Abstract
The gene encoding resistance to β-lactam antibiotics in the staphylococci is found on the chromosome in a genomic island designated staphylococcal cassette chromosome mec, or SCCmec. In addition to the resistance gene mecA, SCCmec also contains site-specific recombinase genes that are capable of catalyzing the chromosomal excision and reintegration of SCCmec. SCCmec is found in five major isotypes partially defined by the recombinase genes present, either ccrAB or ccrC. Of these, SCCmec type IV is presumed to be mobile in the environment, and this mobility may be partially responsible for the rise in community-associated methicillin-resistant staphylococcal infections. In this study, we investigate the presumptive first step in type IV SCCmec mobility: chromosomal excision of the element. CcrAB from a panel of six Staphylococcus aureus and four Staphylococcus epidermidis strains were able to catalyze chromosomal excision of SCCmec types I and II, indicating that these proteins maintain recombinase activity despite varying by up to 3.7% at the amino acid level. Excision of type IV SCCmec was not universally seen, as a subset of S. aureus strains with type IV SCCmec did not excise their element. These strains are all highly related and represent a lineage of successful community-associated pathogens. In addition, the inability to excise SCCmec in these strains is associated with the insertion of a presumptive mobile element containing the gene for staphylococcal enterotoxin H (seh) immediately downstream of SCCmec on the chromosome. Acquisition of this mobile element, containing a known virulence gene, appears to have stabilized the chromosomal integration of the methicillin resistance gene in these strains.
Staphylococcus aureus is a common cause of human disease ranging in severity from skin and soft tissue infections to osteomyelitis, bacteremia, and infective endocarditis. Treatment of these infections is complicated by the organism's resistance to many chemotherapeutic agents. Paramount among these resistance phenotypes is methicillin-resistant Staphylococcus aureus (MRSA) (4). MRSA strains arise by the acquisition of mecA, a gene encoding an alternate penicillin binding protein (PBP2′ or PBP2a). PBP2a is capable of maintaining cell wall synthesis in the presence of β-lactam antibiotics that inhibit the cell's other penicillin binding proteins (3, 7). mecA is found on the chromosome in a genomic island designated staphylococcal cassette chromosome mec (SCCmec) (22). At least five major varieties of SCCmec exist and are defined by the specific mec locus present (composed of mecA and its two regulatory genes, mecI and mecRI) as well as the variety of site-specific recombinase genes present (either ccrAB or ccrC) (20-22, 24).
SCCmec types I to III are larger elements (34 to 67 kb), tend to contain resistance determinants in addition to mecA, and are more frequently found in strains causing infections of hospitalized patients (10, 20). In contrast, MRSA strains are now increasingly recognized as causative agents of community-associated disease in patients that are not affiliated with a hospital or healthcare environment (32). These MRSA strains generally contain the type IV SCCmec element, which is the smallest of the SCCmec elements (21 to 24 kb) and does not usually possess resistance determinants other than mecA (26, 31). SCCmec type IV is found in MRSA strains identical to those susceptible to methicillin that are prevalent in the same community. In addition, some SCCmec type IV-containing MRSA strains in different geographic areas are unrelated. These data suggest that unrelated methicillin-sensitive staphylococcal lineages have acquired this element independently to become MRSA strains (8, 12). These observations, along with its small size, have led to the idea that type IV SCCmec is mobile in the environment.
The horizontal mobility of SCCmec is presumed to involve the site-specific recombinase genes found in each element, either ccrAB or ccrC. These proteins are serine recombinases of the invertase/resolvase family and resemble the large phage integrases (16, 30). Serine recombinases are thought to function as dimers and in the case of CcrAB integrate SCCmec into the staphylococcal chromosome by binding to two core recognition sequences, or attachment sites, one found on SCCmec and the other found on the staphylococcal chromosome (attSCC and attB, respectively). attB is found on the chromosome in the end of an open reading frame of unknown function (orfX), and this is the universal site of SCCmec integration. Following SCCmec integration, two hybrid attachment sites are formed at either end of the inserted SCCmec element, known as attL and attR. It has been shown that both CcrA and CcrB are necessary and capable of catalyzing the site-specific chromosomal excision of SCCmec types I and II (20, 22). Similarly, CcrC has been shown to catalyze the site-specific excision and reintegration of SCCmec type V (21). Spontaneous chromosomal excision, presumably mediated by CcrAB, has also been detected for SCCmec type III (20).
There is considerable sequence variation among the ccr genes of the same type. For example, the ccrAB genes found in SCCmec types II and IV can vary up to 5% at the nucleotide level (18). This is in contrast to mecA, which is known to be highly conserved (99 to 100% nucleotide identity) (17). It is possible that some of the mutations in CcrAB cause a loss of recombinase function, thereby stabilizing SCCmec in the chromosome. In this study, we investigate the sequence diversity and recombinase ability of CcrAB from a panel of strains containing SCCmec type IV and test for site-specific excision of the SCCmec type IV element.
MATERIALS AND METHODS
Bacterial strains and media.
All bacterial strains are listed in Table 1. Strains N315, COL, and MW2 were obtained from the Network on Antimicrobial Resistance in Staphylococcus aureus; strains C98-370 and C99-529 were kindly provided by Paul Fey at the University of Nebraska Medical Center, Omaha, NE; and strains J28, J35, J39, and J52 were kindly provided by Henry Chambers at the San Francisco General Hospital, San Francisco, CA.
TABLE 1.
Bacterial strains
| Strain | Species | Sequence typea | SCCmec type | Descriptionb | Reference(s) |
|---|---|---|---|---|---|
| MW2 | S. aureus | 1 | IV | CA-MRSA | 1, 6, 19 |
| N315 | S. aureus | 5 | II | HA-MRSA | 23 |
| COL | S. aureus | 250 | I | HA-MRSA | 14 |
| C98-370 | S. aureus | 1 | IV | CA-MRSA | 6, 13 |
| C99-529 | S. aureus | 1 | IV | CA-MRSA | 6, 13 |
| J28 | S. aureus | 1 | IV | CA-MRSA | 8 |
| J35 | S. aureus | 12 | IV | CA-MRSA | 8 |
| J39 | S. aureus | 8 | IV | CA-MRSA | 8 |
| J52 | S. aureus | 59 | IV | CA-MRSA | 8 |
| SE5 | S. epidermidis | IV | PVE MRSE | 31 | |
| SE7 | S. epidermidis | IV | PVE MRSE | 31 | |
| SE50 | S. epidermidis | IV | PVE MRSE | 31 | |
| SE63 | S. epidermidis | IV | PVE MRSE | 31 |
Based on multilocus sequence typing.
CA-MRSA, community-associated MRSA; HA-MRSA, hospital-associated MRSA; PVE MRSE, methicillin-resistant S. epidermidis isolated from prosthetic valve endocarditis infections.
Bacteria were grown in brain heart infusion (BHI) broth or agar (Becton Dickinson, Sparks, MD) at 37°C with shaking at 220 rpm. When selection was necessary, ampicillin (100 μg/ml), chloramphenicol (10 μg/ml), or oxacillin (6 μg/ml) was added to the culture medium (Sigma-Aldrich, St. Louis, MO).
ccrAB cloning.
ccrAB was PCR amplified from the genomic DNA of S. aureus strains N315, MW2, C98-370, C99-529, J28, J35, and J52 and Staphylococcus epidermidis strains SE5, SE7, SE50, and SE63 with primers ccuprev and ccdwnfor using the Taq PCR Master Mix kit (QIAGEN, Valencia CA.). PCR products were purified using the QIAquick gel extraction kit, ligated into the ligase-independent cloning site of the PCR 2.1-TOPO vector (Invitrogen, Carlsbad, CA), and transformed into chemically competent Top10 Escherichia coli (Invitrogen, Carlsbad, CA). A staphylococcal origin of replication was introduced by cloning plasmid pRN5543 into the unique XbaI site on PCR 2.1-TOPO, and the constructs were moved into S. aureus RN4220 by electroporation (9). Phage 80α was used to transduce the constructs from RN4220 into all other S. aureus strains used in this study.
Sequence determination and alignments.
ccrAB sequencing was performed on overlapping 600- to 700-bp PCR fragments generated using the sequencing primers listed in Table 2. All sequencing was performed by the Nucleic Acid Research Facility at Virginia Commonwealth University (Richmond, VA). CcrAB open reading frames were translated using Vector NTI software (InforMax, Frederick, MD). Amino acid alignments were generated using ClustalX, and CloureD software was used to generate the dot alignments.
TABLE 2.
Primers used
| Primer | Sequence (5′-3′) | Description | Reference or source |
|---|---|---|---|
| ccdwnfor | CGAGATATTAGCCGATTTGG | ccrAB cloning; amplifies ccrAB and upstream region | This study |
| ccuprev | CCTTCTGTTTCTTCGAATCTGC | ||
| cc1for | CGTACCATGTTCATATCTTAAGC | ccrAB sequencing | This study |
| cc2for | GCAACACTTACAAATATGACC | ||
| cc3for | CGTGTCGGTATCTATGTACG | ||
| cc4for | GCTATGTCACTAAAAAGGGTAAACC | ||
| cc5for | GGACTTAACATCAGTAATCAGACC | ||
| cc1rev | CGTAATGTCATTGAGTTGC | ||
| cc2rev | CGATATAGAATGGGTTAGC | ||
| cc3rev | CGATACTATAACCTTCTGTGC | ||
| cc4rev | GCATTCATGTTTTTAGGACAGACG | ||
| cc5rev | GAGTCGGTCGAAAGCTTGATCC | ||
| cL1 | ATTTAATGTCCACCATTTAACA | Excised chromosomal junction in N315 | 22 |
| cR1 | AAGAATTGAACCAACGCATGA | ||
| mL1 | GAATCTTCAGCATGTGATTTA | Circular SCCmec in N315 | 22 |
| mR8 | ATGAAAGACTGCGGAGGCTAACT | ||
| cL1 | ATTTAATGTCCACCATTTAACA | Excised chromosomal junction in COL | 20 |
| cR2 | AAACGACATGAAAATCACCAT | ||
| co1A | GTTCCCAGTAGCAACCTTCC | Circular SCCmec in COL | This study |
| co1B | CAATGAAAGCTTGGAAGAAGGGC | ||
| jxn1 | ATAGACATCATAGAAGTTACAGACGA | Excised chromosomal junction in MW2, C98-370, C99-529, J28, and MSSA476 | This study |
| jxn2 | ATAGGCAACAATACATGGCAACTCAGA | ||
| mw2A | GTCTATCATTTGAAATTCCCTCC | Circular SCCmec in MW2, C98-370, C99-529, and J28 | This study |
| mw2B | GATAGACAACTTTAAACAGGTCC | ||
| mR8 | ATGAAAGACTGCGGAGGCTAACT | Circular SCCmec in J35 and J52 | 22 |
| mwattRfor | GATCCTCGAGCATCCTCCACGTTATGGAGGTGC | This study | |
| Mw2B | GATAGACAACTTTAAACAGGTCC | Detects MW0048 | This study |
| Imw2R | GCTTTTAATAGAGAACAACCAATGG | ||
| Trafor | GGTACTTCTTGGTATTTTTAGGAC | Detects region from putative transposase to seh | This study |
| sehrev | CTGCTTTCGCATATGATGTG | ||
| I1-F | GTTCCAGACGAAAAAGCACCAG | Binds outside of either end of SCCmec in numerous strains | This study |
| I1-R | CATTTTATGAGTCTCGCAAATTGTCAG |
PCR-based SCCmec excision.
Briefly, ccrAB constructs were transduced into the appropriate S. aureus strain. BHI broth cultures were started from isolated colonies on the transductant plates and grown overnight. The presence of the construct was confirmed by restriction analysis of plasmid DNA (prepared using the QIAprep Spin Miniprep kit), and total genomic DNA was isolated using the QIAamp DNA Mini kit (QIAGEN, Valencia, CA) to serve as a template for PCRs. Detection of the SCCmec excision from the chromosome was done using primer sets designed to amplify across the chromosomal junction and a second set designed to detect the excised, circular SCCmec element. Primer sets used were strain specific and are listed in Table 2. All PCR primers were synthesized by the Nucleic Acid Research Facility at Virginia Commonwealth University. PCR was performed using the Taq PCR Master Mix kit; annealing temperature was generally 50°C, and extensions were done at 72°C for 1.5 min/kb of amplicons. All PCR products were confirmed by nucleotide sequence determination.
Nucleotide sequence accession numbers.
ccrAB sequences have been submitted to GenBank, and the accession numbers are as follows: DQ514327 for S. aureus C98-370, DQ514328 for S. aureus C99-529, DQ514329 for S. aureus J28, DQ514330 for S. aureus J35, DQ514331 for S. aureus J52, DQ514332 for S. epidermidis SE5, DQ514333 for S. epidermidis SE7, DQ514334 for S. epidermidis SE50, and DQ514335 for S. epidermidis SE63.
RESULTS
CcrAB sequence.
The nucleotide sequences of ccrA and ccrB from nine strains carrying type IV SCCmec were determined, and their amino acid sequences were aligned with that of strain N315 (see Fig. S1 in the supplemental material). N315 was used in these comparisons because it harbors SCCmec type II, and CcrAB from N315 are known to catalyze SCCmec excision (22). CcrA from the type IV strains varied from 0.7 to 5.1% from the amino acid sequence of N315. The S. aureus strain MW2 CcrA sequence was most divergent from N315. S. aureus strains C98-370, C99-529, J35, and J52 all shared identical CcrA sequences. When the CcrB amino acid sequences of these strains were compared to that of N315, less variability was seen. S. epidermidis strain SE50 showed the greatest divergence from N315 (3.1%). The CcrB sequence of strain J28 was identical to that of N315. C98-370, C99-529, and J35 also have identical CcrB sequences (CcrA and CcrB amino acid sequence alignments are included in the supplemental material).
SCCmec excision.
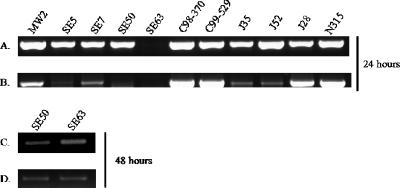
ccrAB contained on a multicopy plasmid has been shown to lead to the excision of SCCmec types I and II by using a PCR-based method of detecting SCCmec excision. This PCR-based approach amplifies the chromosomal junction (attB) where SCCmec has been excised, as has the circularized, extrachromosomal SCCmec (attSCC) (20, 22). In a similar manner, we investigated the recombinase ability of ccrAB from type IV SCCmec. ccrAB from 10 strains containing type IV SCCmec (six S. aureus and four S. epidermidis strains) were cloned onto multicopy plasmids and moved into S. aureus strains N315 (SCCmec type II) and COL (SCCmec type I). The bacteria were grown overnight, and total cellular DNA was extracted. Figure 1 (A and B) shows the results of PCR-based detection of the SCCmec excision in strain N315. An amplicon corresponding to the chromosomal junction was detected when ccrAB from 9 of the 10 type IV strains was present, and an amplicon corresponding to the excised, circular form of SCCmec was detected when ccrAB from 8 of the 10 type IV strains was present. ccrAB from strain SE50 did not lead to excision in N315, while CcrAB from SE63 led to the amplification of the chromosomal junction but no circular form. However, following 48 h of growth, ccrAB from SE50 and SE63 yielded excision in N315 (Fig. 1C and D). Figure 2 (A and B) shows the results of PCR-based detection of SCCmec excision in strain COL. ccrB in the COL SCCmec has a premature stop codon, which is believed to produce a nonfunctional recombinase; therefore, all excision is presumably due to ccrAB contained in trans. All ccrAB genes from type IV SCCmec were capable of catalyzing the excision of SCCmec in COL, except for ccrAB from the S. epidermidis strain SE63. Again, after 48 h, SCCmec was excised by SE63 ccrAB (Fig. 2C and D). Based on these results, we conclude that the ccrAB genes contained in the type IV SCCmec element encode functional proteins capable of the site-specific excision of SCCmec despite the differences in amino acid sequences.
FIG. 1.
PCR-based excision in strain N315. (A) PCR-based detection of the chromosomal junction when SCCmec is excised (attB) using primers cL1 and cR1 (22). (B) PCR-based detection of the excised circular form of SCCmec (attSCC) using primers mR8 and mL1 (22). These PCRs were performed using genomic DNA obtained after strains containing ccrAB constructs were grown for 24 h as a template. (C) PCR-based detection of the chromosomal junction. (D) PCR-based detection of the excised circular form of SCCmec using genomic DNA isolated after the strains containing ccrAB constructs were grown for 48 h. Labels represent the strains from which ccrAB originated.
FIG. 2.
PCR-based excision in strain COL. (A) PCR-based detection of the chromosomal junction when SCCmec is excised (attB) using primers cL1 and cR2 (22). (B) PCR-based detection of the excised circular form of SCCmec (attSCC) using primers colA and colB. These PCRs were performed using genomic DNA obtained after strains containing ccrAB constructs were grown for 24 h as a template. (C) PCR-based detection of the chromosomal junction. (D) PCR-based detection of the excised circular form of SCCmec using genomic DNA isolated after the strains containing ccrAB constructs were grown for 48 h. Labels represent the strains from which ccrAB originated.
Excision of SCCmec type IV in strain MW2.
We next sought to test the ability of these ccrAB constructs to cause the excision of type IV SCCmec in S. aureus strain MW2. Primers were designed to detect the excision of type IV SCCmec in a fashion analogous to that employed for the detection of SCCmec types I and II described above (Jxn1 and Jxn2 detect the chromosomal junction, while mw2A and mw2B detect circular, excised SCCmec). All 10 ccrAB constructs were moved into strain MW2, and PCR-based detection of SCCmec was performed. Using this approach, excision of SCCmec type IV was not detected in MW2 containing any of the ccrAB constructs (data not shown). Based on these results, we hypothesize that S. aureus strain MW2 is somehow defective in SCCmec excision.
Structure of attL and upstream sequence in MW2.
Since we were unable to detect chromosomal excision of SCCmec type IV in MW2 under conditions known to catalyze the excision of other SCCmec elements, we sought to determine if there was any abnormality with the left or right attachment site of this type IV SCCmec. These attachment sites are composed of a 15-bp direct repeat as well as degenerated inverted repeats (1, 20-22). By examining the genome sequence of strain MW2 (GenBank accession number NC_003923) and comparing it to those of N315 and COL (accession numbers NC_002745 and NC_002951, respectively), we found that the structural compositions of the left and right attachment sites were very similar and therefore not a likely explanation for the lack of SCCmec excision in MW2 (1, 14, 23). However, an open reading frame, designated MW0048 on the NCBI website, is found immediately preceding attL in MW2. This open reading frame is not found in this position in either N315 or COL. An open reading frame with 97% nucleotide identity is also found within SCCmec, 326 bp upstream of attR in MW2, N315, and COL (designated MW0025 on the NCBI website). Figure 3 diagrams the structure of the SCCmec elements, including the region upstream of attL, in strains N315 and MW2 and the non-mecA-containing SCC element found in MSSA476. The region between attR and MW0025 in MW2 contains another copy of the 15-bp direct repeat found in the SCCmec attachment sites. This direct repeat is also found in N315. The role of the duplicated direct repeat is not clear, but the presence of this direct repeat preceded by MW0025 is highly reminiscent of the structure found at the left end of SCCmec in MW2, where attL is preceded by MW0048. It is clear from repeated sequencing of the SCCmec-excised chromosomal junction in N315 that excision takes place at the direct repeat in attR and not at the direct repeat found between attR and MW0025 (data not shown). We therefore feel that the presence of MW0048 preceding attL in MW2 somehow prevents CcrAB-mediated recombination at this site.
FIG. 3.
Comparison of SCC476 found in S. aureus strain MSSA476, SCCmec type IV found in S. aureus strain MW2, and SCCmec type II found in S. aureus strain N315. The diagram is based on the sequenced genomes of each strain (GenBank accession numbers NC_002953, NC_003923, and NC_002745, respectively) (1, 19, 23). SCC476 from strain MSSA476 contains genes encoding a type I restriction/modification system and a fusidic acid resistance cassette. MW0025 and MW0048 are duplicated genes, as annotated on the MW2 genome, and the MW2 designations are used for the homologues in MSSA476 and N315. Flanking the left SCCmec boundary in MW2 is a gene cluster containing staphylococcal enterotoxin H (seh), a truncated staphylococcal enterotoxin O (Δseo), and a putative transposase gene. This gene cluster is also found flanking the left boundary of SCC476. attL, left SCC attachment site; attR, right SCC attachment site; orf, open reading frame.
Structure and excision of type IV SCCmec from other S. aureus strains.
We next investigated whether the findings in MW2 were also true for other S. aureus strains harboring SCCmec type IV. SCCmec excision could not be demonstrated in strains C98-370, C99-529, and J28 by overexpressing ccrAB (data not shown) but could be shown for J35 and J52. Figure 4 shows amplification of the circular SCCmec element in J35 and J52 as well as amplification of attR. However, we could not detect the SCCmec-excised chromosomal junction or attL because our primer sets did not bind to the unknown sequence downstream of SCCmec in these strains. attB, corresponding to the SCCmec excised chromosomal junction, was amplified from strain J39, indicating SCCmec excision in this strain (data not shown). A 2,048-bp region extending from MW0048 to the downstream putative transposase, with a sequence identical to those found in MW2 and MSSA476, could be PCR amplified in strains C98-370, C99-529, and J28 (Fig. 5A). Preceding the putative transposase in MW2 and MSSA476 are seh and a truncated seo, which may comprise what was once a mobile genetic element. Figure 5B shows amplification of a 725-bp region from the putative transposase to seh in all strains containing MW0048. Therefore, all of the strains in which SCCmec excision was not detected contain a structure flanking the left boundary of SCCmec that is very similar to that of MW2. Those strains lacking this structure are capable of SCCmec excision.
FIG. 4.
Detection of attB, attSCC, attL, and attR in strains J35 and J52. Attempts to detect the SCCmec excised chromosomal junction (attB) using primers I1-R and I1-F, the excised circular SCCmec element (attSCC) using primers mR8 and MW2attRfor, the right chromosomal SCCmec junction (attR) using primers I1-R and MW2attRfor, and the left chromosomal SCCmec junction (attL) using primers I1-F and mR8 were made.
FIG. 5.
Detection of MW0048, a putative transposase, and seh using PCR. (A) PCR-based detection of a 2,048-bp region from MW0048 to the downstream putative transposase open reading frame using primers Imw2R and mw2B. (B) PCR-based detection of a 725-bp region from the putative transposase open reading frame to seh using primers trafor and sehrev. PCR was performed on total genomic DNA from the strains listed above the figure.
Phenotypic and genotypic confirmation of SCCmec excision.
To confirm the PCR-based SCCmec excision results described above, we passaged each of these strains containing ccrAB in multiple copies for 7 days. We then plated the cultures and picked isolated colonies. These colonies were used to inoculate cultures grown overnight from which 100 μl of each culture was plated onto BHI agar containing 6 μg/ml of oxacillin, and growth was assessed after 72 h. Also, total genomic DNA was isolated from each of these cultures and used as a template in PCRs designed to detect mecA. In this manner, N315, J35, and J52 produced oxacillin-sensitive colonies that were negative for mecA by PCR, indicating that these strains had excised and subsequently lost SCCmec during passage. In contrast, MW2, C98-370, C99-529, and J28 maintained oxacillin resistance and the mecA-positive genotype after passage, indicating that these strains did not excise SCCmec (data not shown). These data are in agreement with data from the PCR-based means of detecting SCCmec excision described above.
DISCUSSION
The variability seen in CcrAB from this collection of strains indicates that ccrAB is much less conserved than the other key component of SCCmec, mecA. PBP2a proteins from S. aureus, S. epidermidis, and S. sciuri differ by only one or two amino acids, whereas the CcrAB sequences described here can vary by 5% within the same species (17). We hypothesized that the changes in ccrAB may result in a loss of recombinase function and may therefore be advantageous to the organism by stabilizing the methicillin resistance gene in the chromosome. However, this was not the case, since all of the ccrAB genes investigated in this study were capable of excising SCCmec.
We have identified a different mechanism by which S. aureus has stabilized the chromosomal integration of SCCmec. Several S. aureus isolates, representing a successful community-associated clonal type (sequence type 1 [ST-1]), have a structure preceding the left SCCmec attachment site that prevents CcrAB-catalyzed excision of the element. These results are not likely due to problems with PCR primers because the primers used are capable of amplifying the left and right chromosomal SCCmec junctions in each of these strains (data not shown), and the PCR-based results were confirmed by phenotypic and genotypic detection of mecA following ccrAB overexpression.
The stable integration of SCCmec in these strains may have implications for the presumed mobility of type IV SCCmec. Multilocus sequence typing has shown that SCCmec type IV is found in several unrelated S. aureus clonal types, suggesting horizontal movement of the element (11, 12, 27). Because of its small size and its presence in distantly related S. aureus clonal types, type IV SCCmec is thought to be a highly promiscuous genetic element. It is also thought that this mobility is at least partially responsible for the increasing recognition of MRSA in the community. It is interesting that all of the S. aureus strains that are deficient in SCCmec excision belong to sequence type 1. If ccrAB-mediated SCCmec excision is truly necessary for the subsequent horizontal mobility of the methicillin resistance gene, it is very unlikely that strains from ST-1 are contributing to the spread of methicillin resistance. ST-8 is emerging as the dominant MRSA type in the community, replacing ST-1. The sole representative of ST-8 in this study, strain J39, was capable of SCCmec excision, and so SCCmec mobility may play a role in the success of this lineage.
The genome sequences of MW2 and MSSA476 (GenBank accession numbers NC_002953 and NC_003923, respectively) reveal that downstream of MW0048 (and its MSSA476 homologue) is a putative transposase followed by staphylococcal enterotoxin (SE) H and a truncated enterotoxin O (seh and seo, respectively). SEs are a family of heat-stable enterotoxins that are responsible for staphylococcal food poisoning. In addition to their gastrointestinal effects, SEs are superantigen toxins capable of causing toxic shock-like diseases (2, 25). SEs are almost exclusively found on mobile genetic elements including plasmids, prophages, and pathogenicity islands (25). The gene cluster located downstream of SCCmec may in itself represent a once-mobile element whose insertion has disrupted the mobility of SCCmec. We have shown here that strains C98-370, C99-529, and J28 contain MW0048 but that J35, J39, and J52 do not. We have also amplified the region from the putative transposase to seh in each of these strains, confirming that all of the strains that do not excise SCCmec have this entire gene cluster located downstream. This suggests that the acquisition of an additional virulence factor has resulted in the maintenance of the methicillin-resistant phenotype.
These data strongly suggest that a DNA sequence outside of the core recognition regions (att sites) is necessary for proper CcrAB-mediated recombination. This observation is not without precedent. The serine recombinases Tn3 resolvase and Sin are both dependent upon accessory DNA binding sites that lie outside of the core recognition sequence where strand exchange occurs. Two subunits of Tn3 resolvase bind to the core recognition site, but catalysis is dependent upon the binding of another four subunits to accessory binding sites located nearly 100 bp outside of the strand exchange site (5, 15). In the case of Sin recombinase, four subunits of Sin bind to the core recognition sequence, but catalysis is dependent upon the binding of the nonspecific DNA-bending protein Hbsu to a site 70 bp outside of the strand cleavage site (28, 29). Although the strand exchange site for CcrAB-mediated recombination has been inferred by the results of SCCmec excision and chromosomal integration, the precise DNA elements necessary for this process are not yet known. It is clear from this study that CcrAB-mediated strand exchange is dependent upon a sequence outside of the core recognition sites and that this sequence is absent or not functional in MRSA strains belonging to sequence type 1.
Supplementary Material
Acknowledgments
This work was supported by grant 5R01AI35705-13 from the National Institute of Allergy and Infectious Diseases.
We thank Paige Fox, Bill Craig, Qixun Zhao, Adriana Rosato, and Alastair Monk for their technical assistance and input.
Footnotes
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. [DOI] [PubMed] [Google Scholar]
- 2.Balaban, N., and A. Rasooly. 2000. Staphylococcal enterotoxins. Int. J. Food Microbiol. 61:1-10. [DOI] [PubMed] [Google Scholar]
- 3.Berger-Bachi, B. 1999. Genetic basis of methicillin resistance in Staphylococcus aureus. Cell. Mol. Life Sci. 56:764-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger-Bachi, B. 1997. Resistance not mediated by beta-lactamase (methicillin resistance), p. 158-174. In K. B. Crossley and G. L. Archer (ed.), The staphylococci in human disease. Churchill Livinstone Inc., New York, N.Y.
- 5.Burke, M. E., P. H. Arnold, J. He, S. V. Wenwieser, S. J. Rowland, M. R. Boocock, and W. M. Stark. 2004. Activating mutations of Tn3 resolvase marking interfaces important in recombination catalysis and its regulation. Mol. Microbiol. 51:937-948. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 1999. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus—Minnesota and North Dakota, 1997-1999. Morb. Mortal. Wkly. Rep. 48:707-710. [PubMed] [Google Scholar]
- 7.Chambers, H. F. 1997. Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin. Microbiol. Rev. 10:781-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charlebois, E. D., F. Perdreau-Remington, B. Kreiswirth, D. R. Bangsberg, D. Ciccarone, B. A. Diep, V. L. Ng, K. Chansky, B. R. Edlin, and H. F. Chambers. 2004. Origins of community strains of methicillin-resistant Staphylococcus aureus. Clin. Infect. Dis. 39:47-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Climo, M. W., V. K. Sharma, and G. L. Archer. 1996. Identification and characterization of the origin of conjugative transfer (oriT) and a gene (nes) encoding a single-stranded endonuclease on the staphylococcal plasmid pGO1. J. Bacteriol. 178:4975-4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diekema, D. J., M. A. Pfaller, F. J. Schmitz, J. Smayevsky, J. Bell, R. N. Jones, and M. Beach. 2001. Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997-1999. Clin. Infect. Dis. 32(Suppl. 2):S114-S132. [DOI] [PubMed] [Google Scholar]
- 11.Enright, M. C. 2003. The evolution of a resistant pathogen—the case of MRSA. Curr. Opin. Pharmacol. 3:474-479. [DOI] [PubMed] [Google Scholar]
- 12.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fey, P. D., B. Said-Salim, M. E. Rupp, S. H. Hinrichs, D. J. Boxrud, C. C. Davis, B. N. Kreiswirth, and P. M. Schlievert. 2003. Comparative molecular analysis of community- or hospital-acquired methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 47:196-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gill, S. R., D. E. Fouts, G. L. Archer, E. F. Mongodin, R. T. Deboy, J. Ravel, I. T. Paulsen, J. F. Kolonay, L. Brinkac, M. Beanan, R. J. Dodson, S. C. Daugherty, R. Madupu, S. V. Angiuoli, A. S. Durkin, D. H. Haft, J. Vamathevan, H. Khouri, T. Utterback, C. Lee, G. Dimitrov, L. Jiang, H. Qin, J. Weidman, K. Tran, K. Kang, I. R. Hance, K. E. Nelson, and C. M. Fraser. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187:2426-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grindley, N. D. 1993. Analysis of a nucleoprotein complex: the synaptosome of gamma delta resolvase. Science 262:738-740. [DOI] [PubMed] [Google Scholar]
- 16.Groth, A. C., and M. P. Calos. 2004. Phage integrases: biology and applications. J. Mol. Biol. 335:667-678. [DOI] [PubMed] [Google Scholar]
- 17.Gurtler, V., and B. C. Mayall. 2001. Genetic transfer and genome evolution in MRSA. Microbiology 147:3195-3197. [DOI] [PubMed] [Google Scholar]
- 18.Hanssen, A. M., G. Kjeldsen, and J. U. Sollid. 2004. Local variants of staphylococcal cassette chromosome mec in sporadic methicillin-resistant Staphylococcus aureus and methicillin-resistant coagulase-negative staphylococci: evidence of horizontal gene transfer? Antimicrob. Agents Chemother. 48:285-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holden, M. T., E. J. Feil, J. A. Lindsay, S. J. Peacock, N. P. Day, M. C. Enright, T. J. Foster, C. E. Moore, L. Hurst, R. Atkin, A. Barron, N. Bason, S. D. Bentley, C. Chillingworth, T. Chillingworth, C. Churcher, L. Clark, C. Corton, A. Cronin, J. Doggett, L. Dowd, T. Feltwell, Z. Hance, B. Harris, H. Hauser, S. Holroyd, K. Jagels, K. D. James, N. Lennard, A. Line, R. Mayes, S. Moule, K. Mungall, D. Ormond, M. A. Quail, E. Rabbinowitsch, K. Rutherford, M. Sanders, S. Sharp, M. Simmonds, K. Stevens, S. Whitehead, B. G. Barrell, B. G. Spratt, and J. Parkhill. 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. USA 101:9786-9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito, T., X. X. Ma, F. Takeuchi, K. Okuma, H. Yuzawa, and K. Hiramatsu. 2004. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob. Agents Chemother. 48:2637-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katayama, Y., T. Ito, and K. Hiramatsu. 2000. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 44:1549-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 24.Ma, X. X., T. Ito, C. Tiensasitorn, M. Jamklang, P. Chongtrakool, S. Boyle-Vavra, R. S. Daum, and K. Hiramatsu. 2002. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 46:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Novick, R. P. 2003. Mobile genetic elements and bacterial toxinoses: the superantigen-encoding pathogenicity islands of Staphylococcus aureus. Plasmid 49:93-105. [DOI] [PubMed] [Google Scholar]
- 26.Okuma, K., K. Iwakawa, J. D. Turnidge, W. B. Grubb, J. M. Bell, F. G. O'Brien, G. W. Coombs, J. W. Pearman, F. C. Tenover, M. Kapi, C. Tiensasitorn, T. Ito, and K. Hiramatsu. 2002. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 40:4289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson, D. A., and M. C. Enright. 2003. Evolutionary models of the emergence of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 47:3926-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rowland, S. J., M. R. Boocock, and W. M. Stark. 2005. Regulation of Sin recombinase by accessory proteins. Mol. Microbiol. 56:371-382. [DOI] [PubMed] [Google Scholar]
- 29.Rowland, S. J., W. M. Stark, and M. R. Boocock. 2002. Sin recombinase from Staphylococcus aureus: synaptic complex architecture and transposon targeting. Mol. Microbiol. 44:607-619. [DOI] [PubMed] [Google Scholar]
- 30.Smith, M. C., and H. M. Thorpe. 2002. Diversity in the serine recombinases. Mol. Microbiol. 44:299-307. [DOI] [PubMed] [Google Scholar]
- 31.Wisplinghoff, H., A. E. Rosato, M. C. Enright, M. Noto, W. Craig, and G. L. Archer. 2003. Related clones containing SCCmec type IV predominate among clinically significant Staphylococcus epidermidis isolates. Antimicrob. Agents Chemother. 47:3574-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zetola, N., J. S. Francis, E. L. Nuermberger, and W. R. Bishai. 2005. Community-acquired methicillin-resistant Staphylococcus aureus: an emerging threat. Lancet Infect. Dis. 5:275-286. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.