Abstract
β-Lactam antibiotics are extremely effective in disrupting the synthesis of the bacterial cell wall in both gram-positive and gram-negative bacteria. However, they are ineffective against Mycobacterium tuberculosis, due to the production of a β-lactamase enzyme encoded on the chromosome of M. tuberculosis that degrades these antibiotics. Indeed, recent studies have demonstrated that deletion of the blaC gene, the only gene encoding a β-lactamase in M. tuberculosis, or inhibition of the encoded enzyme resulted in significantly increased sensitivity to β-lactam antibiotics. In this paper we present a biochemical and structural characterization of M. tuberculosis BlaC. Recombinant BlaC shows a broad range of specificity with almost equal penicillinase and cepholothinase activity. While clavulanate is a mechanism-based inhibitor to class A β-lactamase with high potency (typically Ki < 0.1 μM), it is a relatively poor inhibitor of the M. tuberculosis BlaC (Ki = 2.4 μM). The crystal structure of the enzyme, determined at a resolution of 1.7 Å, shows that the overall fold of the M. tuberculosis enzyme is similar to other class A β-lactamases. There are, however, several distinct features of the active site, such as the amino acid substitutions N132G, R164A, R244A, and R276E, that explain the broad specificity of the enzyme, relatively low penicillinase activity, and resistance to clavulanate.
Penicillin G was the first β-lactam antibiotic discovered, and it was used extensively to treat bacterial infections from the 1940s to 1960s (5). Over the past 3 decades, many other potent β-lactam antibiotics, such as cephems, monobactams, and carbapenems, have been developed and are currently being utilized to treat a wide range of bacterial infections. In fact, the high efficacy, high specificity, and low rate of adverse reactions make β-lactam antibiotics the most prescribed antimicrobial agents worldwide (12). All β-lactam antibiotics show structural mimicry to the natural substrates of the bacterial dd-transpeptidases. dd-Transpeptidases, members of the penicillin-binding protein family, catalyze the cross-linking of two peptidoglycan strands, which is an essential step in bacterial cell wall biosynthesis. Inhibition of dd-transpeptidases by β-lactam antibiotics leads to disruption of the synthesis of the bacteria's cell wall (13).
A significant increase in the number of β-lactam-resistant strains of pathogenic bacteria has been reported over the past few years (35). Resistance to the β-lactams primarily occurs through the horizontal transfer of β-lactamase genes contained on plasmids (9). β-Lactamases inactivate β-lactam antibiotics by efficiently hydrolyzing the amide group of the β-lactam ring, which is the prevalent mechanism of bacterial resistance to β-lactam antibiotics (13). β-Lactamases are grouped into four classes, A, B, C, and D, based on their amino acid sequence homology and general enzyme catalytic properties (for a review of β-lactamases, see reference 13). Historically, class A β-lactamases were found to have much greater penicillinase activity than cephalosporinase activity. However, several new class A β-lactamases that are active against new generations of cephalosporins and carbapenems have been discovered. They are typically grouped as extended-spectrum β-lactamases (ESBLs) to differentiate them from the non-ESBLs.
There have been few studies on the antitubercular effect of β-lactam antibiotics, probably due to the early discovery that Mycobacterium tuberculosis possesses intrinsic resistance to these antibiotics (25). The production of β-lactamase has been proposed to be the most significant reason for mycobacterial resistance (7, 45, 55). Other factors that were believed to contribute to the ineffectiveness of β-lactam in M. tuberculosis include cell envelope permeability and low penicillin-binding protein binding affinity for β-lactams (29, 53). Although M. tuberculosis's outer cell wall, formed by a waxy mycolic acid layer, was thought to make a virtually impenetrable barrier, β-lactams have recently been shown to readily permeate the M. tuberculosis cell wall and bind the penicillin-binding proteins (7).
One approach that holds promise to counter M. tuberculosis's inherent resistance is to utilize β-lactamase inhibitors in combination with β-lactam antibiotics to increase their efficacy (7). For example, class A β-lactamases are generally sensitive to serine β-lactamase inhibitors such as clavulanate and sulbactam (2, 17). These mechanism-based inhibitors covalently cross-link two serines in the active site that are essential to the hydrolysis of β-lactams and permanently inactivate β-lactamase. The combinations of β-lactam antibiotics and β-lactamase inhibitors have been used clinically to treat a wide range of bacterial infections (35). These combinations have also been shown to be somewhat effective against most strains of mycobacteria, including multidrug-resistant (MDR) strains in vitro (11, 48, 56). In fact, MICs of β-lactams are less than 0.5 μg/ml for MDR strains of M. tuberculosis when used in combination with both β-lactamase inhibitors and the antitubercular drug ethambutol (1). These results offer the possibility for treatment of tuberculosis with highly potent inhibitors in combination with β-lactam antibiotics.
blaC (Rv2068c) is the only gene in the M. tuberculosis genome that shows any detectable homology to any of the known β-lactamase genes. Recently, the Pavelka laboratory showed that the deletion of the blaC gene resulted in an increased susceptibility of M. tuberculosis to β-lactam antibiotics by 8- to 256-fold (14, 15), which confirmed the primary role of blaC in M. tuberculosis resistance. In previous studies, partially purified M. tuberculosis BlaC was classified as a class A β-lactamase type 2b based on its relatively high level of penicillinase activity (6, 46). To fully characterize M. tuberculosis BlaC and facilitate the design of new inhibitors, we have characterized the substrate and inhibitor profile for BlaC and determined the structure of BlaC to high resolution.
MATERIALS AND METHODS
Cloning, expression, and purification.
Escherichia coli expression plasmids of the wild type and a truncated form (codons 41 to 307) of M. tuberculosis blaC were cloned from genomic DNA (CSU N01-AI-75320). The amplified product was inserted into pET28b (EMD Biosciences no. 69865-3) using the NdeI and HindIII restriction sites. This plasmid was subsequently transformed into E. coli BL21(DE3) (EMD Bioscience no. 69387-3) and cultured in Luria-Bertani-Miller media containing 50 μg/ml kanamycin at 37°C until the optical density at 600 nm reached 0.8. Expression of the blaC gene was induced for 20 h at 16°C by addition of 1 mM isopropyl β-d-thiogalactopyranoside (IPTG). Cells were harvested by centrifugation and resuspended in 25 mM Tris-HCl (pH 8.0), 500 mM NaCl, and 2 mM β-mercaptoethanol and were lysed by French press. After treatment with 1 mM DNase I, the insoluble material was removed by centrifugation. By sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), full-length BlaC goes into the insoluble pellet, while ∼90% of truncated BlaC (41-307) remained soluble. Therefore, further purification was performed only on truncated BlaC (41-307). The supernatant containing soluble protein was applied to a Ni2+-loaded HiTrap chelating column (AP Biotech) using a fast protein liquid chromatography system and eluted through an imidazole gradient (0 to 0.5 M). BlaC (41-307) was eluted at an imidazole concentration of 120 mM. Fractions that contain BlaC (41-307) were pooled and dialyzed against 25 mM Tris-HCl (pH 8.0), 40 mM NaCl, and 1 mM dithiothreitol. The samples were further purified by gel filtration on a Superdex 200 column to separate the monomeric protein from aggregated material using dialysis buffer. The protein was concentrated to approximately 10 mg/ml. BlaC (41-307) was obtained at 45 mg/liter of E. coli culture and appeared homogeneous by SDS-PAGE and Coomassie blue staining.
Kinetic assays.
Enzyme kinetic evaluation was completed for the substrates ampicillin (Δɛ235 = −820 M−1 cm−1), cephalothin (Δɛ262 = −7,660 M−1 cm−1), cefoxitin (Δɛ270 = −8,380 M−1 cm−1), ceftazidime (Δɛ260 = −10,200 M−1 cm−1), and meropenem (Δɛ299 = −2,500 M−1 cm−1) (24). All assays were repeated in triplicate and carried out on a Cary 100 Bio spectrophotometer at room temperature by monitoring the UV absorbance change of β-lactams (24). The M. tuberculosis BlaC activity assays were performed in 100 mM phosphate buffer (pH 7.5) by adding 10 nM enzyme into substrate solution. Km and Vmax were determined by analyzing a Hanes plot (57). Inhibition studies of clavulanate were performed by adding enzyme into solutions containing the substrate ampicillin and inhibitor clavulanate with increasing concentrations (0.1 to 600 μM). During the measurement time, no time-dependent loss of enzyme activity could be detected. Values for Ki were determined from the x intercepts of a Dixon plot, assuming competitive inhibition.
Crystallization of BlaC (41-307).
Crystallization of the truncated enzyme, missing the first 40 amino acids, was accomplished by the hanging-drop vapor diffusion method (33). Initial crystallization conditions were discovered by sparse matrix screen using crystal screens from Hampton Research. Hanging drops containing 2 μl of protein solution at 10 mg/ml and 2 μl of buffer (0.1 M Tris-HCl, pH 8.0, 2.0 M NH4H2PO4) were equilibrated at 16°C in Linbro plates against 1 ml of the same buffer. Protein crystals appeared in 4 days as thin plate-shaped clusters and were improved to larger rod-like clusters by cocrystallization with its substrate ampicillin. Macroseeding was then used to obtain high-quality single crystals.
Data collection and processing.
A nearly complete diffraction data set to 1.7-Å resolution was collected from a single crystal at 121°K using a cryoprotection solution consisting of reservoir solution (0.1 M Tris-HCl, pH 8.0, 2.0 M NH4H2PO4) with the addition of 20% glycerol. Beam line 14BMC at the Advanced Photon Source, Argonne National Laboratory, was used to collect the data set using 1-degree oscillation widths over a range of 180°. The data were integrated and reduced using HKL2000 (42). The completeness of low-resolution data is slightly lower, due to the ice rings caused by the high rejection rate. Crystals of BlaC belong to the orthorhombic space group P212121 with the following cell dimensions: a = 42.74 Å, b = 71.28 Å, c = 85.17 Å, α = 90°, β = 90°, γ = 90°, with one molecule in each asymmetric unit.
Structure determination and model refinement.
Initial phases were obtained by molecular replacement with the computer program MolRep, part of the CCP4 package (3), using the related structure of β-lactamase from Streptomyces albus G (Protein Data Bank [PDB] identification [ID] 1BSG) (10) as a search model. The best solution gave a correlation coefficient of 34.2% and a Rfactor of 50.9% using data between 25- and 3-Å resolution. The structure from the molecular replacement solution was refined with REFMAC, again in the CCP4 suite (3). The first step in the refinement was to use rigid-body optimization, and this resulted in a slightly improved model (Rfactor, 49.9%; Rfree, 51.8%; 54.6 to 1.7 Å). Electron density maps were produced and manual rebuilding was carried out using XtalView (34). The model was further refined with cycles of model building and REFMAC restrained refinement (3). During the final cycles of the refinement, water molecules were added based on peaks above 3-σ in the Fo − Fc electron density maps that were within hydrogen-bonding distances from the appropriate protein atom. The resulting structure had good geometry (88% of the nonglycine residues were in the most favorable conformation, and none were in a disallowed conformation) and an Rfactor of 17.9% and Rfree of 21.4%. PROCHECK (3) revealed no disallowed φ and ψ, and the overall structure G factor, a measure of a given stereochemical property, was consistently better than expected for this resolution.
Structure analysis.
DALI (22) was used to search the PDB for proteins having folds similar to M. tuberculosis BlaC, in the order of structural similarity. SwissPDB viewer was used to make structural alignments (19). The model was evaluated and analyzed using computer graphic program SPOCK (8).
PDB identification.
The atomic coordinates of BlaC have been deposited in the PDB with the entry code 2GDN.
RESULTS AND DISCUSSION
Sequence analysis of BlaC.
M. tuberculosis BlaC is a 307-amino-acid protein that is predicted to have a signal peptide for translocation into the periplasm and a lipid attachment site (residues 14 to 24) using the computer program ScanProsite (18). This analysis suggests that the mature BlaC is linked via the covalently attached lipid to the outer leaflet of the inner membrane after translocation to the periplasmic space. β-Lactamase lipoproteins have been reported for several gram-positive bacteria (39, 50). However, the enzyme was also predicted using the SignalP program (4) to have a signal peptidase II cleavage site at residue 28, which would eliminate the lipid attachment site in the mature protein. It is therefore not clear if BlaC exists free or as a membrane-bound protein in the periplasm.
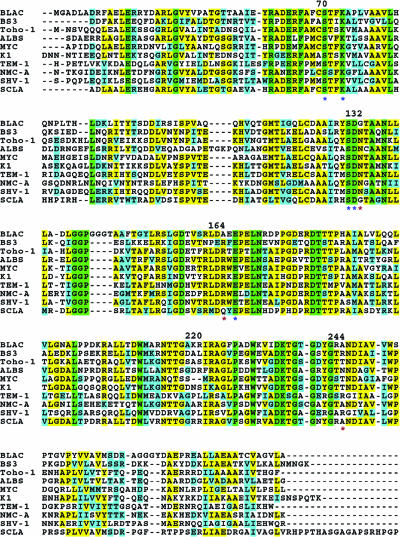
M. tuberculosis BlaC shares approximately 40% amino acid sequence identity with other known class A β-lactamases (Fig. 1). Interestingly, it has the highest sequence identity to the class A β-lactamase of Streptomyces clavuligerus (54%), the bacterium that produces the antibiotic cephamycin and the β-lactamase inhibitor clavulanate.
FIG. 1.
Sequence alignment of β lactamases. Sequence alignment of β lactamases from M. tuberculosis (BLAC), B. licheniformis (BS3), S. albus (ALBS), E. coli (Toho-1 and TEM-1), E. cloacae (NMC-A), P. vulgaris (K1), M. fortuitum (MYC), K. pneumoniae (SHV-1). Green, completely conserved residues; yellow, identical residues; cyan, similar residues. The important residues mentioned in the context are labeled. The residues labeled by asterisks are the equivalents of those indicated significant to the activity.
Three highly conserved regions of the active site, designated motifs I, II, and III, have been identified by multiple sequence alignment of class A β-lactamases from different bacteria. Motif I has a consensus sequence of Arg61-(Xaa)2-Glu-Xaa-Phe-(Xaa)3-Ser-Xaa-Xaa-Lys73 (the residues of M. tuberculosis BlaC were numbered according to a consensus ABL numbering scheme [2]), which extends from the end of the β-strand B2 through a segment of the α-helix H2. Motif II corresponds to the sequence Ser130-Asp-Asn132 and is located between α-helices H6 and H7. Motif III, located on β-strand B3, has the consensus sequence Asp233-Lys-Thr-Gly236. Additionally, all class A β-lactamases have an Ω loop that in BlaC contains residues Val159 to Thr180. While these regions are highly conserved, BlaC from M. tuberculosis has several notable amino acid substitutions. The substitution of a Gly for Asn at residue 132 occurs only in class A β-lactamase from M. tuberculosis and its closest homolog, S. clavuligerus β-lactamase. The substitutions R244A and R169A in M. tuberculosis β-lactamase are also rare in class A β-lactamases. Another important difference identified from amino acid sequence alignment is the four-residue insertion Gly145A to Ala150 and the two-residue insertion Gly271 and Tyr272 in the M. tuberculosis BlaC sequence.
Kinetic study.
Initial attempts to express the full-length protein were unsuccessful. However, by truncating the codons for the first 40 amino acids in our expression construct, we could produce large quantities of active enzyme. This truncation was not predicted to interfere with the enzyme's catalytic function based on what was known about homologous enzymes. The molecular weight of the truncated protein was confirmed by matrix-assisted laser desorption ionization (actual mass, 30,629 Da; calculated mass, 30,666 Da).
Kinetic measurements of the hydrolysis of the substrates ampicillin, cephalothin, cefoxitin, ceftazidime, and meropenem, as well as the inhibition of the enzyme activity with clavulanate, were carried out in order to compare the enzyme activity to other class A β-lactamases (Table 1). Overall, the results show that M. tuberculosis BlaC has a broad-spectrum substrate profile compared to other class A β-lactamases. While the penicillinase activity of BlaC (kcat/Km = 293 mM−1 s−1) is about 10-fold lower than most class A β-lactamases, the enzyme showed relatively good activity against cephalothin (kcat/Km = 103 mM−1 s−1) in contrast to previous studies where M. tuberculosis was shown to possess only minor cephalothin activity (59). Indeed, we found that the cephalosporinase activity of M. tuberculosis BlaC is comparable to its penicillinase activity. Furthermore, BlaC shows modest activity (kcat/Km ∼ 10 mM−1 s−1) against cefoxitin, ceftazidime, and meropenem. Inhibition studies indicated that clavulanate, a very potent inhibitor of most of class A β-lactamases (Ki < 0.1 μM), is a relatively poor inhibitor of M. tuberculosis BlaC (Ki = 2.4 μM). The substrate profile of BlaC is quite similar to that of Mycobacterium fortuitum β-lactamase, which also has relatively high cephalosporinase activity (45).
TABLE 1.
Kinetic parameters of M. tuberculosis class A β-lactamase with different antibiotics as substrates
| Class | Antibiotic | kcat (s−1) | Km (μM) | kcat/Km (mM−1 s−1) | Ki (μM) |
|---|---|---|---|---|---|
| Penicillin | Ampicillin | 18.5 ± 0.9 | 63 ± 7 | 293 | |
| Cephalosporin | Cephalothin | 12.1 ± 1.1 | 117 ± 14 | 103 | |
| Cefoxitin | 1.1 ± 0.3 | 195 ± 15 | 5.6 | ||
| Ceftazidime | 0.2 ± 0.1 | 593 ± 29 | 0.4 | ||
| Carbapenem inhibitor | Meropenem | 0.9 ± 0.3 | 279 ± 23 | 3.2 | |
| Clavulanate | 2.4 ± 1.1 |
Overall structure.
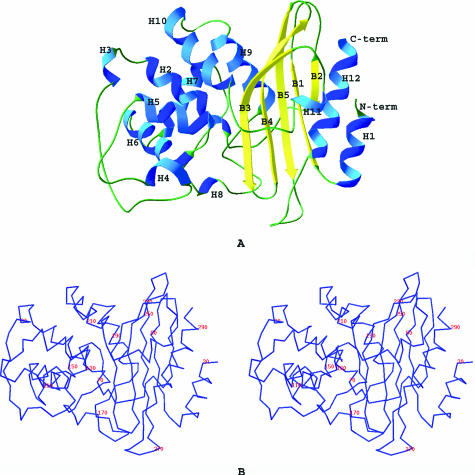
The crystal structure of BlaC was solved using molecular replacement and refined to 1.7-Å resolution. The final Rfactor and Rfree values for all reflections were 0.18 and 0.21, respectively (Table 2). BlaC crystals are orthorhombic, belonging to space group P212121, with one molecule in the asymmetric unit. The refined structure of BlaC includes 265 of the 267 residues in the truncated form of BlaC (41-307) and 248 ordered water molecules. The structure of the N-terminal six-histidine tag along with the first two residues, Gly-Ala, was not defined due to disorder of this region in the electron density map. Like other class A β-lactamases, the BlaC monomer contains an α domain and an α/β domain. The α domain consists of six long α helices (H2, H5, H6, H7, H9, and H10) and three short α helices (H3, H4, and H8). The α/β domain includes five antiparallel β-strands (B1 to B5), the N- and C-terminal α helices (H1 and H12), and one short α helix, H11 (Fig. 2). The overall structure of BlaC is very similar to other class A β-lactamase structures. The root mean square differences (RMSD) of the Cα atoms of several class A β-lactamases range between 0.95 to 3.66 Å and are listed in Table 3. The Cα alignments of BlaC with other class A β-lactamases of known structure indicate that M. tuberculosis BlaC is more similar to K1 (RMSD = 0.95 Å) and NMC-A (RMSD = 1 Å) (enzymes from Proteus vulgaris and Enterobacter cloacae, respectively) compared to SHV-1 and TEM-1 (RMSD > 2.5 Å) (enzymes from Klebsiella pneumoniae and E. coli, respectively). ALBS (PDB ID 1BSG) was used as a search model because it displayed significant sequence homology to BlaC. Surprisingly, it showed a fairly large RMSD value (3.66 Å) compared to BlaC. Indeed, several other class A β-lactamases with lower sequence homology are structurally more similar to BlaC.
TABLE 2.
Data collection, processing, and refinement statistics
| Parameter | Valueb for BlaC |
|---|---|
| Data collection | |
| Maximum resolution (Å) | 1.72 |
| Space group | P212121 |
| a (Å) | 42.7 |
| b (Å) | 71.3 |
| c (Å) | 85.2 |
| Unique reflections | 25,908 (2,808) |
| Rsyma (%) | 11.4 (30.6) |
| Completeness (%) | 91.1 (100) |
| Redundancy | 6.8 (6.9) |
| I/σ | 23.2 (6.8) |
| Refinement statistics | |
| Resolution range (Å) | 54.64-1.72 |
| Number of reflections | 24,570 |
| Number of atoms/subunit | |
| Protein | 2,230 |
| Solvent | 248 |
| Rcryst (%) | 17.9 |
| Rfree (%) | 21.4 |
| Avg B factors (Å2) | |
| Protein | 20.7 |
| Solvent | 32.8 |
| RMSD from ideal values | |
| Bonds (Å) | 0.011 |
| Angles (°) | 1.44 |
Rsym = ∑hkl∑iIi(hkl)−〈I(hkl)〉/∑hkl∑iIi(hkl), where Ii is the intensity of observation and 〈I〉 is the mean intensity of reflection.
The numbers given in parentheses denote the respective values of the highest-resolution shell.
FIG. 2.
A. BlaC structure and its secondary structures. The helices are represented as H1 to H12 and the strands as B1 to B5. B. Stereoscopic view of Cα of BlaC labeled every 20 residues.
TABLE 3.
Structure comparison of class A β-lactamases from different species
| Enzyme | Source | PDB ID | Sequence identity (%) | RMSD (Å) |
|---|---|---|---|---|
| BlaC | M. tuberculosis | |||
| BS3 | B. licheniformis | 1I2S | 47.3 | 1.57 |
| ALBS | S. albus | 1BSG | 45.0 | 3.66 |
| Toho-1 | E. coli | 1IYS | 46.5 | 1.87 |
| SME-1 | S. marcescens | 1DY6 | 42.5 | 0.97 |
| NMC-A | E. cloacae | 1BUE | 41.1 | 1.00 |
| K1 | P. vulgaris | 1HZO | 42.9 | 0.95 |
| MYC | M. fortuitum | 1MFO | 40.7 | 1.54 |
| SHV-1 | K. pneumoniae | 1SHV | 40.0 | 2.74 |
| TEM-1 | E. coli | 1BT5 | 38.0 | 2.85 |
Enzyme active-site structure.
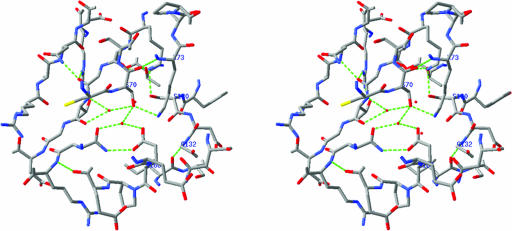
In all class A β-lactamases, the hydrolysis of the β-lactam substrates is accomplished by a nucleophilic attack initiated by active-site serine residue Ser70. It has been proposed that Glu166, a general base in the active site, is the serine70-activating residue based on the ultra-high-resolution crystal structure (0.91 Å for SHV-2 and 0.85 Å for TEM-1) studies (37, 41). However, others believe that Lys73 is involved in the activation (49). Although the mechanism of Ser70 activation is contentious (36), the deacylation mechanism has been established. Glu166 has been demonstrated to be critical for the deacylation of the acyl-enzyme intermediate based on site-directed mutagenesis studies (23). The enzyme active site is located at the interface between the α and the α/β domains (Fig. 2). Ser70 and Lys73, located on α-helix H2 in the center of the active site (Fig. 3), are completely conserved in all class A β-lactamases and are surrounded by other key residues on β-strand B3 (Lys234, Thr235, and Thr237) and the loop region between H5 and H6 (Ser130 and Gly132), as well as the Ω loop (Glu166). These eight residues are all involved in direct hydrogen bonding interactions with β-lactam substrates. Two bound water molecules, WAT36 and WAT65, are also highly conserved in the structures of all class A β-lactamases determined to date. WAT65 is part of a hydrogen bond network with the side chain oxygen of Ser70, as well as the side chain oxygen of Glu166, both at a distance of 2.5 Å. This hydrolytic water functions to deacylate Ser70 from the product after hydrolysis of β-lactam substrates (21, 31, 40). WAT36 is in the oxyanion hole, forming hydrogen bonds with the backbone nitrogen of Ser70 and the backbone nitrogen of Thr237 at distances of 2.6 Å and 2.7 Å, respectively. Although ampicillin improved the diffraction quality of the crystals, neither the substrate nor any products could be identified in the active site.
FIG. 3.
Stereo view of the substrate-binding site of M. tuberculosis BlaC.
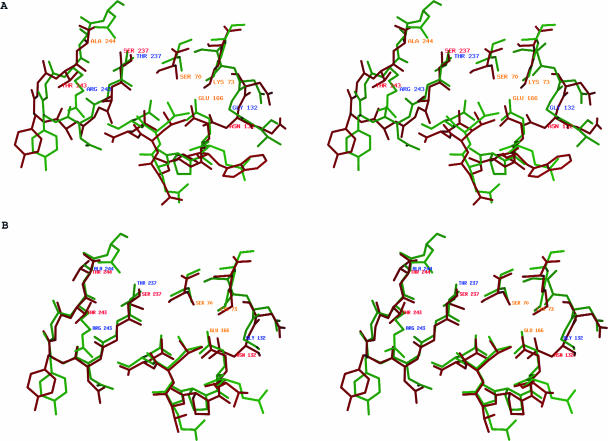
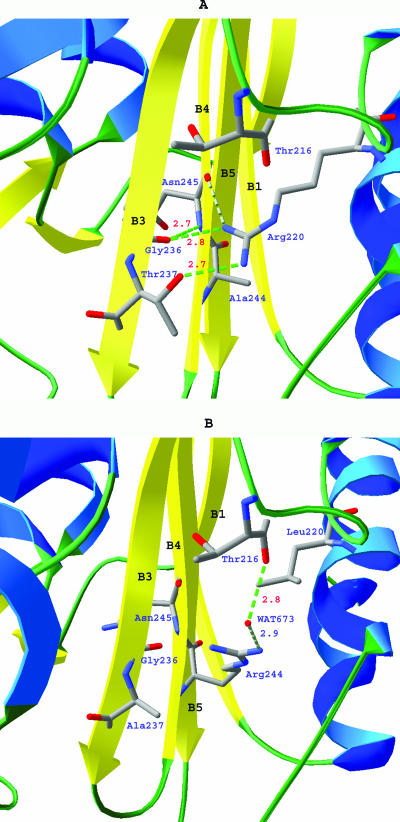
Although the overall structure of M. tuberculosis BlaC is similar to other class A β-lactamases, some differences are clear, the most obvious being the size of the active site (Fig. 4). The active site of M. tuberculosis BlaC is significantly larger than the other class A β-lactamases with known structures. Three factors that contribute to the increased size of the active site are (i) amino acid substitution of several key residues, (ii) rearrangement of conserved side chains, and (iii) a four-residue insertion in the active-site region. The β-lactam-binding motif Ser-Asp-Asn is highly conserved among the class A β-lactamases. In M. tuberculosis BlaC, glycine occupies the position of asparagine. In the class A β-lactamases with structures determined thus far, the side chain of this asparagine points toward the center of the active site, forming a hydrogen bond with the side chain nitrogen of Lys73. The carbamide group of the asparagine is in close contact with the ester carbonyl groups of penicillin and cephems in the complex structures published (47, 49). The 6α-1R-hydroxyl group of imipenem was also observed to hydrogen bond with the side chain oxygen of Asn132 in the structure of E. coli TEM-1 (PDB ID: 1BT5) (32) and E. cloacae NMC-A (PDB ID: 1BUE) (51). It was proposed that the extra space at this location, created by about a 1-Å shift of Asn132 (equivalent to Gly132 in M. tuberculosis BlaC), and that its side chain pointing away from catalytic center would accommodate the substituents at the R6 position of carbapenem and therefore increase carbapenemase activity (38, 51). In fact, this is true for all the carbapenemases of known structures. Having a glycine instead of asparagine in this position makes the BlaC substrate-binding pocket wider and likely more flexible. Therefore, this substitution in BlaC offers an alternative approach to enlarge the active site and could allow M. tuberculosis BlaC to specifically bind carbapenems or cephems with large α substituents. Furthermore, on the same side of the substrate-binding site as Gly132, the side chain of Ser104 is unusual in that it is extended away from the substrate-binding site. In other class A β-lactamases, this position is typically an asparagine or an aspartate and the side chain points into the active site. Like the Asn132-to-Gly132 substitution, the conformation of the Ser104 side chain creates more space in the active site and reduces the potential hydrogen bonding interactions with the substrate. On β-strand B3 of BlaC, Thr237 is unusual for class A β-lactamases. This location is most often occupied by an alanine, as found in non-ESBLs, or a serine in the ESBLs. The hydroxyl group of Thr237 points to the same direction as the comparable serine in the ESBLs. Ser237 (equivalent to Thr237 in M. tuberculosis BlaC) is believed to play a critical role in the extended-spectrum activity of K1 (P. vulgaris) β-lactamase. Superimposition of BlaC structure with substrate-bound complex forms of Bs3 from Bacillus licheniformis (PDB ID: 1I2W) and a mutant of the RTEM-1 from E. coli (PDB ID: 1FQG) (16, 49) indicates that Thr237 potentially forms three hydrogen bonding interactions with the substrate: backbone oxygen to carbamide nitrogen of the α substituent of β-lactam (∼3.5 Å), backbone nitrogen to ester carbonyl oxygen of β-lactam (∼2.8 Å), and side chain oxygen to carboxyl oxygen of the β substituent of β-lactam (∼3 Å). The side chain of Arg220 extends into the top part of the active site, as illustrated in Fig. 5, and forms hydrogen bonds with the side chain oxygen of Thr237, the backbone oxygen of Gly236, and the ester carboxyl oxygen of bound β-lactam. A similar hydrogen bond is seen only in the carbapenemase NCM-A (E. cloacae). The position of Arg220 is usually occupied by a serine in ESBL type enzymes. The hydrogen bonding between Thr237 and Arg220 of M. tuberculosis BlaC pulls the side chain of Thr237 about 2.5 Å away from the active site. This conformation helps to contain the relatively large substituents, such as the 6α-1R-hydroxyl group of carbapenems, which could be critical for their binding. On the other side of the β sheets, Arg243 is unusual for class A β-lactamases. This position is typically occupied by a threonine. The two nitrogens of the guanidinyl group of Arg243 form hydrogen bonds with the backbone CO group of Asp172 and Asn170 at distances of 2.8 Å and 3.1 Å, respectively, and serve to link B4 with the Ω loop. Another observed difference of potential importance is that Arg171 in M. tuberculosis BlaC is usually either a threonine or a serine in other class A β-lactamases. The long side chain of Arg171 extends toward the side of the entrance to the active site. On the opposite site of the Ω loop, the region Ile102 to Ile105 of BlaC is quite different from other class A β-lactamases. While all other class A β-lactamases have either tyrosine or histidine with their aromatic side chains covering the entrance, Ile105 at the same position makes BlaC's active site ∼3 Å wider than other class A β-lactamases. A four-residue insertion, consisting of Gly145A to Thr145D, at the beginning of α helix H7 makes H7 ∼6 Å longer than that of other class A β-lactamases.
FIG. 4.
Stereo view of active sites in class A β-lactamases. A. Superimposition of active sites of BlaC (green) and NMC-A (red). B. Superimposition of active sites of BlaC (green) and Toho-1 (red).
FIG. 5.
Comparison of hydrogen bond pattern involving residues 244 and 220 in M. tuberculosis BlaC (A) and class A β-lactamase of Bacillus licheniformis 749/C (B).
Structural basis for broad substrate profile.
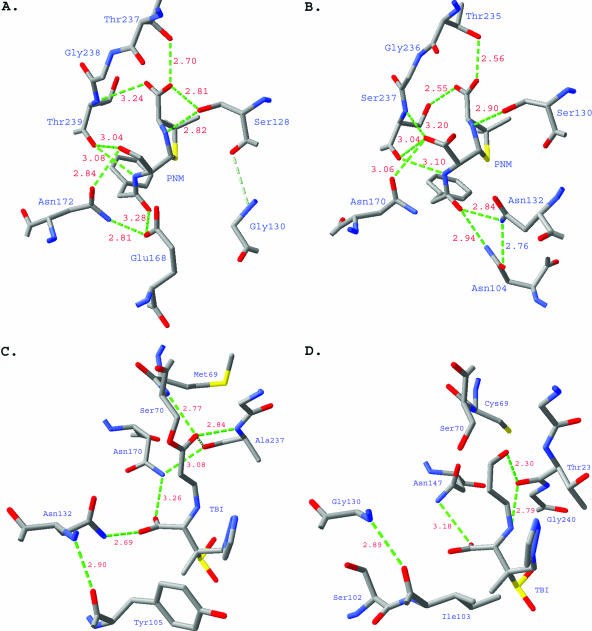
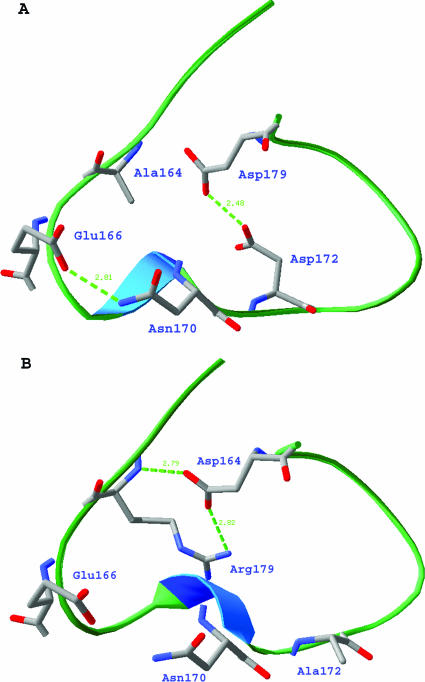
M. tuberculosis BlaC has a broad-spectrum substrate profile. The penicillinase activity of BlaC is about 10-fold lower than most of class A β-lactamases. However, the enzyme showed moderate activity against cephalothin and some activity against cefoxitin, ceftazidime, and meropenem. The broad substrate profile is due to the enzyme's large and flexible substrate-binding site, which allows it to accommodate different types of substrates. While a large active site can provide broader substrate specificity, it can also lead to weak enzyme-ligand interactions. This could explain the generally low activity of M. tuberculosis BlaC in hydrolyzing substrates and inhibitors. The substrate profile of M. tuberculosis BlaC is quite similar to the β-lactamase of S. clavuligerus; however, the M. tuberculosis enzyme has better activity against all β-lactams we have tested. The structure suggests that the low penicillinase activity is in part due to the Asn-to-Gly substitution at position 132. As a conserved residue in other class A β-lactamases, the nitrogen atom of the Asn132 side chain forms a hydrogen bond with the carbonyl oxygen of the α substituent of β-lactam substrates. Glycine at this position would result in a loss of hydrogen bonding to the substrate, as shown by the superimposition of Toho-1-benzylpenicillin (47) and M. tuberculosis BlaC structures (Fig. 6A and B). The hydrogen bond is apparently important for stabilizing the substrate in the active site. Therefore, the loss of hydrogen bonding interactions could explain the low activity of M. tuberculosis BlaC, especially to penicillin type substrates. The critical role of Asn132 in the activity of class A β-lactamase has been proven by the study of an N132A mutant of S. albus G β-lactamase (28), which showed a 100- to 200-fold decrease of penicillinase and cephalosporinase activities. Unlike the N132A mutant of S. albus G β-lactamase, M. tuberculosis BlaC has relatively good activity against cephalothin. The notable R164A substitution in the neck of the Ω loop is likely the reason why BlaC maintains good cephalothin activity. In other class A β-lactamases with known structures, this position is always occupied by Arg164. Normally, Arg164 hydrogen bonds to another highly conserved residue, Asp179, to form a salt bridge in the neck of the Ω loop, which holds the Ω loop closed and rigid (Fig. 7B). Loss of this hydrogen bond could potentially destabilize the Ω loop (Fig. 7A), which has been shown to play an important role in the expansion of activity against cephalosporins, based on a mutagenesis study (54). Indeed, compared to many other class A β-lactamases, part of the Ω loop region, Asn170 to Pro173, of BlaC is curved away from B4 by ∼2 Å.
FIG. 6.
A. Active site of Toho-1 in complex with benzylpenicillin. B. Active site of M. tuberculosis BlaC with a docked ligand benzylpenicillin, using SwissPDB. C. Active site of E166A mutant SHV-1 in complex with the trans-enamine intermediate of tazobactam. D. Active site of M. tuberculosis BlaC with a docked intermediate of tazobactam, using SwissPDB.
FIG. 7.
Comparison of the Ω loops comprising residues 160 to 179 of M. tuberculosis BlaC (A) and SHV-1 (B).
Several other residue substitutions could contribute to the boost of cephalosporinase activity. Superimposition to structures of ESBLs in complex with cephalosporins indicated that the hydroxyl group of Thr237 (equivalent to Ala237 or Ser237 in other class A β-lactamases) of M. tuberculosis BlaC occupies the same position as the side chain of Ser237 in the ESBLs and that the hydroxyl group is within the hydrogen bonding distance (2.5 Å) for the carboxyl group of β substituents of cephalosporins. This could help to stabilize the acyl intermediate of cephalosporins and increase the activity. This is consistent with the finding that the A237T mutation increased the relative cephalosporinase activity in TEM β-lactamase (non-ESBL) (20). Since S. albus G β-lactamase is a non-ESBL at the equivalent position, it has alanine, not serine or threonine. The loss of the hydrogen bonding interactions at both sites could be the reason why the N132A mutant totally lost cephalosporinase activity.
Class A β-lactamase inhibitors.
Clavulanate is a suicide inhibitor specific for class A β-lactamases. This potent (Ki < 0.1 μM) inhibitor forms a covalent acyl enzyme complex with class A β-lactamase. After acylation, the intermediate can either tautomerize to form a transient inhibitor to the enzyme or acylate another active-site serine, Ser130, to form a cross-linked, irreversibly inactivated enzyme (58). Our inhibition studies show that clavulanate is a relatively poor inhibitor of M. tuberculosis BlaC (Ki = 2.4 μM) with a 24-fold-higher Ki compared to TEM-1. Class A β-lactamases with amino acid substitutions at Arg244, Asn276, Arg275, Met69, Met182, and Trp165 have been reported to be resistant to clavulanate and other mechanism-based inhibitors (52). Among these residues, the substitutions R244A, N278E, M69C, and M182T are all found in BlaC. It is interesting that clavulanate is produced by S. clavuligerus, whose β-lactamase is the closest homolog of M. tuberculosis BlaC. Not surprisingly, S. clavuligerus β-lactamase is resistant to clavulanate (Ki = 12 μM) (44). The superimposition of an E166A variant of SHV-1 with tazobactam bound (43) and M. tuberculosis BlaC structures reveals that the N132G substitution would also cause the loss of a hydrogen bond between the side chain of Asn132 and 3-β-carboxyl group of the inhibitor (Fig. 6C and D). In the complex structure of wild-type SHV-1 with tazobactam bound, Asn132 was also observed hydrogen bonding with the sulfinic acid group of the trans-enamine intermediate (30). Based on the structure and activity relationship, it is quite clear that M. tuberculosis and S. clavuligerus share very similar strategies in the resistance of general class A β-lactamase inhibitors, that is, to weaken the binding of the enzyme to those inhibitors mainly through the single amino acid substitution N132G, even though this will also sacrifice enzyme activity to other β-lactam substrates.
BlaC has Ala244 on β strand B4. This position is usually occupied by arginine in non-ESBL enzymes and threonine in ESBL enzymes. Interestingly, in M. tuberculosis BlaC, the side chain of Arg220 is almost in the same position usually occupied by the side chain of Arg244 in other class A β-lactamases. This Arg220-Ala244 residue pair exists only in M. tuberculosis BlaC, carbapenemase NMC-A (E. cloacae), and SCLA (S. clavuligerus). Mutation of Arg244 to Ser in the TEM-1 β-lactamase has been shown to produce resistance to inactivation by clavulanate in the mutant enzyme and resistance to ampicillin plus clavulanate in a strain of E. coli producing this enzyme (27). The modeling studies indicated that the guanidinyl group of Arg244 and an ordered water molecule stabilize the carboxyl group of clavulanate and its acylated intermediate through hydrogen bonding (26). In the R244S mutant, these hydrogen bonds would be lost, which could potentially cause resistance to clavulanate. It is tempting to speculate that the R244A substitution in BlaC would have a similar mechanism of resistance to clavulanate. However, the position of the Arg244 side chain in TEM-1 is occupied by the guanidinyl group of Arg220 in BlaC, which does not exist in TEM-1 (Fig. 5); this complicates the situation. Even though the water molecule in the TEM-1 structure that is believed to be important to clavulanate inhibition is not observed in the BlaC structure, and the guanidinyl group of Arg220 in BlaC does not have the same orientation as that of Arg244 in TEM-1, it is possible that the presence of the guanidinyl group of Arg220 could to some degree restore the sensitivity of BlaC to clavulanate. Indeed, the resistance of BlaC to clavulanate (Ki = 2.4 μM) is about 10-fold less than the R244S mutant of TEM-1 (Ki = 33 μM) (26). However, the clavulanate inhibition is biphasic, and Ki reflects only the first phase. Arg244 in TEM-1 has been shown to be important to both the binding of clavulanate and the chemistry of inactivation (26). Arg220 in BlaC may be able to help the binding of clavulanate by a hydrogen bonding interaction in the first phase but is not likely to facilitate the chemistry of inactivation in the second phase.
Although clavulanate is not an ideal inhibitor to M. tuberculosis BlaC, it has been demonstrated that the combinations of β-lactam antibiotics and mechanism-based inhibitors such as clavulanate are effective in the treatment of M. tuberculosis, even MDR strains. This makes M. tuberculosis BlaC a promising target for the design of new inhibitors that are specific for and potent against M. tuberculosis BlaC. Based on the structure of M. tuberculosis BlaC, it is now possible to either modify current inhibitors to increase inhibitory potency or design novel inhibitors that are specific for the enzyme. In combination with the highly potent M. tuberculosis BlaC inhibitors, β-lactam antibiotics could become a new regimen to treat tuberculosis.
Acknowledgments
This work was supported by the National Institutes of Health and by funds from the Robert A. Welch Foundation.
REFERENCES
- 1.Abate, G., and H. Miorner. 1998. Susceptibility of multidrug-resistant strains of Mycobacterium tuberculosis to amoxycillin (sic) in combination with clavulanic acid and ethambutol. J. Antimicrob. Chemother. 42:735-740. [DOI] [PubMed] [Google Scholar]
- 2.Ambler, R. P., A. F. Coulson, J. M. Frere, J. M. Ghuysen, B. Joris, M. Forsman, R. C. Levesque, G. Tiraby, and S. G. Waley. 1991. A standard numbering scheme for the class A beta-lactamases. Biochem. J. 276:269-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anonymous. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. Sect. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 4.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 5.Bentley, R. 2005. The development of penicillin: genesis of a famous antibiotic. Perspect. Biol. Med. 48:444-452. [DOI] [PubMed] [Google Scholar]
- 6.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for beta-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chambers, H. F., D. Moreau, D. Yajko, C. Miick, C. Wagner, C. Hackbarth, S. Kocagoz, E. Rosenberg, W. K. Hadley, and H. Nikaido. 1995. Can. penicillins and other beta-lactam antibiotics be used to treat tuberculosis? Antimicrob. Agents Chemother. 39:2620-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christopher, J. A. 1998. SPOCK: the Structural Properties Observation and Calculation Kit. Program manual. Center for Macromolecular Design, Texas A&M University, College Station, TX.
- 9.Davies, J. 1994. Inactivation of antibiotics and the dissemination of resistance genes. Science 264:375-382. [DOI] [PubMed] [Google Scholar]
- 10.Dideberg, O., P. Charlier, J. P. Wery, P. Dehottay, J. Dusart, T. Erpicum, J. M. Frere, and J. M. Ghuysen. 1987. The crystal structure of the beta-lactamase of Streptomyces albus G at 0.3 nm resolution. Biochem. J. 245:911-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dincer, I., A. Ergin, and T. Kocagoz. 2004. The in vitro efficacy of beta-lactam and beta-lactamase inhibitors against multidrug resistant clinical strains of Mycobacterium tuberculosis. Int. J. Antimicrob. Agents 23:408-411. [DOI] [PubMed] [Google Scholar]
- 12.Edwards, J. R., and M. J. Betts. 2000. Carbapenems: the pinnacle of the beta-lactam antibiotics or room for improvement? J. Antimicrob. Chemother. 45:1-4. [DOI] [PubMed] [Google Scholar]
- 13.Fisher, J. F., S. O. Meroueh, and S. Mobashery. 2005. Bacterial resistance to beta-lactam antibiotics: compelling opportunism, compelling opportunity. Chem. Rev. 105:395-424. [DOI] [PubMed] [Google Scholar]
- 14.Flores, A. R., L. M. Parsons, and M. S. Pavelka, Jr. 2005. Characterization of novel Mycobacterium tuberculosis and Mycobacterium smegmatis mutants hypersusceptible to beta-lactam antibiotics. J. Bacteriol. 187:1892-1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flores, A. R., L. M. Parsons, and M. S. Pavelka, Jr. 2005. Genetic analysis of the beta-lactamases of Mycobacterium tuberculosis and Mycobacterium smegmatis and susceptibility to beta-lactam antibiotics. Microbiology 151:521-532. [DOI] [PubMed] [Google Scholar]
- 16.Fonze, E., M. Vanhove, G. Dive, E. Sauvage, J. M. Frere, and P. Charlier. 2002. Crystal structures of the Bacillus licheniformis BS3 class A beta-lactamase and of the acyl-enzyme adduct formed with cefoxitin. Biochemistry 41:1877-1885. [DOI] [PubMed] [Google Scholar]
- 17.Frere, J. M. 1995. Beta-lactamases and bacterial resistance to antibiotics. Mol. Microbiol. 16:385-395. [DOI] [PubMed] [Google Scholar]
- 18.Gattiker, A., E. Gasteiger, and A. Bairoch. 2002. ScanProsite: a reference implementation of a PROSITE scanning tool. Appl. Bioinformatics 1:107-108. [PubMed] [Google Scholar]
- 19.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 20.Hall, A., and J. R. Knowles. 1976. Directed selective pressure on a beta-lactamase to analyse molecular changes involved in development of enzyme function. Nature 264:803-804. [DOI] [PubMed] [Google Scholar]
- 21.Herzberg, O., and J. Moult. 1987. Bacterial resistance to beta-lactam antibiotics: crystal structure of beta-lactamase from Staphylococcus aureus PC1 at 2.5 Å resolution. Science 236:694-701. [DOI] [PubMed] [Google Scholar]
- 22.Holm, L., and C. Sander. 1995. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 20:478-480. [DOI] [PubMed] [Google Scholar]
- 23.Ibuka, A., A. Taguchi, M. Ishiguro, S. Fushinobu, Y. Ishii, S. Kamitori, K. Okuyama, K. Yamaguchi, M. Konno, and H. Matsuzawa. 1999. Crystal structure of the E166A mutant of extended-spectrum beta-lactamase Toho-1 at 1.8 Å resolution. J. Mol. Biol. 285:2079-2087. [DOI] [PubMed] [Google Scholar]
- 24.Ibuka, A. S., Y. Ishii, M. Galleni, M. Ishiguro, K. Yamaguchi, J. M. Frere, H. Matsuzawa, and H. Sakai. 2003. Crystal structure of extended-spectrum beta-lactamase Toho-1: insights into the molecular mechanism for catalytic reaction and substrate specificity expansion. Biochemistry 42:10634-10643. [DOI] [PubMed] [Google Scholar]
- 25.Iland, C. N. 1946. The effect of penicillin on the tubercle bacillus: tubercle penicillinase. J. Pathol. Bacteriol. 58:495-500. [DOI] [PubMed] [Google Scholar]
- 26.Imtiaz, U., E. Billings, J. R. Knox, E. K. Manavathu, S. A. Lerner, and S. Mobashery. 1993. Inactivation of class A beta-lactamases by clavulanic acid: the role of arginine-244 in a proposed nonconcerted sequence of events. J. Am. Chem. Soc. 115:4435-4442. [Google Scholar]
- 27.Imtiaz, U., E. K. Manavathu, S. A. Lerner, and S. Mobashery. 1993. Critical hydrogen bonding by serine 235 for cephalosporinase activity of TEM-1 beta-lactamase. Antimicrob. Agents Chemother. 37:2438-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jacob, F., B. Joris, S. Lepage, J. Dusart, and J. M. Frere. 1990. Role of the conserved amino acids of the 'SDN′ loop (Ser130, Asp131 and Asn132) in a class A beta-lactamase studied by site-directed mutagenesis. Biochem. J. 271:399-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jarlier, V., L. Gutmann, and H. Nikaido. 1991. Interplay of cell wall barrier and beta-lactamase activity determines high resistance to beta-lactam antibiotics in Mycobacterium chelonae. Antimicrob. Agents Chemother. 35:1937-1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuzin, A. P., M. Nukaga, Y. Nukaga, A. Hujer, R. A. Bonomo, and J. R. Knox. 2001. Inhibition of the SHV-1 beta-lactamase by sulfones: crystallographic observation of two reaction intermediates with tazobactam. Biochemistry 40:1861-1866. [DOI] [PubMed] [Google Scholar]
- 31.Lim, D., F. Sanschagrin, L. Passmore, L. De Castro, R. C. Levesque, and N. C. Strynadka. 2001. Insights into the molecular basis for the carbenicillinase activity of PSE-4 beta-lactamase from crystallographic and kinetic studies. Biochemistry 40:395-402. [DOI] [PubMed] [Google Scholar]
- 32.Maveyraud, L., L. Mourey, L. P. Kotra, J. Pedelacq, V. Guillet, S. Mobashery, and J. Samama. 1998. Structure basis for clinical longevity of carbapenem antibiotics in the face of challenge by the common class A beta-lactamases from the antibiotic-resistant bacteria. J. Am. Chem. Soc. 120:9748-9752. [Google Scholar]
- 33.McPherson, A. 1982. Preparation and analysis of protein crystals. Waverly, Baltimore, Md.
- 34.McRee, D. E. 1999. XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125:156-165. [DOI] [PubMed] [Google Scholar]
- 35.Medeiros, A. A. 1997. Evolution and dissemination of beta-lactamases accelerated by generations of beta-lactam antibiotics. Clin. Infect. Dis. 24(Suppl. 1):S19-S45. [DOI] [PubMed] [Google Scholar]
- 36.Meroueh, S. O., J. F. Fisher, H. B. Schlegel, and S. Mobashery. 2005. Ab initio QM/MM study of class A beta-lactamase acylation: dual participation of Glu166 and Lys73 in a concerted base promotion of Ser70. J. Am. Chem. Soc. 127:15397-15407. [DOI] [PubMed] [Google Scholar]
- 37.Minasov, G., X. Wang, and B. K. Shoichet. 2002. An ultrahigh resolution structure of TEM-1 beta-lactamase suggests a role for Glu166 as the general base in acylation. J. Am. Chem. Soc. 124:5333-5340. [DOI] [PubMed] [Google Scholar]
- 38.Mourey, L., K. Miyashita, P. Swaren, A. Bulychev, J. Samama, and S. Mobashery. 1998. Inhibition of the NMC-A beta-lactamase by a penicillanic acid derivative and the structural bases for the increase in substrate profile of this antibiotic resistance enzyme. J. Am. Chem. Soc. 120:9382-9383. [Google Scholar]
- 39.Nielsen, J. B., and J. O. Lampen. 1982. Glyceride-cysteine lipoproteins and secretion by gram-positive bacteria. J. Bacteriol. 152:315-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nukaga, M., K. Mayama, G. V. Crichlow, and J. R. Knox. 2002. Structure of an extended-spectrum class A beta-lactamase from Proteus vulgaris K1. J. Mol. Biol. 317:109-117. [DOI] [PubMed] [Google Scholar]
- 41.Nukaga, M., K. Mayama, A. M. Hujer, R. A. Bonomo, and J. R. Knox. 2003. Ultrahigh resolution structure of a class A beta-lactamase: on the mechanism and specificity of the extended-spectrum SHV-2 enzyme. J. Mol. Biol. 328:289-301. [DOI] [PubMed] [Google Scholar]
- 42.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 43.Padayatti, P. S., M. S. Helfand, M. A. Totir, M. P. Carey, A. M. Hujer, P. R. Carey, R. A. Bonomo, and F. van den Akker. 2004. Tazobactam forms a stoichiometric trans-enamine intermediate in the E166A variant of SHV-1 beta-lactamase: 1.63 Å crystal structure. Biochemistry 43:843-848. [DOI] [PubMed] [Google Scholar]
- 44.Perez-Llarena, F., J. F. Martin, M. Galleni, J. J. Coque, J. L. Fuente, J. M. Frere, and P. Liras. 1997. The bla gene of the cephamycin cluster of Streptomyces clavuligerus encodes a class A beta-lactamase of low enzymatic activity. J. Bacteriol. 179:6035-6040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quinting, B., J. M. Reyrat, D. Monnaie, G. Amicosante, V. Pelicic, B. Gicquel, J. M. Frere, and M. Galleni. 1997. Contribution of beta-lactamase production to the resistance of mycobacteria to beta-lactam antibiotics. FEBS Lett. 406:275-278. [DOI] [PubMed] [Google Scholar]
- 46.Segura, C., M. Salvado, I. Collado, J. Chaves, and A. Coira. 1998. Contribution of beta-lactamases to beta-lactam susceptibilities of susceptible and multidrug-resistant Mycobacterium tuberculosis clinical isolates. Antimicrob. Agents Chemother. 42:1524-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shimamura, T., A. Ibuka, S. Fushinobu, T. Wakagi, M. Ishiguro, Y. Ishii, and H. Matsuzawa. 2002. Acyl-intermediate structures of the extended-spectrum class A beta-lactamase, Toho-1, in complex with cefotaxime, cephalothin, and benzylpenicillin. J. Biol. Chem. 277:46601-46608. [DOI] [PubMed] [Google Scholar]
- 48.Sorg, T. B., and M. H. Cynamon. 1987. Comparison of four beta-lactamase inhibitors in combination with ampicillin against Mycobacterium tuberculosis. J. Antimicrob. Chemother. 19:59-64. [DOI] [PubMed] [Google Scholar]
- 49.Strynadka, N. C., H. Adachi, S. E. Jensen, K. Johns, A. Sielecki, C. Betzel, K. Sutoh, and M. N. James. 1992. Molecular structure of the acyl-enzyme intermediate in beta-lactam hydrolysis at 1.7 Å resolution. Nature 359:700-705. [DOI] [PubMed] [Google Scholar]
- 50.Sutcliffe, I. C., and R. R. Russell. 1995. Lipoproteins of gram-positive bacteria. J. Bacteriol. 177:1123-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swaren, P., L. Maveyraud, X. Raquet, S. Cabantous, C. Duez, J. D. Pedelacq, S. Mariotte-Boyer, L. Mourey, R. Labia, M. H. Nicolas-Chanoine, P. Nordmann, J. M. Frere, and J. P. Samama. 1998. X-ray analysis of the NMC-A beta-lactamase at 1.64-Å resolution, a class A carbapenemase with broad substrate specificity. J. Biol. Chem. 273:26714-26721. [DOI] [PubMed] [Google Scholar]
- 52.Therrien, C., and R. C. Levesque. 2000. Molecular basis of antibiotic resistance and beta-lactamase inhibition by mechanism-based inactivators: perspectives and future directions. FEMS Microbiol. Rev. 24:251-262. [DOI] [PubMed] [Google Scholar]
- 53.Trias, J., V. Jarlier, and R. Benz. 1992. Porins in the cell wall of mycobacteria. Science 258:1479-1481. [DOI] [PubMed] [Google Scholar]
- 54.Vakulenko, S. B., P. Taibi-Tronche, M. Toth, I. Massova, S. A. Lerner, and S. Mobashery. 1999. Effects on substrate profile by mutational substitutions at positions 164 and 179 of the class A TEM(pUC19) beta-lactamase from Escherichia coli. J. Biol. Chem. 274:23052-23060. [DOI] [PubMed] [Google Scholar]
- 55.Voladri, R. K., D. L. Lakey, S. H. Hennigan, B. E. Menzies, K. M. Edwards, and D. S. Kernodle. 1998. Recombinant expression and characterization of the major beta-lactamase of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 42:1375-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wong, C. S., G. S. Palmer, and M. H. Cynamon. 1988. In-vitro susceptibility of Mycobacterium tuberculosis, Mycobacterium bovis and Mycobacterium kansasii to amoxycillin (sic) and ticarcillin in combination with clavulanic acid. J. Antimicrob. Chemother. 22:863-866. [DOI] [PubMed] [Google Scholar]
- 57.Wong, J. T. 1985. Kinetics of enzyme mechanisms. Academic Press, New York, N.Y.
- 58.Yang, Y., B. A. Rasmussen, and D. M. Shlaes. 1999. Class A beta-lactamases—enzyme-inhibitor interactions and resistance. Pharmacol. Ther. 83:141-151. [DOI] [PubMed] [Google Scholar]
- 59.Zhang, Y., V. A. Steingrube, and R. J. Wallace, Jr. 1992. Beta-lactamase inhibitors and the inducibility of the beta-lactamase of Mycobacterium tuberculosis. Am. Rev. Respir. Dis. 145:657-660. [DOI] [PubMed] [Google Scholar]