in the carriage of antibiotic resistance genes by commensal bacteria in the human and animal gut (8). In general it was observed that most bacteria causing disease carry the same tetracycline resistance genes as environmental or commensal bacteria. This supports the suggestion that environmental and commensal bacteria act as a reservoir for tetracycline and other antibiotic resistance genes found in pathogens. It is therefore very important to elucidate how antibiotic resistance genes are maintained and spread through commensal bacterial communities.
The ribosome protection-type tetracycline resistance gene tet(W) (3, 33) is one of the most widespread tetracycline resistance genes in environmental samples (1, 38). This 1.9-kb gene was originally identified in the rumen commensal anaerobe Butyrivibrio fibrisolvens, where it was present on a large mobile chromosomal element (3, 32). The partial sequence of this element, designated TnB1230, was published recently (21) (GenBank accession number AJ222769). Copies of tet(W) have also been found in other isolates of B. fibrisolvens and in isolates of Selenomonas spp., Mitsuokella spp., Clostridium spp., Roseburia spp., Bifidobacterium longum, and Megasphaera elsdenii from bovine and sheep rumens as well as porcine and human feces (3, 33, 35, 37). The conservation of the tet(W) gene sequences from different isolates is remarkably high (3, 33). The tet(W) gene was also reported to occur in isolates of the animal pathogen Arcanobacterium pyogenes, where it is also carried on a mobile genetic element (5).
The aim of the present study was to determine whether the widespread distribution of tet(W) genes can be ascribed to one or a small number of mobile genetic elements or transfer cassettes. We conclude that the genetic context of tet(W) varies widely between different bacteria, but the immediate flanking regions reveal several conserved features, suggesting that they might function as a minielement or transfer cassette.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The unrelated bacterial strains, isolated from various human and animal hosts over a period of years (33), that were used in this study are described in Table 1. All strains were routinely grown at 37°C in anaerobic M2SGC broth (24) containing tetracycline (10 μg/ml) or rifampin (100 μg/ml) (antibiotics obtained from Sigma). For conjugation experiments, M2GSC agar plates (2.5% [wt/vol] agar) supplemented with appropriate antibiotics were used. Strains were cultured in Bellco tubes under 100% CO2 or in an anaerobic cabinet Concept Plus (Ruskin Technology Limited) in an atmosphere of 10% CO2, 10% H2, and 80% N2.
TABLE 1.
Bacterial strains used in this study
| Strain(s) | Source | Drug resistance | Reference and/or provider |
|---|---|---|---|
| Bifidobacterium longum F8a | Human feces; RRIb | Tcr | 33 |
| Butyrivibrio fibrisolvens JK51a | Sheep rumen; Australia | Tcr | Keith Gregg and Jan Kopečný |
| Butyrivibrio fibrisolvens 1.230a | Bovine rumen 1; RRI | Tcr | 32 |
| Clostridium sp. strain K10a | Human feces 1; RRI | Tcr | 33 |
| Mitsuokella multiacida P208-58a | Pig feces; Japan | Tcr | 23 |
| Roseburia sp. strain A2-183a | Human feces 2; RRI | Tcr | 4 |
| Selenomonas ruminantium FB32, FB34, and FB322a | Bovine rumen 2; RRI | Tcr | 12 |
| Bifidobacterium adolescentis L2-32 | Human feces 3; RRI | Tcs | 4 |
| Bifidobacterium adolescentis L2-32Rc | Spontaneous Rifr mutant of strain L2-32 | Tcs | This work |
| Butyrivibrio fibrisolvens 2221Rc | Spontaneous Rifr mutant of type strain 2221 | Tcs | 21 |
| Mitsuokella multiacida F120Rc | Spontaneous Rifr mutant of strain A405-1 | Tcs | Jennifer Martin |
| Roseburia inulinivorans A2-194Rc | Human feces 2; RRI; spontaneous Rifr mutant | Tcs | 4; Jennifer Martin |
| Selenomonas ruminantium HD4Rc | Sheep rumen; United States; spontaneous Rifr mutant | Tcs | 19 |
| Butyrivibrio fibrisolvens Tc8 to Tc12, Tc21 | Transconjugants from matings between 1.230 and 2221R | Tcr | 32 |
| Bifidobacterium adolescentis TcKK1 to TcKK5 | Transconjugants from matings between F8 and L2-32R | Tcr | This work |
| Bifidobacterium adolescentis LMG11579d | Bovine rumen; Gent, Belgium | Tcr | Liesbeth Masco |
| Bifidobacterium animalis subsp. lactis LMG11580d | Chicken feces; Gent, Belgium | Tcr | Liesbeth Masco |
| Bifidobacterium bifidum LM588d | Probiotic product; Gent, Belgium | Tcr | Liesbeth Masco |
| Bifidobacterium pseudocatenulatum LMG11593d | Sewage; Gent, Belgium | Tcr | Liesbeth Masco |
Isolate for which flanking sequences were studied in detail in this work.
RRI, Rowett Research Institute.
Strain which was used as a recipient in mating experiments.
Only DNA isolated from the strain was used in this study.
Conjugation experiments.
Bacterial matings were based on the method of Hespell and Whitehead (16). Cells were grown overnight anaerobically in M2GSC medium (24) containing appropriate antibiotics. The following morning bacteria were subcultured into fresh medium without antibiotic selection, grown to mid-exponential phase (optical density at 600 nm of 0.4), and pelleted at 1000 × g for 15 min at 18°C. The pellet was washed twice anaerobically in 1/4 of the original volume of RGM broth lacking a carbohydrate source (RGM-C) (17) and finally resuspended in 1/10 of the original volume of RGM-C broth. The donor and recipient cultures were then mixed (1:1 ratio, assuming the same cell density) and centrifuged gently. Most of the supernatant was decanted, and the cells were resuspended in the remainder and placed on the center of the 0.2-μm-pore-size Millipore filter disc on an M2GSC agar plate. After incubation on the filter for 16 h, cells were washed in RGM-C buffer and dilutions were plated onto selective M2GSC agar plates containing the appropriate double-antibiotic selection. For matings between B. longum F8 and Butyrivibrio adolescentis L2-32R, basal M2 medium with 0.5% starch as a sole carbon source was used. Potential transconjugants grew in 2 days, and their resistance phenotype was confirmed by replating selected colonies on antibiotic plates containing tetracycline and rifampin.
DNA isolation and molecular techniques.
Total genomic DNA was isolated from overnight cultures by using the Wizard Genomic DNA purification kit (Promega, Southampton, United Kingdom). Southern blotting and nucleic acid hybridizations were performed following standard procedures (30).
Genome walking of the regions flanking tet(W) was carried out using a Universal GenomeWalker kit (BD Biosciences Clontech), following the manufacturer’s recommendations. Total genomic DNA digested with a blunt-cutting enzyme was purified and concentrated using Centri-Sep spin columns (Cambio, United Kingdom) prior to ligation to the GenomeWalker adaptor. The ligation was carried out overnight at 16°C, followed by heat inactivation (70°C for 5 min) of the ligase enzyme. The ligation mix was used as the template in a PCR. PCR products for sequencing flanking regions were obtained using BD Advantage polymerase mix (BD Biosciences Clontech). Primer AP1 and nested primer AP2 (provided in the Universal GenomeWalker kit) were used in combination with primer 5′tetW or nested primer 5′GSPtet, and with primer 3′tetW and nested primer 3′GPtet, (Table 2) to amplify regions upstream and downstream of the gene, respectively.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′→3′) |
|---|---|
| 5′tetW | AGGGCATAAAAATCCCCAGCAGTAAA |
| 5′GSPtet | CCTGTTTGTGATTGCTGTTTTTGGG |
| 3′tetW | TACCTTTCCAGGGCTTATCATGATGC |
| 3′GPtet | TACAGAGCTGAAAGGATATCAGGCCG |
| tetW_out | GGCATATAGCAGGCTCTCC |
| meth_out | GGCAATTATGGATATTACGG |
Standard PCR products for sequencing were obtained using BioTaq DNA polymerase (BIOLINE, United Kingdom), optimizing the annealing temperature and extension time according to the specific primers and the size of the expected product. PCR products were sequenced using a Taq ABI Prism kit (Perkin-Elmer, Warrington, United Kingdom) and separated on an ABI377 automated sequencer. Sequences were assembled using UWGCG software (10), which was available through the HGMP facility (Human Genome Mapping Project, Cambridge, United Kingdom).
The primer combinations tetW_out-3′GPtet and tetW_out-meth_out (Table 2) were used to amplify any circular intermediates containing tet(W).
Nucleotide sequence accession numbers.
The nucleotide sequences described in this paper have been deposited in the GenBank database with the following accession numbers: Roseburia sp. strain A2-183, AJ421625; B. fibrisolvens JK51, AJ427421; Clostridium sp. strain K10, AY601650; Mitsuokella multiacida P208-58, AY603069; Selenomonas ruminantium FB322, DQ294295; S. ruminantium FB32, DQ294296; S. ruminantium FB34, DQ294297; and B. longum F8, DQ294299.
RESULTS
Sequence conservation in tet(W) flanking regions.
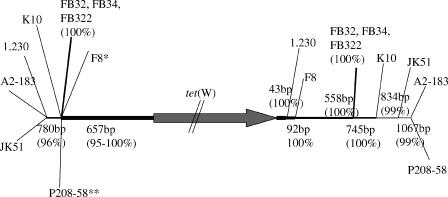
The regions flanking tet(W) in representatives from six different genera of gut commensal bacteria (Table 1) were sequenced, and these sequences were compared to the recently published partial sequence of the B. fibrisolvens 1.230 transposon TnB1230 (21) (GenBank accession number AJ222769). A 657-bp region upstream from the start codon showed 95 to 100% sequence conservation in all analyzed strains and was also present upstream of the TnB1230 tet(W) gene (21) (Fig. 1). Putative regulatory regions, including a short, 14-amino-acid (aa) open reading frame (ORF) possibly involved in transcriptional attenuation, were present in the first 330 bp upstream of tet(W) (21). Clostridium sp. strain K10 was the most divergent, with 22 to 25 randomly spaced base pair differences in the upstream region compared to other strains. In B. longum F8 an inserted sequence of 1,047 bp was present 354 bp upstream of the tet(W) start codon. The conserved upstream sequence continued to 790 bp in Roseburia sp. strain A2-183, B. fibrisolvens JK51, and B. fibrisolvens 1.230, while the three S. ruminantium strains (FB32, FB324, and FB322) were 100% identical throughout the available sequence (5.4 kb) (Fig. 1).
FIG. 1.
Diagram showing the points at which the sequences upstream and downstream of tet(W) in the nine analyzed isolates diverge. The tet(W) ORF is indicated by an arrow. The thickness of the line represents the number of species for which sequence is conserved. As the sequence diverges, the line becomes thinner. This figure is not drawn to scale. *, B. longum F8 has insertion of 1,047 bp into the conserved region upstream of tet(W). **, M. multiacida P208-58 contains an insertion of a 23-bp directly repeated sequence 291 bp upstream from the tet(W) start codon.
Downstream of tet(W) the sequences diverged more rapidly, and only 43 bp was 100% conserved between all analyzed isolates (Fig. 1). Thus, it was possible to distinguish a conserved core region of 2.6 kb, including the tet(W) gene, in all strains studied (Fig. 1).
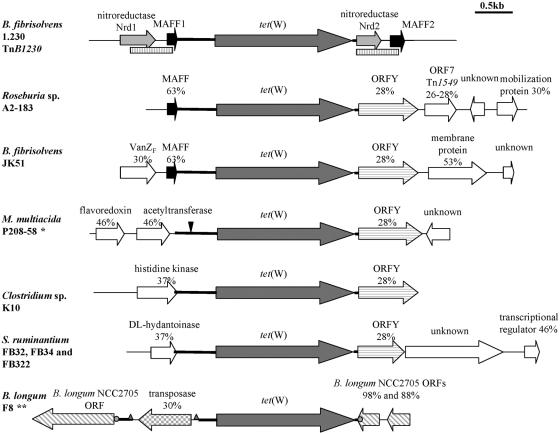
Analysis of ORFs in regions flanking tet(W).
Potential ORFs were identified in the sequences flanking tet(W) (Fig. 2). The deduced amino acid sequences were compared to those present in the nonredundant protein database at the National Center for Biotechnology Information website by using the TBLASTN and BLASTP programs.
FIG. 2.
Organization of the regions upstream and downstream of tet(W). ORFs are pattern coded and those conserved between different species are presented in the same shading. The percent identities to the closest match in the database are given above the ORFs. The rectangular boxes underneath the B. fibrisolvens diagram indicate the positions of the DRs. *, M. multiacida P208-58 contains an inserted 23-bp directly repeated sequence 291 bp upstream from the tet(W) start codon (indicated by the arrowhead). **, B. longum F8 has tandem repeats flanking the transposase inserted into the conserved core of 657 bp (represented by triangles) and duplicated 6 nucleotides flanking the insertion of the tet(W) and transposase genes (represented by circles).
(i) MAFF protein-encoding ORFs.
It was previously reported that in TnB1230, tet(W) is flanked by two identical direct-repeat (DR) DNA sequences (707 bp in length) that encode proteins with significant similarity (38% identity) to bacterial nitroreductases (Nrd) (Fig. 2) (21). An alternative start codon present upstream of DR1 potentially encodes a larger nitroreductase, while the shorter nrd2 gene within DR2 is truncated at the 5′ end.
Sequences upstream of tet(W) in Roseburia sp. strain A2-183, B. fibrisolvens JK51, and B. fibrisolvens 1.230 all contained a short ORF (here designated MAFF to represent the first four amino acids) capable of encoding a 46-aa protein (Fig. 2). This protein showed 87 to 93% amino acid sequence conservation between these three strains and also had 61 to 63% identity to a protein of unknown function found in Enterococcus faecalis V583 (GenBank accession number AE016954).
The 46-aa MAFF protein was encoded at the 3′ end of the TnB1230 DR. The MAFF ORF downstream of the nrd2 gene continued outside the DR to encode a 70-aa protein (designated MAFF2) (Fig. 2). The closest database match for this longer protein (79% amino acid sequence identity) was a previously unidentified ORF encoding a hypothetical protein of 71 aa located on the E. faecalis conjugative transposon Tn1549 (14) (GenBank accession number AF192329). In summary, the sequences upstream of tet(W) in TnB1230 encoded a full-length Nrd and a C-terminally truncated MAFF protein, while the sequences downstream encoded an N-terminally truncated Nrd and a full-length MAFF.
(ii) ORFY-type sequences.
A second conserved ORF was found immediately downstream of tet(W) in five of the analyzed species from different genera. This ORF encoded a protein of 184 aa (S. ruminantium FB32, FB34, and FB322), 246 aa (Clostridium sp. strain K10), or 255 aa (B. fibrisolvens JK51, Roseburia sp. strain A2-183, and M. multiacida P208-58), depending on the location of the stop codon (Fig. 2). This ORF was 100% conserved at the amino acid level between most of the analyzed isolates except the S. ruminantium strains, where the last 13 residues differed from those in other species. The translated product of the ORF had 28 to 30% identity to a methyltransferase protein (and contained the conserved methyltransferase domain) and to the hypothetical protein OrfY. The orfY gene is present in many mobile plasmids and transposons and is often associated with erythromycin resistance genes (6) (GenBank accession number AF516335). orfY was also found in the A. pyogenes erm(B)-like element, which does not encode tetracycline resistance (18).
(iii) Diversity of the ORFs identified in sequences flanking tet(W).
All other ORFs identified differed between the analyzed species (Fig. 2). Those further downstream of tet(W) in Roseburia sp. strain A2-183 had identities to other ORFs associated with mobile genetic elements, including one with 26 to 28% amino acid identity to ORF7 on Tn5832, Tn1549, Tn5397, and Tn916 (Fig. 2), which itself encodes a hypothetical protein.
The product of the second gene upstream of tet(W) in B. fibrisolvens JK51 had 30% amino acid identity to VanZF, a transmembrane protein from Paenibacillus popilliae that is associated with a vancomycin resistance gene cluster (25).
In S. ruminantium FB32, FB34, and FB322, tet(W) was preceded by an ORF with 37% amino acid identity to Pseudomonas sp. dl-hydantoinase, an enzyme often encoded on plasmids (39) (Fig. 2). Hybridization profiles of DNA extracted following either plasmid or chromosomal purification methods indicated that the tet(W) genes in S. ruminantium FB32 and FB34 were plasmid encoded, while that in strain FB322 was chromosomally encoded (data not shown) (2).
Transposase adjacent to tet(W) in B. longum F8.
The upstream conserved region in B. longum F8 was interrupted by an inserted sequence of 1,047 bp (Fig. 2), as noted earlier. Terminal direct repeats with a single-nucleotide deletion of 29 bp (5′-TACAATAAGGGGAAGAAAAATTTCTTTTA-3′, right arm) and 30 bp (5′-TACATATAAGGGGAAGAAAAATTTCTTTTA-3′, left arm) defined the ends of this insertion sequence (IS). The left-arm sequence repeat was present within the conserved 657-bp sequence.
This insertion sequence contained an 818-bp ORF capable of encoding a 272-aa protein with up to 30% identity to transposases found on different mobile elements, including IS1002 from Bordetella sp., the Vibrio cholerae pathogenicity island, and the Micrococcus sp. strain 28 plasmid pSD10. The C-terminal end of the F8 transposase, from residue P135, contained a conserved integrase and transposase catalytic core domain (Fig. 3), including the triad of amino acids with characteristic spacing known as the DDE motif and the associated conserved residues. The spacing between each residue in the triad was D142(X65)D208(X45)E254 (Fig. 3). Each of the amino acids in the triad together with the conserved surrounding residues created three characteristic regions, N2, N3, and C1 (Fig. 3), which are conserved motifs in transposases of the IS4 family of insertion sequences (15, 26).
FIG. 3.
Amino acid sequence of the B. longum F8 transposase. The conserved catalytic domain starts at residue P135. Amino acids creating the DDE motif are indicated in boldface, characteristic residues upstream and/or downstream of the triad are in boldface italic, and regions N2, N3, and C1 are underlined.
The sequences immediately downstream of tet(W) and immediately upstream of the transposase had 98% identity to the same ORF of unknown function from B. longum NCC2705 (GenBank accession number AE014714). Two short directly repeated sequences flanked the insertion of the transposase and tet(W) genes (Fig. 2). These short duplicated sequences were six nucleotides long, CAATGC. Thus, it appeared that a single B. longum ORF had been interrupted by the insertion of a DNA fragment carrying the putative transposase and the tet(W) gene and that duplication of the 6-bp sequence had occurred during the insertion event. The last ORF downstream of tet(W) had 88% identity to another hypothetical ORF from B. longum NCC2705. The relative orientation of these ORFs in B. longum NCC2705 and B. longum F8 is the same.
PCR-based analysis of four additional Bifidobacterium strains known to carry tet(W) (Table 1) indicated that none of them contained the transposase.
Mobility of the tet(W) genes.
The mobility of the tet(W) genes was investigated in filter mating experiments with tetracycline-susceptible (Tcs) recipient strains. We used selected isolates carrying tet(W) as donors, and in each case B. fibrisolvens 2221R and a Tcs isolate belonging to the same species as the donor strain were used as recipients. No transconjugants were obtained in matings involving Roseburia sp. strain A2-183, M. multiacida, or, S. ruminantium as the donor strain. In previous matings, transfer of tet(W) from Clostridium sp. strain K10 to B. fibrisolvens 2221R was not detected, although a second tetracycline resistance gene [tet(O/32/O)] was transferable (22, 36).
Colonies were obtained at a low frequency (<2 × 10−7 per recipient cell) in matings between B. longum F8 and B. adolescentis L2-32R. Total genomic DNA was isolated from these transconjugants, and 16S rRNA sequences confirmed that they were indeed derived from B. adolescentis L2-32R. PCR amplifications using specific primer sets confirmed that tet(W) and the upstream region including the transposase gene were present in all transconjugants analyzed. Furthermore, the same ORF was interrupted by the tet(W) insertion in B. adolescentis L2-32R transconjugants as in the donor strain (Fig. 2). This ORF was again 100% identical to the homologue in B. longum NCC2705. Attempts to transform B. adolescentis L2-32R with genomic DNA extracted from B. longum F8 were unsuccessful (results not shown).
Identification of a tet(W) minielement.
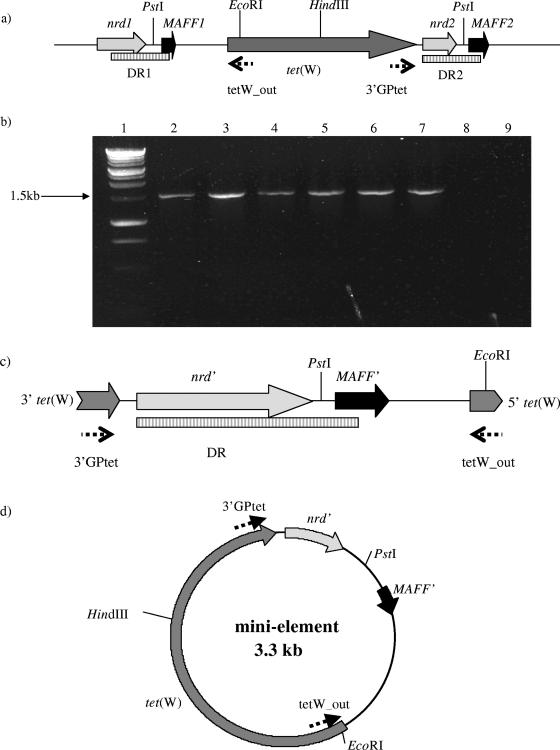
The presence of conserved sequences and of identical ORFs upstream or downstream of the tet(W) gene suggested that there might be a common element(s) involved in the spread of the gene. A PCR-based approach was used in order to detect circular forms of any possible mobile minielements. Primer pairs reading out from the ends of tet(W) (primers tetW_out and 3′GPtet) and also reading out from the 5′ end of tet(W) (tetW_out) and the 3′ end of the conserved ORF located downstream of the tet(W) gene (ORFY; primer meth_out) (Fig. 4a) were tested in all isolates as well as in B. fibrisolvens 1.230 and in six B. fibrisolvens transconjugants containing the transposon TnB1230 (Table 1) (32). A specific PCR product was obtained only when DNAs isolated from B. fibrisolvens transconjugants Tc8 to Tc12 and Tc21 were used (Fig. 4b). Sequencing of the 1.5-kb product revealed that it contained the conserved regions upstream (657 bp) and downstream (43 bp) of the tet(W) gene and truncated versions of the nrd and MAFF genes (Fig. 4c). We postulate that this may be part of a transferable circular intermediate involved in the spread of tet(W) in certain hosts (Fig. 4d). Interestingly no product was obtained for the donor strain (B. fibrisolvens 1.230) used in conjugal matings producing these transconjugants.
FIG. 4.
Evidence for the presence of the circular minielement. ORFs are represented as solid arrows, and the locations of the primers used in PCR are shown by dotted arrows. Restriction sites are indicated, and DRs are represented by hatched boxes. (a) Organization of the tet(W) gene and its flanking regions in TnB1230 from B. fibrisolvens. (b) PCR products obtained using primers tetW_out and 3′GPtet reading outwards from the 5′ and 3′ ends of tet(W), respectively. Lane 1, 1-kb ladder (Promega, United Kingdom); lanes 2 to 7, B. fibrisolvens transconjugants Tc8 to Tc12 and Tc21, respectively; lane 8, B. fibrisolvens 1.230 (donor); lane 9, B. fibrisolvens 2221R (recipient). (c) Organization of the PCR product shown in panel b following sequence analysis. MAFF′ and nrd′ are the truncated forms of the respective genes. (d) Diagram representing the circular form of the minielement carrying tet(W) identified in B. fibrisolvens transconjugants.
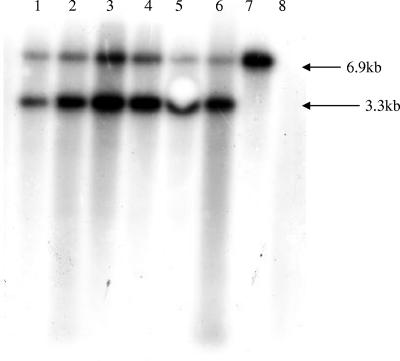
DNAs from the transconjugants and the donor and recipient strains were analyzed after restriction with EcoRI or HindIII in Southern blot hybridization with the tet(W)-specific probe. A single band of the expected size (6.9 kb when cut with EcoRI) was obtained for the donor strain B. fibrisolvens 1.230 (Fig. 5). In all the transconjugants an additional band of 3.3 kb was observed (Fig. 5), which corresponds in size to the postulated circular minielement carrying tet(W) (Fig. 4). Similarly, the HindIII digest gave hybridizing bands of the expected sizes in the donor and all transconjugants, with an additional band visible only in the transconjugants (data not shown). These results confirmed the presence of a second copy of the tet(W) gene in transconjugants Tc8 to Tc12 and Tc21, which can apparently exist as a circular intermediate.
FIG. 5.
Southern blot of EcoRI-digested chromosomal DNAs from B. fibrisolvens strains hybridized to the 1-kb tet(W)-specific probe. Lanes 1 to 6, transconjugants Tc8 to Tc12 and Tc21, respectively; lane 7, Donor strain 1.230; lane 8, recipient strain 2221R. The sizes of the bands are indicated.
DISCUSSION
Mobile genetic elements, bacteriophage-mediated transduction, and natural transformation are all recognized as potential mechanisms for gene transfer between bacteria that inhabit the complex gut communities of the rumen and large intestine in mammals. There is, however, very little information on the mechanisms responsible for natural gene transfer in the groups of predominant anaerobic gut bacteria considered here. Plasmids are known to be abundant in Butyrivibrio and Selenomonas (11, 13), but little is known about the prevalence of chromosomal conjugative elements or natural transformation in these bacteria (31-33).
Direct evidence for tet(W) transfer, in laboratory matings, has been obtained from two of the bacterial strains examined here, B. fibrisolvens 1.230 and B. longum F8. tet(W) transfer has also been detected at low frequencies between A. pyogenes strains (5). tet(W) is transferred at high frequencies from B. fibrisolvens 1.230 to other B. fibrisolvens strains, in association with a 45- to 50-kb conjugative element designated TnB1230 (3, 21, 32).
The circular minielement detected here in B. fibrisolvens 2221R transconjugants that had acquired TnB1230 was not detected in the donor strain B. fibrisolvens 1.230 or in other B. fibrisolvens strains examined. The minielement contains truncated nrd and MAFF genes, found in TnB1230, as well as the tet(W) gene itself, strongly suggesting that the DRs that flank tet(W) in TnB1230 give rise to the circular intermediate via homologous recombination. The appearance of the minielement may depend on a trans-acting factor from the recipient strain, thus explaining its absence in the donor strain. While there is no evidence that formation of the minielement is essential for tet(W) transfer between strains, it may help to explain secondary tet(W) insertions observed in the transconjugants (3).
We previously reported that proteins encoded by the DRs show significant amino acid sequence identity (38%) with bacterial nitroreductases (21). The proteins encoded by both the nrd genes also contain residues (D8D38D78) that could represent the catalytic core of an IS-associated transposase (20). Although the third core residue is more typically glutamic acid, the spacing between these acidic residues, and the key flanking amino acids, appears to be consistent with previously characterized IS transposases. Functional studies will be required to clarify the true role and origin of the DR ORFs. The small ORF (MAFF) that occurs in the B. fibrisolvens DR regions, both upstream and downstream of tet(W), was also found upstream of tet(W) in two other strains, but the significance of this is unknown.
We have also shown here that tet(W) transfers at low frequencies in laboratory matings between B. longum F8 and B. adolescentis L2-32R. The putative transposase gene inserted into the region upstream of tet(W) in B. longum F8 is assumed to be responsible for its mobilization in this strain. The catalytic core domain characteristic of IS-encoded transposases, which coordinates the divalent cations required for catalysis and excision of the minielement (15, 20), is present in the putative transposase of B. longum F8. The site for chromosomal insertion of the tet(W)/transposase element in B. adolescentis L2-32 transconjugants was identical to that in the donor strain B. longum F8, consistent with a site-specific insertion event. On the other hand, we cannot rule out the possibility of a larger mobile element or that this transfer occurred by transformation followed by homologous recombination.
No transfer of tet(W) was observed from any of the other strains tested (Roseburia sp. strain A2-183, B. fibrisolvens, M. multiacida, S. ruminantium, and Clostridium sp. strain K10). In each of these strains tet(W) was followed by a highly conserved (although truncated in the S. ruminantium strains) ORFY sequence. We hypothesize that the conserved tet(W)/ORFY core unit might constitute some form of mobile cassette, but the absence of tet(W) transfer from these strains suggests that other factors may be required for mobilization. It has been demonstrated that some bacteria that are able to act as recipients for conjugative transposons cannot themselves act as donors (7). In some cases (e.g., Roseburia sp. strain A2-183 and B. fibrisolvens JK51), sequences of the variable flanking sequences outside this core region suggest that the cassette is, or has been, embedded in a larger mobile element. In other cases (e.g., M. multiacida), the flanking regions may represent chromosomal genes.
Evidence for plasmid carriage of the tet(W) gene has been found so far only in S. ruminantium FB32 and FB34. The related strain of S. ruminantium (FB322) carried an identical tet(W)/ORFY sequence that appeared to be chromosomally located. The similarity in sequence of the flanking regions of these three strains, together with the hybridization results, implies that this plasmid, or part of it, is capable of chromosomal integration.
In conclusion, this analysis demonstrates the transfer of tet(W) by distinct mechanisms in B. fibrisolvens 1.230 and B. longum F8. The possible small transposable element in B. longum consists only of tet(W) and an adjacent transposase, whereas the transposable element in B. fibrisolvens 1.230 is about 50 kb in size. In the other strains studied here transfer was not detected in the laboratory, and flanking regions varied, but with strong conservation of a core sequence including tet(W) and ORFY. This implies the presence of a cassette in which ORFY might contribute to the acquisition of tet(W) through as-yet-unknown mechanisms. The highly abundant tet(W) gene has evidently become distributed via many different mechanisms, with one or more small core cassettes becoming incorporated into larger mobile elements that include both conjugative transposons and plasmids.
Acknowledgments
This work was supported by SEERAD (Scottish Executive Environment and Rural Affairs Department).
We thank Pauline Young and Donna Henderson for DNA sequencing and Peter Mullany and Teresa Barbosa for useful discussions and critical reading of the manuscript. Genomic DNAs from a selection of Bifidobacterium strains were kindly gifted by Liesbeth Masco.
REFERENCES
- 1.Aminov, R. I., N. Garrigues-Jeanjean, and R. I. Mackie. 2001. Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl. Environ. Microbiol. 67:22-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbosa, T. M. 1998. Tetracycline resistance transfer among obligate anaerobes from the ruminant gut. PhD thesis. University of Aberdeen, Aberdeen, United Kingdom.
- 3.Barbosa, T. M., K. P. Scott, and H. J. Flint. 1999. Evidence for recent intergeneric transfer of a new tetracycline resistance gene, tet(W), isolated from Butyrivibrio fibrisolvens, and the occurence of tet(O) in ruminal bacterial. Environ. Microbiol. 1:53-64. [DOI] [PubMed] [Google Scholar]
- 4.Barcenilla, A., S. E. Pryde, J. C. Martin, S. H. Duncan, C. S. Stewart, C. Henderson, and H. J. Flint. 2000. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl. Environ. Microbiol. 66:1654-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Billington, S. J., J. G. Songer, and B. H. Jost. 2002. Widespread distribution of a tet W determinant among tetracycline-resistant isolates of the animal pathogen Arcanobacterium pyogenes. Antimicrob. Agents Chemother. 46:1281-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boerlin, P., A. P. Burnens, J. Frey, P. Kuhnert, and J. Nicolet. 2001. Molecular epidemiology and genetic linkage of macrolide and aminoglycoside resistance in Staphylococcus intermedius of canine origin. Vet. Microbiol. 79:155-169. [DOI] [PubMed] [Google Scholar]
- 7.Bringel, F., G. L. Van Alstine, and J. R. Scott. 1991. A host factor absent from Lactococcus lactis subspecies lactis MG1363 is required for conjugative transposition. Mol. Microbiol. 5:2983-2993. [DOI] [PubMed] [Google Scholar]
- 8.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Connell, S. R., D. M. Tracz, K. H. Nierhaus, and D. E. Taylor. 2003. Ribosomal protection proteins and their mechanism of tetracycline resistance. Antimicrob. Agents Chemother. 47:3675-3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fliegerova, K., O. Benada, and H. J. Flint. 1998. Large plasmids in ruminal strains of Selenomonas ruminantium. Lett. Appl. Microbiol. 26:243-247. [DOI] [PubMed] [Google Scholar]
- 12.Flint, H. J., and J. Bisset. 1990. Genetic diversity in Selenomonas ruminantium isolated from the rumen. FEMS Microbiol. Ecol. 73:351-359. [Google Scholar]
- 13.Flint, H. J., and K. P. Scott. 2000. Genetics of rumen micro-organisms: gene transfer, genetic analysis, and strain manipulation, p. 389-408. In P. B. Cronjé (ed.), Ruminant physiology: digestion, metabolism, growth and reproduction. CAB International, Cambridge, Mass.
- 14.Garnier, F., S. Taourit, P. Glaser, P. Courvalin, and M. Galimand. 2000. Characterization of transposon Tn1549, conferring VanB-type resistance in Enterococcus spp. Microbiology 146:1481-1489. [DOI] [PubMed] [Google Scholar]
- 15.Haren, L., B. Ton-Hoang, and M. Chandler. 1999. Integrating DNA: transposases and retroviral integrases. Annu. Rev. Microbiol. 53:245-281. [DOI] [PubMed] [Google Scholar]
- 16.Hespell, R. B., and T. R. Whitehead. 1991. Conjugal transfer of Tn916, Tn916ΔE, and pAMβ1 from Enterococcus faecalis to Butyrivibrio fibrisolvens strains. Appl. Environ. Microbiol. 57:2703-2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hespell, R. B., R. Wolf, and R. J. Bothast. 1987. Fermentation of xylans by Butyrivibrio fibrisolvens and other ruminal bacteria. Appl. Environ. Microbiol. 53:2849-2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jost, B. H., H. T. Trinh, J. G. Songer, and S. J. Billington. 2004. Ribosomal mutations in Arcanobacterium pyogenes confer a unique spectrum of macrolide resistance. Antimicrob. Agents Chemother. 48:1021-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krumholz, L. R., M. P. Bryant, W. J. Brulla, J. L. Vicini, J. H. Clark, and D. A. Stahl. 1993. Proposal of Quinella ovalis gen. nov., sp. nov., based on phylogenetic analysis. Int. J. Syst. Bacteriol. 43:293-296. [DOI] [PubMed] [Google Scholar]
- 20.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melville, C. M., R. Brunel, H. J. Flint, and K. P. Scott. 2004. The Butyrivibrio fibrisolvens tet(W) gene is carried on the novel conjugative transposon TnB1230, which contains duplicated nitroreductase coding sequences. J. Bacteriol. 186:3656-3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melville, C. M., K. P. Scott, D. K. Mercer, and H. J. Flint. 2001. Novel tetracycline resistance gene, tet(32), in the Clostridium-related human colonic anaerobe K10 and its transmission in vitro to the rumen anaerobe Butyrivibrio fibrisolvens. Antimicrob. Agents Chemother. 45:3246-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitsuoka, T., A. Terada, K. Watanabe, and K. Uchida. 1974. Bacteroides multiacidus, a new species from faeces of humans and pigs. Int. J. Syst. Bacteriol. 24:35-41. [Google Scholar]
- 24.Miyazaki, K., J. C. Martin, R. Marinsek-Logar, and H. J. Flint. 1997. Degradation and utilization of xylans by the rumen anaerobe Prevotella bryantii (formerly P. ruminicola subsp brevis) B14. Anaerobe 3:373-381. [DOI] [PubMed] [Google Scholar]
- 25.Patel, R., K. Piper, F. R. Cockerill III, J. M. Steckelberg, and A. A. Yousten. 2000. The biopesticide Paenibacillus popilliae has a vancomycin resistance gene cluster homologous to the enterococcal VanA vancomycin resistance gene cluster. Antimicrob. Agents Chemother. 44:705-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rezsohazy, R., B. Hallet, J. Delcour, and J. Mahillon. 1993. The Is4 family of insertion sequences-evidence for a conserved transposase motif. Mol. Microbiol. 9:1283-1295. [DOI] [PubMed] [Google Scholar]
- 27.Roberts, M. C. 2005. Update on acquired tetracycline resistance genes. FEMS Microbiol. Lett. 245:195-203. [DOI] [PubMed] [Google Scholar]
- 28.Roberts, M. C. 2003. Tetracycline therapy: update. Clin. Infect. Dis. 36:462-467. [DOI] [PubMed] [Google Scholar]
- 29.Ross, J. I., E. A. Eady, J. H. Cove, and W. J. Cunliffe. 1998. 16S rRNA mutation associated with tetracycline resistance in a gram-positive bacterium. Antimicrob. Agents Chemother. 42:1702-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Scott, K. P. 2002. The role of conjugative transposons in spreading antibiotic resistance between bacteria that inhabit the gastrointestinal tract. Cell Mol. Life Sci. 59:2071-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scott, K. P., T. M. Barbosa, K. J. Forbes, and H. J. Flint. 1997. High-frequency transfer of a naturally occurring chromosomal tetracycline resistance element in the ruminal anaerobe Butyrivibrio fibrisolvens. Appl. Environ. Microbiol. 63:3405-3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scott, K. P., C. M. Melville, T. M. Barbosa, and H. J. Flint. 2000. Occurrence of the new tetracycline resistance gene tet(W) in bacteria from the human gut. Antimicrob. Agents Chemother. 44:775-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Speer, B. S., L. Bedzyk, and A. A. Salyers. 1991. Evidence that a novel tetracycline resistance gene found on two Bacteroides transposons encodes an NADP-requiring oxidoreductase. J. Bacteriol. 173:176-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stanton, T. B., and S. B. Humphrey. 2003. Isolation of tetracycline-resistant Megasphaera elsdenii strains with novel mosaic gene combinations of tet(O) and tet(W) from swine. Appl. Environ. Microbiol. 69:3874-3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stanton, T. B., S. B. Humphrey, K. P. Scott, and H. J. Flint. 2005. Hybrid tet genes and tet gene nomenclature: request for opinion. Antimicrob. Agents Chemother. 49:1265-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stanton, T. B., J. S. McDowall, and M. A. Rasmussen. 2004. Diverse tetracycline resistance genotypes of Megasphaera elsdenii strains selectively cultured from swine feces. Appl. Environ. Microbiol. 70:3754-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Villedieu, A., M. L. az-Torres, N. Hunt, R. McNab, D. A. Spratt, M. Wilson, and P. Mullany. 2003. Prevalence of tetracycline resistance genes in oral bacteria. Antimicrob. Agents Chemother. 47:878-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watabe, K., T. Ishikawa, Y. Mukohara, and H. Nakamura. 1992. Cloning and sequencing of the genes involved in the conversion of 5-substituted hydantoins to the corresponding l-amino acids from the native plasmid of Pseudomonas sp. strain NS671. J. Bacteriol. 174:962-969. [DOI] [PMC free article] [PubMed] [Google Scholar]