Abstract
The distribution and resistance levels of 189 in vitro-selected rifampin-resistant Mycobacterium tuberculosis mutants of Beijing and other genotypes were determined. Apart from a higher amount of codon 522 point mutations and large deletions, a spread of mutations similar to that reported for clinical isolates was seen. Most mutations were correlated with high-level resistance; a lower level, or a MIC of <16 mg/liter, was associated with codon 522 mutations. Multiple mutations were detected in two Beijing mutants.
Rifampin (RIF) is a key drug in the four-drug treatment regimen of tuberculosis. It interacts with the β-subunit of the RNA polymerase and inhibits the early steps of transcription. Resistance to RIF is almost entirely coupled (>97%) to mutations within an 81-bp region of the rpoB gene, called the RIF resistance-determining region (RRDR) (5, 23). Mutations in the rpoB gene of Mycobacterium tuberculosis clinical isolates were analyzed by Telenti et al. (23), and many studies have since followed (1, 4, 7, 13, 21, 22). A few studies of in vitro-generated RIF resistance mutations in M. tuberculosis have also been conducted; however, these were limited to the H37Rv reference strain and a single clinical isolate (3, 14, 16).
Strains of the M. tuberculosis Beijing genotype have been spreading increasingly and are often associated with multidrug-resistant (resistance to at least RIF and isoniazid) tuberculosis (6, 15, 17-19, 24, 25). Isolates of this genotype have been shown not to have a higher in vitro mutation rate for the development of RIF resistance (27). A few clinical studies have analyzed RIF-resistant Beijing isolates, for which no clear differences in occurring RRDR mutations compared to those of other genotypes have been seen (8, 18, 19, 24, 25). There are, however, no data on in vitro-selected rpoB mutations in Beijing strains.
In the present study, we wanted to investigate the frequency and distribution of rpoB mutations occurring in vitro among several independent M. tuberculosis parental strains and subsequently compare these to available data on clinical isolates. Additionally, we determined whether Beijing genotype mutants display differences in the types and/or the frequencies of rpoB mutations compared to those of mutants of non-Beijing genotypes. Finally, we wanted to measure the levels of resistance associated with the various RRDR mutations.
By use of the Luria and Delbrück fluctuation assay (12), 189 RIF-resistant mutants were previously selected (27). By setting a series of parallel cultures from a single parent strain, this method selects spontaneously occurring, independent mutational events in nonselective broth. Then, by inoculation of the cultures on selective media (2 mg/liter RIF), the mutants are obtained. In all, nine fully drug-susceptible parent strains with unique DNA fingerprint patterns (26) were used to select RIF-resistant mutants. Eight were clinical isolates, of which one was a Dutch Harlingen isolate (11) and one was the H37Rv (ATCC 25618) reference strain. Based on their spoligotype pattern (10), four parent isolates were identified to be of the Beijing genotype. In total, 89/189 in vitro-selected mutants had a Beijing genotype, while 100/189 belonged to other genotypes.
Following mutant selection, DNA was extracted and a 382-bp fragment containing the RRDR was sequenced in subsequent mutants. With primers OPRIF-F, 5′ CGG TCG GCG AGC TGA TCC 3′, and OPRIF-R, 5′ TGG ACC CGC GCG TAC ACC 3′, the PCR and sequencing of the fragment were carried out using a previously described method (9). With BLAST2 sequence software, the sequences were aligned to wild-type H37Rv's rpoB gene (Rv0667). The nucleotides and codons were designated based on Escherichia coli codon numbering.
Altogether, 172 mutants (91%) harbored single nucleotide substitutions, comprising 12 different substitutions within seven codons. For each group of isogenic mutants, the most common single nucleotide mutation sites were codons 526, 531, and 522, and in total these codons' relative frequencies were 40%, 34%, and 13%, respectively. The frequencies of codon 526 and 531 mutations were similar to the distribution reported among RIF-resistant clinical isolates, while codon 522 mutations have rarely been seen clinically (1, 4, 7, 13, 21, 22). Mutations in the H37Rv-derived mutants showed the narrowest span, with only five different types recorded (three being single nucleotide substitutions) (data not shown). This finding is consistent with the range obtained from the two earlier in vitro selection studies of H37Rv-derived mutants (3, 16).
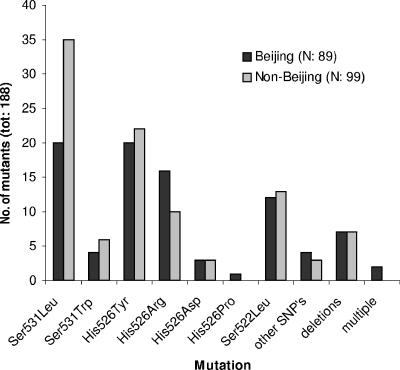
Eleven types of in-frame deletions, whose lengths varied between 3 and 18 nucleotides, were seen among 14 (7.4%) mutants. Many of these are larger and altogether more frequent than what is seen clinically (7, 13, 21, 22). Multiple mutations occurred in two mutants from different Beijing parent strains. A single mutant (Harlingen genotype) with a high level of RIF resistance showed a wild-type RRDR. As seen in Fig. 1, the in vitro-selected mutants of Beijing and non-Beijing genotypes showed similar rpoB mutation profiles.
FIG. 1.
Distribution of in vitro-selected, rpoB mutation sites in 89 Beijing and 99 non-Beijing genotype M. tuberculosis mutants. The mutant lacking an RRDR mutation is not included. tot, total; SNP's, single nucleotide polymorphism.
The level of RIF resistance for each of the 189 mutants was determined as the MIC on Middlebrook 7H10 agar supplemented with oleic acid-albumin-dextrose-catalase and amphotericin B (8 mg/liter) (the levels of resistance are listed in Table 1). Briefly, by use of a 96-stick replicator, bacteria were inoculated in a twofold-dilution series of RIF (0.0625 mg/liter to 256 mg/liter), and the MICs were determined after 4 weeks of incubation. All parent strains were confirmed to be RIF susceptible, showing MICs of ≤1 mg/liter.
TABLE 1.
rpoB mutations and MICs of the 189 in vitro-selected RIF-resistant M. tuberculosis mutants
| Base pair mutation(s) (no. of strains) | Amino acid mutation(s) | Total no. of strains with mutation(s) (n = 189) | No. of strains
|
|||
|---|---|---|---|---|---|---|
| Beijing (n = 89)
|
Non-Beijing (n = 100)
|
|||||
| MIC of ≤16 | MIC of ≥32 | MIC of ≤16 | MIC of ≥32 | |||
| Single nucleotide substitutions (n = 172) | ||||||
| TCG → TTG | Ser531Leu | 55 | 20 | 35 | ||
| CAC → TAC | His526Tyr | 42 | 20 | 22 | ||
| TCG → TTG | Ser522Leu | 25 | 5 | 7 | 5 | 8 |
| CAC → CGC | His526Arg | 26 | 16 | 10 | ||
| TCG → TGG | Ser531Trp | 10 | 4 | 6 | ||
| CAC → GAC | His526Asp | 6 | 3 | 3 | ||
| CAA → AAA | Gln513Lys | 2 | 1 | 1 | ||
| GAC → GTC | Asp516Val | 2 | 1 | 1 | ||
| AAC → AAG | Asn519Lys | 1 | 1 | |||
| CAC → CCC | His526Pro | 1 | 1 | |||
| CTG → CCG | Leu533Pro | 1 | 1 | |||
| CGA → CCA | Arg529Pro | 1 | 1 | |||
| Deletions (n = 14) | ||||||
| ΔGAC | 516 | 2 | 1 | 1 | ||
| ΔAAC | 518 | 2 | 2 | |||
| ΔCAG AAC | 517-518 | 1 | 1 | |||
| ΔTG G | 515-516 | 1 | 1 | |||
| ΔG GA | 515-516 | 1 | 1 | |||
| ΔG CTG AGC CA | 510-512 | 1 | 1 | |||
| ΔTC ATG G | 514-516 | 1 | 1 | |||
| ΔAGC CAA TTC ATG | 512-515 | 1 | 1 | |||
| ΔCAA TTC ATG GAC | 513-516 | 2 | 2 | |||
| ΔG TTG ACC CA | 523-526 | 1 | 1 | |||
| ΔGC CAA TTC ATG GAC CAG A | 512-517 | 1 | 1 | |||
| Multiple mutations (n = 2) | ||||||
| CAC → CCC, AAG → CAG | His526Pro, Lys527Gln | 1 | 1 | |||
| TTG → TGG, ACC → CCC, ΔACA | Leu524Trp, Thr525Pro, 526-527 | 1 | 1 | |||
| None | Wild-type RRDR | 1 | 1 | |||
| Total | 189 | 6 | 83 | 10 | 90 | |
Generally, all RRDR mutations were coupled with a high level of RIF resistance (MIC of 32 to ≥256 mg/liter). However, the Ser522Leu substitution was, in 10 mutants arising from six parent strains, correlated with a lower level of resistance (MIC of 8 to 16 mg/liter). This codon was the third most prevalent among our in vitro mutants (13%), while it is relatively rare among clinical isolates (7, 13, 21). It is known that resistance mutations within bacteria impair the biological fitness to different extents (2), and it has been seen that, in vitro, this particular mutation is linked to a considerable fitness deficit (14). This observation and the lower resistance level we measured could explain Ser522Leu's lower frequency in the clinical setting.
No significant difference in mutations or in levels of resistance between the Beijing and non-Beijing mutants was seen. Interestingly, the only multiple mutations we obtained were limited to two Beijing mutants. Although not significant, this correlation is in line with Tracevska et al.'s observation that such mutations occurred in 8.2% of their analyzed Beijing clinical isolates, compared to 2% in strains of the other genotype (25). This could be an indication of a hampered DNA repair system in the Beijing genotype that would lead to a higher accumulation of mutations (including drug resistance mutations). Mutations in putative mutator (mut) genes, involved in the regulation of mutation frequencies by repairing DNA lesions, have indeed been reported to occur in strains of the Beijing genotype (20). The parent strains of the two mutants harboring multiple mutations in our study did not, however, show an increased mutation rate (27).
In the present study, we could detect no significant difference in the number or type of rpoB mutations between the Beijing and non-Beijing mutants. The Beijing genotype's high correlation with multidrug-resistant tuberculosis could be the result of a less affected fitness when drug resistance is acquired. Competition assays comparing synonymous mutations of different genotypes (i.e., Beijing versus non-Beijing) could be used to elucidate this point. Finally, further investigation is needed to clarify the association between multiple mutations, mutator genes, and the Beijing family.
Acknowledgments
This study was partly supported by the European Commission project no. LHSP-CT-2004-516028.
REFERENCES
- 1.Ahmad, S., and E. Mokaddas. 2005. The occurrence of rare rpoB mutations in rifampicin-resistant clinical Mycobacterium tuberculosis isolates from Kuwait. Int. J. Antimicrob. Agents 26:205-212. [DOI] [PubMed] [Google Scholar]
- 2.Andersson, D. I. 2003. Persistence of antibiotic resistant bacteria. Curr. Opin. Microbiol. 6:452-456. [DOI] [PubMed] [Google Scholar]
- 3.Billington, O. J., T. D. McHugh, and S. H. Gillespie. 1999. Physiological cost of rifampin resistance induced in vitro in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 43:1866-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bobadilla-del-Valle, M., A. Ponce-de-Leon, C. Arenas-Huertero, G. Vargas-Alarcon, M. Kato-Maeda, P. M. Small, P. Couary, G. M. Ruiz-Palacios, and J. Sifuentes-Osornio. 2001. rpoB gene mutations in rifampin-resistant Mycobacterium tuberculosis identified by polymerase chain reaction single-stranded conformational polymorphism. Emerg. Infect. Dis. 7:1010-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cole, S. T., and A. Telenti. 1995. Drug resistance in Mycobacterium tuberculosis. Eur. Respir. J. 20(Suppl.):701s-713s. [PubMed] [Google Scholar]
- 6.Glynn, J. R., J. Whiteley, P. J. Bifani, K. Kremer, and D. van Soolingen. 2002. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerg. Infect. Dis. 8:843-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heep, M., B. Brandstatter, U. Rieger, N. Lehn, E. Richter, S. Rusch-Gerdes, and S. Niemann. 2001. Frequency of rpoB mutations inside and outside the cluster I region in rifampin-resistant clinical Mycobacterium tuberculosis isolates. J. Clin. Microbiol. 39:107-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hillemann, D., T. Kubica, S. Rusch-Gerdes, and S. Niemann. 2005. Disequilibrium in distribution of resistance mutations among Mycobacterium tuberculosis Beijing and non-Beijing strains isolated from patients in Germany. Antimicrob. Agents Chemother. 49:1229-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jureen, P., J. Werngren, and S. E. Hoffner. 2004. Evaluation of the line probe assay (LiPA) for rapid detection of rifampicin resistance in Mycobacterium tuberculosis. Tuberculosis (Edinburgh) 84:311-316. [DOI] [PubMed] [Google Scholar]
- 10.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kiers, A., A. P. Drost, D. van Soolingen, and J. Veen. 1997. Use of DNA fingerprinting in international source case finding during a large outbreak of tuberculosis in The Netherlands. Int. J. Tuberc. Lung Dis. 1:239-245. [PubMed] [Google Scholar]
- 12.Luria, S., and M. Delbrück. 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28:491-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mani, C., N. Selvakumar, V. Kumar, S. Narayanan, and P. R. Narayanan. 2003. Comparison of DNA sequencing, PCR-SSCP and PhaB assays with indirect sensitivity testing for detection of rifampicin resistance in Mycobacterium tuberculosis. Int. J. Tuberc. Lung Dis. 7:652-659. [PubMed] [Google Scholar]
- 14.Mariam, D. H., Y. Mengistu, S. E. Hoffner, and D. I. Andersson. 2004. Effect of rpoB mutations conferring rifampin resistance on fitness of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 48:1289-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mokrousov, I., H. M. Ly, T. Otten, N. N. Lan, B. Vyshnevskyi, S. Hoffner, and O. Narvskaya. 2005. Origin and primary dispersal of the Mycobacterium tuberculosis Beijing genotype: clues from human phylogeography. Genome Res. 15:1357-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morlock, G. P., B. B. Plikaytis, and J. T. Crawford. 2000. Characterization of spontaneous, in vitro-selected, rifampin-resistant mutants of Mycobacterium tuberculosis strain H37Rv. Antimicrob. Agents Chemother. 44:3298-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Narvskaya, O., T. Otten, E. Limeschenko, N. Sapozhnikova, O. Graschenkova, L. Steklova, A. Nikonova, M. L. Filipenko, I. Mokrousov, and B. Vyshnevskiy. 2002. Nosocomial outbreak of multidrug-resistant tuberculosis caused by a strain of Mycobacterium tuberculosis W-Beijing family in St. Petersburg, Russia. Eur. J. Clin. Microbiol. Infect. Dis. 21:596-602. [DOI] [PubMed] [Google Scholar]
- 18.Park, Y. K., S. Shin, S. Ryu, S. N. Cho, W. J. Koh, O. J. Kwon, Y. S. Shim, W. J. Lew, and G. H. Bai. 2005. Comparison of drug resistance genotypes between Beijing and non-Beijing family strains of Mycobacterium tuberculosis in Korea. J. Microbiol. Methods 63:165-172. [DOI] [PubMed] [Google Scholar]
- 19.Qian, L., C. Abe, T. P. Lin, M. C. Yu, S. N. Cho, S. Wang, and J. T. Douglas. 2002. rpoB genotypes of Mycobacterium tuberculosis Beijing family isolates from East Asian countries. J. Clin. Microbiol. 40:1091-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rad, M. E., P. Bifani, C. Martin, K. Kremer, S. Samper, J. Rauzier, B. Kreiswirth, J. Blazquez, M. Jouan, D. van Soolingen, and B. Gicquel. 2003. Mutations in putative mutator genes of Mycobacterium tuberculosis strains of the W-Beijing family. Emerg. Infect. Dis. 9:838-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramaswamy, S., and J. M. Musser. 1998. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber. Lung Dis. 79:3-29. [DOI] [PubMed] [Google Scholar]
- 22.Sougakoff, W., M. Rodrigue, C. Truffot-Pernot, M. Renard, N. Durin, M. Szpytma, R. Vachon, A. Troesch, and V. Jarlier. 2004. Use of a high-density DNA probe array for detecting mutations involved in rifampicin resistance in Mycobacterium tuberculosis. Clin. Microbiol. Infect. 10:289-294. [DOI] [PubMed] [Google Scholar]
- 23.Telenti, A., P. Imboden, F. Marchesi, D. Lowrie, S. Cole, M. J. Colston, L. Matter, K. Schopfer, and T. Bodmer. 1993. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet 341:647-650. [DOI] [PubMed] [Google Scholar]
- 24.Toungoussova, O. S., D. A. Caugant, P. Sandven, A. O. Mariandyshev, and G. Bjune. 2004. Impact of drug resistance on fitness of Mycobacterium tuberculosis strains of the W-Beijing genotype. FEMS Immunol. Med. Microbiol. 42:281-290. [DOI] [PubMed] [Google Scholar]
- 25.Tracevska, T., I. Jansone, V. Baumanis, O. Marga, and T. Lillebaek. 2003. Prevalence of Beijing genotype in Latvian multidrug-resistant Mycobacterium tuberculosis isolates. Int. J. Tuberc. Lung Dis. 7:1097-1103. [PubMed] [Google Scholar]
- 26.Van Embden, J., M. Cave, J. Crawford, J. Dale, K. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, and T. Shinnick. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Werngren, J., and S. E. Hoffner. 2003. Drug-susceptible Mycobacterium tuberculosis Beijing genotype does not develop mutation-conferred resistance to rifampin at an elevated rate. J. Clin. Microbiol. 41:1520-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]