Abstract
The objectives of this study were to elucidate spatial and temporal dynamics in source-specific Bacteroidales 16S rRNA genetic marker data across a watershed; to compare these dynamics to fecal indicator counts, general measurements of water quality, and climatic forces; and to identify geographic areas of intense exposure to specific sources of contamination. Samples were collected during a 2-year period in the Tillamook basin in Oregon at 30 sites along five river tributaries and in Tillamook Bay. We performed Bacteroidales PCR assays with general, ruminant-source-specific, and human-source-specific primers to identify fecal sources. We determined the Escherichia coli most probable number, temperature, turbidity, and 5-day precipitation. Climate and water quality data collectively supported a rainfall runoff pattern for microbial source input that mirrored the annual precipitation cycle. Fecal sources were statistically linked more closely to ruminants than to humans; there was a 40% greater probability of detecting a ruminant source marker than a human source marker across the basin. On a sample site basis, the addition of fecal source tracking data provided new information linking elevated fecal indicator bacterial loads to specific point and nonpoint sources of fecal pollution in the basin. Inconsistencies in E. coli and host-specific marker trends suggested that the factors that control the quantity of fecal indicators in the water column are different than the factors that influence the presence of Bacteroidales markers at specific times of the year. This may be important if fecal indicator counts are used as a criterion for source loading potential in receiving waters.
Natural waters contaminated with feces harbor pathogens and are a significant risk to human health (3, 15, 27). Fecal pollution also leads to economic losses due to closure of shellfish-harvesting and recreational areas. Federally regulated fecal detection methods rely on enumeration of fecal indicator bacteria in the water column (Escherichia coli and enterococci for recreational waters [30] and fecal coliforms for shellfish waters). However, fecal indicator counts do not discriminate between different animal sources, and therefore the origin of fecal contamination cannot be determined. Point and nonpoint sources of contamination include agricultural practices, storm drainage, failing septic tanks, overloads at sewage treatment facilities, recreational water use, and native wildlife. Understanding the sources of fecal pollution has become critical for assessing associated health risks, for developing management plans to protect recreational waters, and for preserving the integrity of drinking water supplies.
The need to identify the sources of fecal pollution in affected watersheds has led to intense study of fecal source tracking (FST) methods (for reviews, see references 16, 24, 27, and 28). FST methods are now being used to develop total maximum daily load definitions mandated by the Clean Water Act of 1972 and to evaluate the best management practices across the United States. One of the most widely used FST approaches is a PCR-based method that targets host-specific Bacteroidales 16S rRNA genes (4, 5). This technique has been used successfully to discriminate between ruminant and human fecal sources in fresh and marine waters (5, 8, 17, 19), and it can also identify fecal sources such as dogs, pigs, and horses (12, 14).
However, it is still not clear how FST methods such as the host-specific Bacteroidales 16S rRNA gene technique relate to measurements of fecal indicators in natural waters. Building databases that can compare these two water quality-monitoring strategies requires a large number of water samples collected over an extended period of time. The Tillamook basin in Oregon has a long history of water impairment by fecal bacteria from ruminant and human sources and contains both well-defined point sources and nonpoint sources of contamination (4, 5, 7, 25). Genetic markers targeting ruminant-specific and human-specific Bacteroidales 16S rRNA genes were first developed using fecal and water samples collected from this watershed (4, 5).
The primary objectives of this study were to elucidate spatial and temporal dynamics in source-specific Bacteroidales 16S rRNA genetic marker data across a watershed; to compare these dynamics to fecal indicator counts, general measurements of water quality, and climatic data; and to identify sites with intense exposure to specific sources of contamination. Insights obtained from inclusion of source-specific genetic marker data identified factors that influence fecal pollution trends in the Tillamook basin.
MATERIALS AND METHODS
Site description.
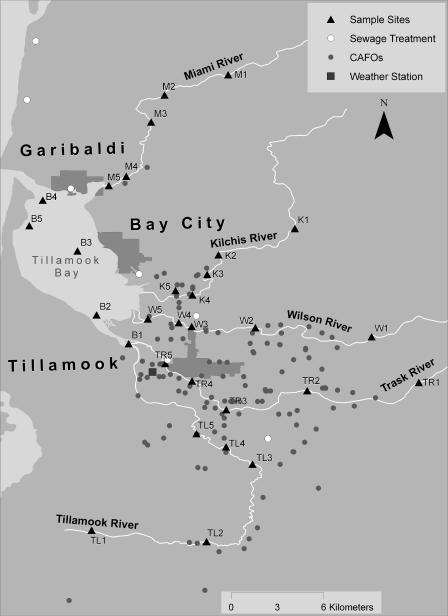
Tillamook Bay is a shallow estuary situated along the north coast of Oregon. Its watershed covers nearly 150,000 ha and consists of six major bodies of water, including the Tillamook Bay and the Miami, Kilchis, Wilson, Trask, and Tillamook rivers (Fig. 1). About 90% of the watershed is forested, and about 8% is used for agriculture. The slopes range from 0 to 90%. The soils are varied and range from deep, well-drained, coarse-textured bottomland soils with high permeability and slow runoff to well-drained, fine-textured upland soils with moderate permeability and medium to rapid runoff. The human population in the watershed has remained relatively stable in the past few decades (62,000 according to the 1994 census), but the dairy cow population has doubled in the last 15 years to approximately 30,000 animals. Fecal bacteria may enter the watershed from the 185 permitted confined-animal-feeding operations (CAFO), numerous other agricultural animal sites, wastewater treatment plants (WWTPs), public campgrounds, rural onsite septic systems, and native animals. The bay includes an estimated 3,590 ha of surface water at high tide and is affected by aquaculture, agriculture, recreational use, and two WWTPs located near Bay City and Garibaldi. Both agriculture and rural residential areas affect each river. Point sources of fecal contamination are found on the Trask River (City of Tillamook and Port of Tillamook WWTPs), the Wilson River (Tillamook Creamery Association outfall, WWTP, and public campground), and the Tillamook Bay (Garibaldi and Bay City WWTPs). Fecal pollution is currently monitored in the watershed with standard E. coli count methods (25). Counts greater than a 5-day geometric mean of 126 organisms per 100 ml are considered unsafe for recreational use (30).
FIG. 1.
Geographic information systems map of the Tillamook basin, showing the locations of sampling sites, sewage treatment plants, CAFO facilities, and a weather station.
Analytical analyses.
Each of the five rivers and the bay had five sampling sites (Fig. 1). The river sites were located where roads or bridges provided access. Water samples (n = 2,912) were collected bimonthly from the 30 sites from 14 March 2001 through 19 March 2003. Samples were collected in triplicate for the first year, from 14 March 2001 to 13 March 2002 (n = 2,194). Samples were collected in sterile 1-liter containers from surface water and were immediately stored on ice during transport to the laboratory.
E. coli counts (most probable number [MPN] per 100 ml of water sample) were obtained using Colilert Quantitray (Idexx, Westbrook, ME). The surface water temperature and pH were recorded, and the turbidity (expressed in formazin turbidity units) for one water sample collected from each site was determined using a HACH DR/2000 spectrophotometer. Precipitation (in inches) was monitored daily at a canyon station on the Wilson River situated at the first orographic lift point on the Coastal Range (Fig. 1).
Previous studies demonstrated that 100-ml water samples gave detection levels similar to those for indicator bacterium assays (5, 6, 13, 17). Therefore, for each sample, 100 ml of water was filtered through 0.2-μl-pore-size Supor-200 filters (Whatman). The filters were placed in sterile 15-ml tubes containing 500 μl of GITC buffer (5 M guanidine isothiocyanate, 100 mM EDTA [pH 8.0], 0.5% Sarkosyl) and stored at −80°C. Water samples were filtered in the Tillamook County Performance Partnership laboratory (Garibaldi, OR) and were transported to Oregon State University (Corvallis, OR) for molecular analysis. We recovered DNA from filters using DNeasy 96 tissue DNA extraction kits (QIAGEN). Seven hundred microliters of AL buffer was added to thawed tubes and vigorously agitated for 1 min. Buffer mixtures were transferred to wells in DNeasy 96 plates; the tubes and filters were discarded. The plates were sealed with AirPore tape and centrifuged at 4,700 rpm for 15 min. The plates were washed with AW1 buffer once and with AW2 buffer twice according to the manufacturer's protocol. DNA was eluted with 100 μl of AE buffer into low-retention 96-well PCR microplates (Axygen Scientific, Inc., Union City, CA) and stored at −20°C.
Amplification reaction mixtures (25 μl) contained 1× ExTaq PCR buffer (Panvera, Madison, WI), 200 μM (each) dATP, dCTP, dGTP, and dTTP, 0.2 μM forward primer CF128, CF193, HF134, HF183, EF447F, or 32F (4, 5, 14), 0.2 μM reverse primer 708R (4) or EF990R (14), 0.06% bovine serum albumin, 0.625 U of ExTaq enzyme, and 1 μl of sample DNA. The reaction mixtures were denatured at 94°C for 2 min, and this was followed by 35 cycles of 94°C for 1 min, 53°C (primer 32F), 62°C (primers CF128, CF193, and EF447F), or 63°C (primers HF134 and HF183) for 1 min, and 72°C for 1.5 min. One-microliter portions of purified 32F/708 PCR products (QiaQuick PCR product clean-up kit [QIAGEN]) amplified from water samples collected on 27 June 2001, 25 July 2001, 8 August 2001, 22 August 2001, 3 October 2001, 2 January 2002, 16 January 2002, 13 February 2002, 27 February 2002, and 13 March 2002 were used as templates for human- and ruminant-host-specific amplification. All samples that were positive for either ruminant marker were analyzed by PCR with elk-specific primers EF447F and EF990R (14). Amplification was performed in 96-well PCR microplates by using Hybaid HBPX110 and HBPX2 thermal cyclers (Thermo Electron Corporation, Somerset, NJ). PCR products were separated by electrophoresis on 96-well Precast Ready-to-Run 2.2% agarose gels in a Ready-to-Run separation unit (Amersham Biosciences, Piscataway, NJ), and the results were recorded with a UVP gel imager (UVP, Upland, CA).
PCR limits of detection were determined using ruminant- and human-specific 16S rRNA Bacteroidales gene fragments amplified from a Tillamook Bay water sample. Fragments were inserted into pCR2.1 TOPO vectors (Invitrogen, Carlsbad, CA) and used to transform E. coli TOPO SureShot cells (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. The resulting recombinant plasmids (pGenBac, pHF134, pHF183, pCF128, and pCF193) were diluted in TE buffer (10 mM Tris, 0.1 mM EDTA; pH 8.0) to generate samples that contained approximately 1, 10, 102, 103, and 104 molecules of template DNA (104 molecules of pCR2.1 [∼3,900 bp] was equivalent to 4.2 × 10−5 ng of DNA).
PCR inhibition was assessed by using assay mixtures that included 100 copies of pHF183 as the template and 2 μl of DNA extracts that were negative for human-specific Bacteroidales 16S rRNA markers. Water samples used for DNA extraction (n = 19) were collected from the Tillamook River (9 September 2003 through 4 December 2003) and contained various amounts of E. coli (range, 1 to >2,400 MPN/100 ml).
Contamination controls.
Each stage of the research was carried out in separate laboratories, either at Tillamook County Performance Partnership or Oregon State University. Contaminating DNA from equipment, other samples, and amplification products was limited by using physical barriers and dedicated equipment. For each sample batch analyzed, we included at least one mock extraction, purification, and amplification using buffer alone. For every 96-well PCR, we performed at least three no-template amplifications with purified water substituted for template DNA.
Statistical analysis.
Triplicate data for each sampling event were averaged to create a single value for each variable on a given sampling date. The pooled data set (n = 1,500) was used for all statistical analyses unless indicated otherwise. Host-specific genetic markers for ruminants (CF128 and CF193) and humans (HF134 and HF183) were combined by group within a sampling event into two new variables, RUM and HUM, for ruminants and humans, respectively. The RUM and HUM presence/absence values were 1 if at least one genetic marker for the group was found. Sampling stations in each river were coded upstream to downstream; bay stations were coded from south to north and from east to west. Statistical analyses to determine the significance of sample location were performed by nesting sampling sites within water bodies. Daily precipitation data were added for 5 days up to and including the day of sampling (5-day precipitation). To determine whether replicate sampling significantly increased the accuracy of host-specific marker and E. coli detection, we used chi-square tests to compare the frequencies of detection after addition of duplicate and triplicate measurements for all samples containing triplicate data (n = 749).
General relationships among continuous variables were identified using Pearson correlations for the water quality variables and 5-day precipitation. Factorial analysis of covariance (ANCOVA) models were used to test for the effects of body of water, month, and sampling location within a body of water on water temperature, pH, and turbidity. Five-day precipitation was used as a covariate. The statistical output included least-square means with adjusted pairwise comparisons using the Tukey option in SAS (SAS, Cary, NC). We used an ANCOVA model to test for the effects of body of water, month, and spatial location within a body of water on E. coli counts, with water temperature, pH, 5-day precipitation, and turbidity as the covariates. RUM and HUM were also included in the model. E. coli counts were log transformed before analysis to meet assumptions of normality.
Logistic regression was used to analyze the RUM and HUM presence/absence data. The logistic regressions related the proportions of RUM and HUM to the independent variables body of water, month, spatial location within a body of water, E. coli count, and the other water quality variables in a stepwise algorithm for testing significant effects. Multicolinearity among continuous variables was examined as suggested by Allison (1). The maximum-likelihood method was used to fit the data with the LOGISTIC procedure in SAS. Wald chi-square statistics (significance, P < 0.05) were used to test the global null hypothesis (i.e., the specified factorial combination had no effect on the proportions of RUM and HUM) and the relative significance of individual factors used in the model. The returned logit estimates from the logistic regressions were used to calculate p̂ as follows:
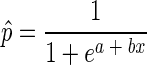 |
where a and b are parameter estimates (one b for each significant factor remaining in the model) and p̂ is the predicted probability of detecting a ruminant or human marker given significant effects remaining in the models.
Chi-square tests were also used to test for significant departures at individual sampling sites from an average marker frequency. This procedure was primarily used to help verify a specific source (RUM and/or HUM), if E. coli counts suggested that there were elevated loads of fecal bacteria at a sampling site.
RESULTS
Method validation.
PCRs performed with primers 32F, HF134, HF183, and CF128, all paired with primer 708R, routinely detected 100 copies of the pGenBac, pHF134, pHF183, and pCF128 templates, respectively. The CF193/708R primer set routinely detected 10 copies of the pCF193 template. No PCR inhibition was detected regardless of the E. coli count. Of the 1,258 no-template and extraction blank PCR control reactions 1,255 (99.8%) were negative. PCR products were detected in one no-template control and one extraction blank using the CF128/708R primer set and in one no-template control using the 32F/708R primer set.
The initial analysis of water samples in this study yielded both positive and negative results for each genetic marker, indicating that the 100-ml sample volume established in previous studies allowed isolation of detectable and nondetectable quantities of target DNA without coextraction of other substances that inhibit the PCR assay.
Replicate sampling in large-scale studies can be expensive and time-consuming. Therefore, we analyzed 1 year of triplicate samples to test whether replication significantly increased the frequency of detection of high E. coli counts (>126 MPN/100 ml) and host-specific markers. We found no significant increase in the frequency of detection of either host-specific markers or samples with an E. coli count greater than 126 MPN/100 ml when replicate samples were analyzed. The 95% confidence intervals for the returned frequencies were ±2.6%, ±3.5%, and ±3.3% for RUM, HUM, and E. coli counts of >126 MPN/ml, respectively, when only the first replicate was used. The confidence intervals decreased by 1.1, 1.5, and 1.5%, respectively, when all three replicates were considered. The frequencies for replicated and single-measurement analyses were not significantly different. Based on these results and given the logistical and time constraints imposed by collecting and performing additional analyses, site by event level replication was discontinued after the first year of the study.
Climate and water quality trends in the Tillamook basin.
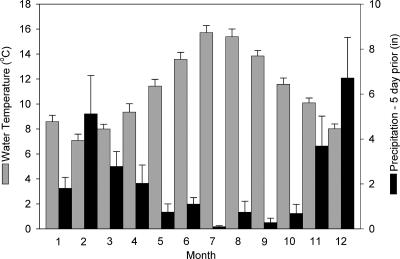
On the Oregon coast the water temperature rises in the summer months; the upward trend begins in May, the maximum occurs in July, and the temperature decreases to a relatively consistent value by November (Fig. 2). Temperature is negatively correlated with precipitation; a wet winter season begins in October and lasts through March, and this is followed by a drier summer season. In the 2 years of this study, rainfall that occurred during the 5 days leading up to a sampling event followed this general climatic trend, with maximums in December and February and a minimum in July. The reciprocal of this pattern was observed for the water temperature data.
FIG. 2.
Mean monthly values for water temperature and cumulative precipitation for the 5 days leading up to and including the day of a sampling event, from the pooled data set including the period from 14 March 2001 to 19 March 2003. The error bars indicate 95% confidence intervals.
ANCOVA results for the continuous water quality variables for a body of water suggested potentially important differences among the variables that may influence microbial dynamics. In the rivers, the water temperature generally increased toward the river mouth, and the Wilson River was significantly warmer than the other rivers (Table 1). The temperatures in the bay were higher than the temperatures in all of the rivers except the Wilson River throughout the year. The water in the bay tended to be colder toward the ocean inlet except during the winter months, when the opposite trend was observed. Turbidity generally increased downstream except in the Tillamook River and the bay, where there was significantly higher turbidity and no clear trend among sampling locations (Table 1). The turbidity in the Miami and Kilchis rivers was significantly lower than that in the other bodies of water (Table 1). In the summer months the turbidity was lower but more variable than the turbidity during the rest of the year.
TABLE 1.
Summary of statistics for water quality and microbial marker variables in different bodies of watera
| Body of water |
E. coli counts (MPN · 100 ml−1)
|
Ruminant markers
|
Human markers
|
Turbidity (formazin turbidity units)
|
Temp (°C)
|
pH
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Geometric mean | Lower 95% CI | Upper 95% CI | Frequency | 95% CI | Frequency | 95% CI | Geometric mean | Lower 95% CI | Upper 95% CI | Mean | 95% CI | Mean | 95% CI | |
| Tillamook Bay | 38.38 A | 8.77 | 11.28 | 0.76 A | ±0.05 | 0.65 A | ±0.06 | 4.38 A | 0.32 | 0.35 | 11.61 A | ±0.50 | 7.26 A | ±0.05 |
| Kilchis River | 38.97 A | 8.40 | 10.64 | 0.72 A | ±0.06 | 0.27 B | ±0.06 | 1.22 B | 0.15 | 0.17 | 10.24 B,C,D | ±0.39 | 6.89 B | ±0.04 |
| Miami River | 37.16 A | 6.82 | 8.31 | 0.76 A | ±0.05 | 0.28 B | ±0.06 | 0.95 C | 0.10 | 0.11 | 9.83 B,C | ±0.28 | 6.90 B | ±0.03 |
| Tillamook River | 242.83 B | 54.81 | 70.69 | 0.81 A | ±0.05 | 0.40 B,C | ±0.06 | 3.13 D | 0.29 | 0.32 | 10.20 B,C,D | ±0.42 | 6.76 C | ±0.04 |
| Trask River | 78.04 C | 13.48 | 16.26 | 0.77 A | ±0.05 | 0.51 C | ±0.06 | 2.75 D | 0.36 | 0.41 | 10.63 B,D | ±0.48 | 6.99 D | ±0.04 |
| Wilson River | 25.13 D | 4.66 | 5.66 | 0.68 A | ±0.06 | 0.25 B | ±0.05 | 2.07 E | 0.28 | 0.32 | 13.20 E | ±0.75 | 7.13 E | ±0.04 |
For continuous variables, the results are the least-square means (back-transformed for E. coli and turbidity). Within a column, values followed by the same letter are not significantly different at an alpha of 0.05 based on corrected multiple ’pairwise’ comparison procedures. CI, confidence interval.
There were significant differences in pH among bodies of water, suggesting that there was an annual cycle. However, the interaction terms were also significant (site by month), which made it difficult to generalize the main effects. The bay had the highest pH, which reflected the higher salinities driven by oceanic exchange (6). The Wilson River had the next highest pH followed by the Trask River (Table 1). The Wilson-4 sampling station (Fig. 1) ranked unusually high in temperature, turbidity, and pH, and this was consistent throughout the year.
E. coli analyses in the Tillamook basin.
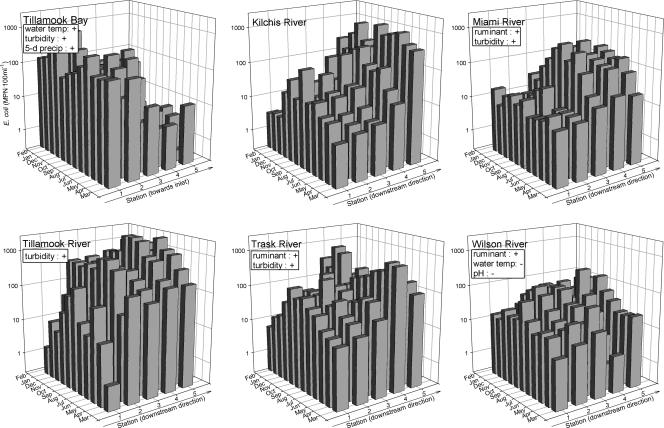
Pearson correlation coefficients suggested significant positive relationships between E. coli counts and both water temperature and turbidity and a negative relationship between E. coli counts and pH. However, E. coli count differences among bodies of water did not simply reflect the dominant water quality trends shown in Table 1. For example, the Tillamook River had the highest geometric mean E. coli count among the bodies of water, while it ranked the second lowest, second highest, and lowest in water temperature, turbidity, and pH, respectively. The ANCOVA test for E. coli counts revealed significant differences among bodies of water (Table 1), indicated a significant month effect, and highlighted the trend toward increasing E. coli counts downstream (Fig. 3). Therefore, we performed separate tests to identify significant effects for each body of water, and we used month, location, the interaction of the month and location, RUM, HUM, water temperature, 5-day precipitation, pH, and turbidity as test factors in the ANCOVA. Month and location were significant for each body of water (Fig. 3). Although not consistent across all rivers, E. coli counts appeared more likely to be linked to a ruminant source than to a human source and to increase with turbidity. In the bay, E. coli counts were significantly related to water temperature and 5-day precipitation.
FIG. 3.
ANCOVA results for E. coli counts. Of the factors month, location, RUM, HUM, water temperature, 5-day precipitation, pH, and turbidity, only month and location were significant for each body of water; month and location are plotted against E. coli counts. The information in boxes indicates additional significant effects and the directions of the effects (a plus sign for a continuous variable indicates an effect that is positively correlated with the E. coli count, and a minus sign indicates an effect that is negatively correlated).
At the uppermost sampling stations of each river, E. coli counts increased with the increase in water temperature from April through October (Fig. 2). This summer pattern decreased in the downstream direction, along with increasing E. coli counts (Fig. 3). In the bay, E. coli counts decreased drastically as the stations progressed from the Tillamook River and Trask River outlets to the ocean inlet. The positive effects of temperature and precipitation in the bay were reflected in the higher E. coli counts in the spring and fall (Fig. 3). Compared to the counts for the rivers, E. coli counts for the bay were lower than expected given its higher average turbidity and water temperature.
Ruminant source markers in the Tillamook basin.
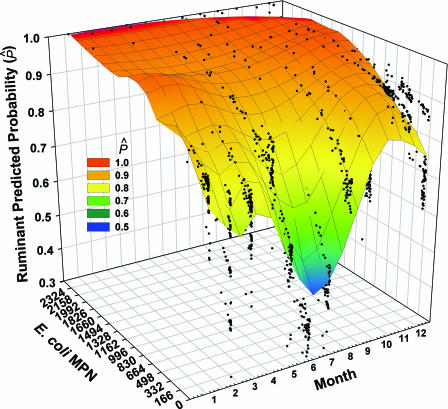
Logistic regression supported the hypothesis that there was tighter linkage between E. coli counts and ruminant source markers than between E. coli counts and human markers. The probability of detecting one of the ruminant markers was relatively high for all bodies of water in the basin; the average frequency of detection ranged from 0.68 to 0.81, and there were no significant differences among bodies of water (Table 2). The logistic regression output for RUM is summarized on a river-wide basis in Fig. 4, in which the significant relationship with the E. coli count and the monthly trend are plotted against the predicted probability of detection. When E. coli counts were more than 1,500 MPN/100 ml, the chance of detecting a ruminant marker was nearly 100%.
TABLE 2.
Stepwise logistic regression results for human markers tabulated by body of water
| Body of water | Quantiles of predicted probability
|
Model fit
|
Test of global null hypothesis
|
Significant parameter(s)a | ||||
|---|---|---|---|---|---|---|---|---|
| 10% | 90% | −2 log likelihood | Rescaled R2 | Chi square | df | P | ||
| Tillamook Bay | 0.39 | 0.9 | 272.26 | 0.2 | 37.97 | 5 | <0.0001 | Location, water temp (−) |
| Kilchis River | 0.05 | 0.58 | 231.01 | 0.3 | 57.09 | 14 | <0.0001 | Month, E. coli (+), pH (−), 5-day precipitation (−) |
| Miami River | 0.03 | 0.6 | 240.336 | 0.22 | 40.03 | 12 | <0.0001 | Month, E. coli (+) |
| Tillamook River | 0.29 | 0.56 | 268.11 | 0.08 | 11.96 | 2 | 0.0025 | pH (+), 5-day precipitation (+) |
| Trask River | 0.25 | 0.93 | 264.65 | 0.29 | 55.54 | 5 | <0.0001 | Location, water temp (−) |
| Wilson River | 0.14 | 0.46 | 249.13 | 0.11 | 18.47 | 1 | <0.0001 | pH (+) |
A minus sign indicates that there was a negative effect, and a plus sign indicates that there was a positive effect.
FIG. 4.
River-wide logistic regression results for RUM. A Loess smoother at 0.3 sampling proportion and first-degree polynomial was applied to the solution of the regression model (rescaled R2, 0.2415; −2 log likelihood ratio chi-square, 203.0006; P < 0.0001).
For the rivers the monthly trend for the frequency of RUM detection differed from the monthly trend for E. coli counts. Ruminant marker detection probabilities were lowest during the hottest and driest summer months, and the maximum probability occurred in the fall, at the beginning of the wet season. The predicted probability fell somewhat during the winter and rose again with temperature in the spring (Fig. 4). The level of ruminant marker detection increased to a maximum at around 4 in. of cumulative rainfall and decreased thereafter; the effect of 5-day precipitation on marker detection was best represented with a quadratic term.
To eliminate the possibility that the observed ruminant fecal contamination came from elk, which occur naturally in the basin, all samples that were positive with either ruminant primer were also tested using a set of primers that amplify elk fecal DNA but do not amplify fecal DNA from cattle (14). None of the ruminant-positive samples showed amplification when the elk primers were used.
Human source markers in the Tillamook basin.
Occurrences of human-specific markers differed significantly among the bodies of water. Overall, across the basin the frequency of detection of HUM was much lower than the frequency of detection of RUM. The frequency of HUM in the rivers ranged from 0.25 to 0.51, and the frequency of detection was significantly greater in the Trask River (0.51) and the Tillamook River (0.40). The Wilson River had the lowest frequency of HUM (0.25), and the bay had the highest frequency of HUM (0.65) (Table 1). Given these differences among bodies of water, we used separate logistic regression models to study significant effects (Table 2). In contrast to the RUM results, for HUM the E. coli count and month were significant only for the Kilchis and Miami rivers, where the probability of detection was fairly low, providing further evidence that E. coli is linked more closely to ruminant sources across the basin.
Sites with elevated levels of fecal exposure.
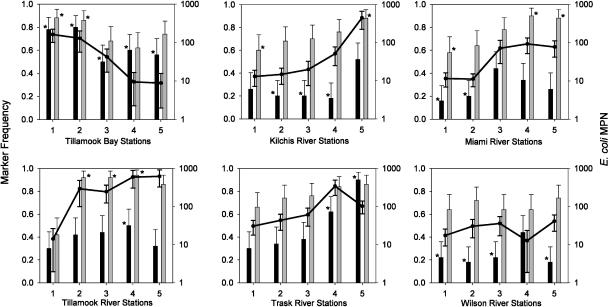
To identify sites with elevated levels of fecal exposure, the observed frequencies of HUM and RUM host-specific markers were calculated for each sampling site and plotted against the corresponding E. coli geometric means (Fig. 5). RUM frequencies were higher than HUM frequencies at all sampling sites except the Trask-5 site. At eight sampling sites the average levels of E. coli did not meet the state contact standard (>126 MPN/100 ml); these were the Bay-1 and -2, Kilchis-5, Trask-4, and Tillamook River-2, -3, -4, and -5 sites. Chi-square tests were used to identify sampling sites with significantly higher or lower levels of exposure to human and ruminant fecal sources than the river-wide frequencies (74.8 for RUM and 34.8 for HUM). For RUM, the sampling sites with frequencies significantly lower than 74.8 included three upstream sites, Kilchis-1, Miami-1, and Tillamook River-1. The sampling sites with significantly higher frequencies included Bay-1 and -2, Kilchis-5, Miami-4 and -5, and Tillamook River-2, -3, and -4. For HUM, the sampling sites with frequencies significantly lower than 34.8 included Kilchis-2, -3, and -4, Miami-1 and -2, and Wilson-2, -3, and -5. The sampling sites with significantly higher frequencies included Trask-4 and -5, Tillamook River-4, and all sites in the bay. Figure 5 also shows the general trend toward increasing frequencies for both source-specific markers in the downstream direction for all rivers, with the exception of the HUM marker in the Wilson River. This coincided with the significant increases in the E. coli count and turbidity in the downstream direction.
FIG. 5.
Geometric mean E. coli counts (lines) and RUM (shaded bars) and HUM (solid bars) frequencies for different bodies of water and sampling stations. The error bars indicate 95% confidence intervals. The asterisks indicate marker frequencies that are significantly higher or lower than the river-wide frequencies (0.75 and 0.35 for the RUM and HUM markers, respectively) at a P value of <0.05, as determined by chi-square tests.
DISCUSSION
The distribution of ruminant genetic markers fits a rainfall runoff pattern in the Tillamook basin.
The probability of detecting a ruminant marker fell to a minimum from April to July and then increased in the fall (Fig. 4), which was probably related to the annual precipitation cycle (Fig. 2). For the least-affected rivers, the Kilchis, Miami, and Wilson rivers, logistic models were best fit with a quadratic term (data not shown), indicating a tendency toward lower detection probabilities for extremes of precipitation. This suggests a source loading potential consistent with rainfall runoff models, in which pollutants build up in the landscape between rain events and are washed off during subsequent rain events. The concentration in the receiving water is then based on the duration between rain events and the quantity of water running off the landscape. This build-up and wash-off scenario has prompted runoff-monitoring programs to collect a sample of the “first-flush,” represented typically by the first 30 min of rainfall-generated runoff.
For waters where point sources were present (Tillamook River, Trask River, and the bay), the quadratic behavior was lost for both ruminant and human marker probabilities, and the marker trends were positively correlated with rainfall over the entire rainfall distribution. This could be explained by decreased wastewater management performance when rainwater infiltrated treatment systems.
Elevated E. coli counts coincide with point and nonpoint sources of fecal contamination.
Relatively low E. coli counts tended to be found in the upper portions of the five rivers (Fig. 3). Sampling sites along the Miami and Wilson rivers were least affected by high E. coli counts, and the values for all sites were below the threshold of detection (126 MPN/100 ml) on average (Fig. 5). Interestingly, although these rivers also differed significantly with respect to the general water quality variables, the differences were not systematic. The Miami River was significantly cooler and had significantly lower turbidity and pH than the Wilson River (Table 1), illustrating the difficulty of discerning dominant environmental factors influencing the counts of indicator bacteria in different rivers.
More than one-quarter of all sampling sites were in violation of the Oregon water quality standard for E. coli counts (Fig. 5). All of these sites are situated near known human point sources or agricultural operations (Fig. 1). For example, the values for four sampling sites along the Tillamook River, affected by rural residential areas and more than 30 CAFO facilities, exceeded the Oregon E. coli standards more than 75% of the time, suggesting that this portion of the river is severely polluted throughout the year. E. coli counts were also very high at two sites that were affected by urban and agriculture activities, including sampling sites that were the farthest downstream along the Kilchis River (Kilchis-5; 446 MPN/100 ml) and the Trask River (Trask-4; 345 MPN/100 ml) near a slough adjacent to the city of Tillamook. The values for two bay sites (Bay-1 and -2) routinely exceeded the recreational use standard; these sites are near the confluence of the Tillamook and Trask rivers, two of the most polluted rivers according to the E. coli counts.
Ruminant fecal pollution trends in the Tillamook basin.
Coupling the E. coli counts with source marker presence/absence methods in a fecal pollution-monitoring program offers the opportunity to tease out relative differences in source potential for “problem areas” that depart from climate-forced natural dynamics of fecal bacteria in receiving waters and that are independent of probable methodological limitations.
The enormous dairy cow population in the basin produces an estimated 325,000 tons of manure each year (25). Assuming that 11.6% of the manure is solid waste (26), the basin is affected by 38,000 tons of solids each year. This source of fecal material from the 185 permitted CAFO facilities (Fig. 1) and other animal-feeding operations consistently affected receiving waters, producing a basin-wide probability of detection of RUM that was near 75%. This percentage rose to more than 90% during periods of moderate precipitation in the spring and fall (Fig. 4), and the values coincided with elevated levels of E. coli as turbidity increased and diffuse sources accumulated in the downstream direction of most rivers. Generally, turbidity in the rivers tended to correlate with the annual precipitation pattern.
Using chi-square tests comparing ruminant-specific and river-wide genetic marker frequencies, we identified eight locations with above average levels of exposure to ruminant fecal bacteria (Fig. 5). For the Tillamook River (Tillamook-2, -3, and -4), Kilchis River (Kilchis-5), and Tillamook Bay (Bay-1) the RUM marker frequencies were greater than 90%, strongly suggesting that elevated E. coli counts originated from ruminant fecal pollution in these areas. It appeared that ruminants also contributed more to fecal loads than humans did in the lower section of the Miami River, where the RUM frequencies were greater than 88% (Miami-5 and -4) (Fig. 5), E. coli levels were near violation levels, and the sampling sites were close to the only CAFO facilities in the surrounding area.
Human fecal pollution trends in the Tillamook basin.
The human population in the basin has remained relatively stable in the past few decades, and WWTPs, public campgrounds, recreational water use, and rural onsite septic systems are the primary sources of human fecal matter. Assuming that the daily waste production is 200 g total solids per person (31), the human population produces roughly 5,000 tons of fecal source material a year, which is approximately one-seventh the amount estimated for the ruminant population. The probability of detecting a human marker (approximately 35%) was less than one-half the probability of detecting a ruminant marker for the rivers. The likelihood doubled in the bay, where known point sources (sewage outfall pipes) near the mouth of the Trask River, from Bay City, and from Garibaldi appeared to have a strong effect on the probability of detecting the human marker (Fig. 1).
By combining water quality data for E. coli with significant departures from river-wide genetic marker frequencies, “problem areas” where human sources most likely contribute to the fecal indicator bacterial loads were identified. Elevated frequencies of detection of a human marker in the downstream section of the Trask River identified a point source from a WWTP. In the Wilson River, where the human marker frequencies at most sites were significantly lower than the average frequency, the human marker frequency at the Wilson-4 site was higher. This illustrates how FST data can extend the amount of information available to make water management decisions. Even though there was no apparent increase in E. coli counts at the Wilson-4 site, the FST data suggested that there was an increased level of exposure to human fecal contamination. This finding and the higher-than-average water temperatures at this site are likely to be linked to the WWTP and factory water discharge at the Tillamook Creamery (a large cheese manufacturer). Recently, the Tillamook Creamery has begun using a cooler and running discharge water through a wetland before it is released to the river; thus, both human fecal marker frequencies and water temperature are likely to have improved since the samples for this study were collected.
Elevated human marker frequencies also occurred in the Kilchis River (Kilchis-5), Miami River (Miami-3), and Tillamook River (Tillamook-4). Interestingly, the Bay-1 site was the only location with elevated E. coli, RUM, and HUM values, suggesting that the impact of the upstream WWTP on the Trask River (Trask-5) and ruminant fecal loads from the Tillamook River blended together at the confluence of these two bodies of water.
Inconsistencies between E. coli counts and Bacteroidales fecal markers.
In upper sections of the rivers, the E. coli counts increased during the summer months, and the upward trend began around April (Fig. 3); the counts trended negatively with precipitation data. At the same time, the probability of detecting a ruminant marker decreased to a minimum from April to July and then increased in the fall (Fig. 4), showing a lack of congruence with the E. coli data. Ruminant source loading was a product of rainfall runoff, while the bacterial indicator concentrations changed independent of runoff events. As sources were concentrated in the downstream direction, the seasonal trend for E. coli was dampened. This lack of congruence could be explained either by differential persistence and survival of E. coli and Bacteroidales or by a shift to primarily nonruminant, non-runoff-based fecal contamination in the summer.
There is increasing evidence that E. coli cells grow in a variety of extraintestinal habitats (2, 11, 18, 32, 33); in some cases, E. coli may persist in the environment long after the organism is introduced into the water column. Environmental persistence or growth during summer months could confound the interpretation of baseline dynamics. In addition, little is known about the long-term persistence of Bacteroidales molecular markers. If Bacteroidales cells were more sensitive to temperature increases than E. coli cells are, higher temperatures in the summer could lead to increased die-off of the ruminant markers.
Alternatively, a summer decrease in ruminant fecal contamination due to decreased runoff may coincide with an increase in fecal contamination due to fauna other than ruminants and humans. These sources would then be significant sources of E. coli, increasing incrementally during the summer. This could reflect a seasonal increase in wildlife, such as migratory birds, during these months.
A second inconsistency is that on average, the probability of detecting a human marker in Tillamook Bay was double the probability of detecting a human marker in the rivers, and for two bay sites the probability of detecting a ruminant marker was significantly different than the river-wide probability. The increases in the probability of detecting a host-specific genetic marker in the bay were not reflected in the E. coli counts, for which the bay appeared to be no different from the least-affected rivers.
Salinity has been shown to negatively affect E. coli survival, while it has less effect on the persistence of certain pathogens (1a, 2, 8, 20, 22). This is one reason why enterococci have replaced E. coli as the fecal indicator in marine waters. However, fecal coliforms are still used as an indicator of fecal contamination in shellfish waters. Regardless of the explanation for the low E. coli counts in the bay, the marker data suggest that there are significant contributions from ruminant and human sources that are above the background level. This contrasts with patterns observed in the rivers, where elevated probabilities of detecting one or both markers coincided with elevated E. coli counts. Hence, the suggested matrix effect on E. coli counts may give a false impression for the relative source loading potential if E. coli counts are considered to be directly proportional to the strength of the source.
The fact that the level of detection of Bacteroidales ruminant and human markers was high compared to the E. coli counts in the bay suggests that the seawater matrix affects these markers less. We previously showed that seawater did not affect the PCR detection limit for HF134, HF183, CF128, and CF193 (6, 12, 17), as well as two other Bacteroidales host-specific markers, PF163 and DF475 (6, 12, 17), but we did not examine the effect of seawater on survival of the marker cells. However, PCR assays could in theory detect dead cells, in contrast to indicator bacteria assays that depend on live cells. To use Bacteroidales source-specific markers to directly estimate the risk to human health, therefore, more information is needed about the survival of the markers and their correlation to pathogens.
Potential benefits of fecal source tracking.
Microbial source tracking studies are frequently undertaken either to identify fecal sources in order to target cleanup or to prioritize areas for cleanup based on perceived human health risk. Counts of fecal indicator bacteria lump together many different potential sources of fecal contamination, which may contain completely different associated pathogens. Human and animal feces both pose threats to human health. The human health risk from domestic and agricultural animal feces is usually assumed to be less than the risk from human feces, in part because viruses, which are the most common cause of human illnesses from exposure to fecal contamination in water, are highly host specific. The disease risk due to contamination by feces of wild animals, such as gulls, is poorly understood.
Water quality standards were established based on the results of epidemiological studies that measured human health outcomes following recreational exposure to human-derived fecal contamination. Few studies have been carried out to determine the risk due to animal feces as a source of waterborne zoonotic infections (29). In a Hong Kong study, the rates of illness for two marine beaches affected by animal (pig) wastes were lower than the rates of illness for seven other beaches (9, 21). In a New Zealand study of seven marine beaches, no substantial differences in the risk of illness were found for human waste- and animal waste-affected beaches, although both beaches differed from the control beaches (23). A recent exposure study performed at Mission Bay in California (10) demonstrated a much lower level of human illnesses than expected considering the levels of fecal indicator bacteria. A follow-up microbial source tracking study showed that the primary source of the fecal indicator bacteria at Mission Bay was nonhuman, most likely water birds (unpublished data). These results underscore the need for larger epidemiological studies to measure human health risks due to animal fecal contamination.
Even if microbial source tracking shows that fecal contamination is completely animal derived, current regulations do not usually allow a higher permitted level of fecal indicator bacteria. Hence, the benefits of microbial source tracking at this time are only that it allows the source or sources of fecal contamination to be accurately assigned, located, and corrected.
In the Tillamook watershed, although in many cases the human marker frequencies increased with elevated E. coli values, the increases never occurred in the absence of high ruminant marker frequency. Nowhere in the watershed did a significantly higher human marker frequency coincide with a significantly lower ruminant marker frequency. Hence, a watershed manager's best strategy for decreasing indicators of fecal pollution in this watershed is to mitigate runoff from ruminant sources.
Acknowledgments
We gratefully acknowledge assistance from volunteers in the Tillamook Bay watershed. Staff members at the Tillamook Bay National Estuary/Tillamook County Performance Partnership contributed administrative and logistical support. Cartography was provided by Molly Reif and Eric O'Neal of SoBran, Inc. We thank Sarah Walters and Linda Dick for helpful discussions.
This work was partially supported by grant 00-S1130-9818 from the U.S. Department of Agriculture, by grant R82-7639 from the U.S. Environmental Protection Agency, and by grant NA76RG0476 (project R/ECO-04) from the National Oceanic and Atmospheric Administration to the Oregon Sea Grant College Program.
REFERENCES
- 1.Allison, P. A. 1999. Logistic regression using the SAS system: theory and application, p. 48-51. SAS Institute, Cary, N.C.
- 1a.Anderson, I. C., M. Rhodes, and H. Kator. 1979. Sublethal stress in Escherichia coli: a function of salinity. Appl. Environ. Microbiol. 38:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, K. L., J. E. Whitlock, and V. J. Harwood. 2005. Persistence and differential survival of fecal indicator bacteria in subtropical waters and sediments. Appl. Environ. Microbiol. 71:3041-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balarajan, R., V. Soni Raleigh, P. Yuen, D. Wheeler, D. Machin, and R. Cartwright. 1991. Health risks associated with bathing in sea water. Br. Med. J. 303:1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernhard, A. E., and K. G. Field. 2000. Identification of nonpoint sources of fecal pollution in coastal waters by using host-specific 16S ribosomal DNA genetic markers from fecal anaerobes. Appl. Environ. Microbiol. 66:1587-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernhard, A. E., and K. G. Field. 2000. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl. Environ. Microbiol. 66:4571-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernhard, A. E., T. Goyard, M. T. Simonich, and K. G. Field. 2003. Application of a rapid method for identifying fecal pollution sources in a multi-use estuary. Water Res. 37:909-913. [DOI] [PubMed] [Google Scholar]
- 7.Blair, T., and K. Michener. 1962. Sanitary survey of Tillamook Bay and sanitary significance of the fecal coliform organism in shellfish growing area waters. Oregon State Board of Health, Portland.
- 8.Boehm, A. B., J. A. Fuhrman, R. D. Morse, and S. B. Grant. 2003. Tiered approach for identification of a human fecal pollution source at a recreational beach: case study at Avalon Bay, Catalina Island, California. Environ. Sci. Technol. 37:673-680. [DOI] [PubMed] [Google Scholar]
- 9.Cheung, W. H. S. (ed.). 1988. Health effects of beach water pollution in Hong Kong, vol. 88. Vincent Blue Copy Co., Hong Kong. [DOI] [PMC free article] [PubMed]
- 10.Colford, J. M., T. J. Wade, K. C. Schiff, C. Wright, J. F. Griffith, S. K. Sandhu, and S. B. Weisberg. 2005. Recreational water contact and illness in Mission Bay, California. Technical report 449. Southern California Coastal Water Research Project, Westminster, Calif.
- 11.Desmarais, T. R., H. M. Solo-Gabriele, and C. J. Palmer. 2002. Influence of soil on fecal indicator organisms in a tidally influenced subtropical environment. Appl. Environ. Microbiol. 68:1165-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dick, L. K., A. E. Bernhard, T. J. Brodeur, J. W. Santo Domingo, J. M. Simpson, S. P. Walters, and K. G. Field. 2005. Host distributions of uncultivated fecal Bacteroidales bacteria reveal genetic markers for fecal source identification. Appl. Environ. Microbiol. 71:3184-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dick, L. K., and K. G. Field. 2004. Rapid estimation of numbers of fecal Bacteroidetes by use of a quantitative PCR assay for 16S rRNA genes. Appl. Environ. Microbiol. 70:5695-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dick, L. K., M. T. Simonich, and K. G. Field. 2005. Microplate subtractive hybridization to enrich for Bacteroidales genetic markers for fecal source identification. Appl. Environ. Microbiol. 71:3179-3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dufour, A. P. 1984. Health effects criteria for fresh recreational waters. U.S. Environmental Protection Agency, Washington, D.C.
- 16.Field, K. G. 2004. Faecal source identification, p. 349-366. In J. A. Cotruvo, A. Dufour, G. Rees, J. Bartram, R. Carr, D. O. Cliver, G. F. Craun, R. Fayer, and V. P. G. Gannon (ed.), Waterborne zoonoses: identification, causes and control. World Health Organization, IWA Publishing Alliance House, London, United Kingdom.
- 17.Field, K. G., E. C. Chern, L. K. Dick, J. Fuhrman, J. Griffith, P. A. Holden, M. G. LaMontagne, J. Le, B. Olson, and M. T. Simonich. 2003. A comparative study of culture-independent, library-independent genotypic methods of fecal source tracking. J. Water 1:181-194. [PubMed] [Google Scholar]
- 18.Fujioka, R., C. Sian-Denton, M. Borja, J. Castro, and K. Morphew. 1999. Soil: the environmental source of Escherichia coli and enterococci in Guam's streams. J. Appl. Microbiol. 85:83S-89S. [DOI] [PubMed] [Google Scholar]
- 19.Gilpin, B., T. James, F. Nourozi, D. Saunders, P. Scholes, and M. Savill. 2003. The use of chemical and molecular microbial indicators for faecal source identification. Water Sci. Technol. 47:39-43. [PubMed] [Google Scholar]
- 20.Harwood, V. J., A. D. Levine, T. M. Scott, V. Chivukula, J. Lukasik, S. R. Farrah, and J. B. Rose. 2005. Validity of the indicator organism paradigm for pathogen reduction in reclaimed water and public health protection. Appl. Environ. Microbiol. 71:3163-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holmes, P. R. 1989. Research into health risks at bathing beaches in Hong Kong. J. Inst. Water Environ. Manag. 3:488-495. [Google Scholar]
- 22.Louis, V. R., E. Russek-Cohen, N. Choopun, I. N. Rivera, B. Gangle, S. C. Jiang, A. Rubin, J. A. Patz, A. Huq, and R. R. Colwell. 2003. Predictability of Vibrio cholerae in Chesapeake Bay. Appl. Environ. Microbiol. 69:2773-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McBride, G. B., C. E. Salmond, D. R. Bandaranayake, S. J. Turner, G. D. Lewis, and D. G. Till. 1998. Health effects of marine bathing in New Zealand. Int. J. Environ. Health Res. 8:173-189 [Google Scholar]
- 24.Meays, C. L., K. Broersma, R. Nordin, and A. Mazumder. 2004. Source tracking fecal bacteria in water: a critical review of current methods. J. Environ. Manag. 73:71-79. [DOI] [PubMed] [Google Scholar]
- 25.Newell, A. 1998. Water quality, p. 4-1-4-55. In R. Hinzman and S. Nelson (ed.), Tillamook Bay environmental characterization: a scientific and technical summary. United States Environmental Protection Agency, Garibaldi, OR.
- 26.Ohio State University. 1992. Ohio livestock manure and wastewater management guide bulletin 604. Ohio State University, Columbus.
- 27.Scott, T. M., J. B. Rose, T. M. Jenkins, S. R. Farrah, and J. Lukasik. 2002. Microbial source tracking: current methodology and future directions. Appl. Environ. Microbiol. 68:5796-5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simpson, J. M., D. J. Reasoner, and J. W. Santo Domingo. 2002. Microbial source tracking: state of the science. Environ. Sci. Technol. 36:5279-5288. [DOI] [PubMed] [Google Scholar]
- 29.Till, D., K. G. Field, and A. P. Dufour. 2004. Managing risk of waterborne zoonotic disease through water quality surveillance, p. 338-348. In J. A. Cotruvo, A. Dufour, G. Rees, J. Bartram, R. Carr, D. O. Cliver, G. F. Craun, R. Fayer, and V. P. G. Gannon (ed.), Waterborne zoonoses: identification, causes and control. World Health Organization, IWA Publishing Alliance House, London, United Kingdom.
- 30.U.S. Environmental Protection Agency. 2003. Bacterial water quality standards for recreational waters (freshwater and marine waters). Status report EPA-823-R-03-008. U.S. Environmental Protection Agency, Washington, D.C.
- 31.U.S. Environmental Protection Agency. 2002. Onsite wastewater treatment systems. Manual EPA-625-R-00-008. U.S. Environmental Protection Agency, Washington, D.C.
- 32.Wheeler Alm, E., J. Burke, and A. Spain. 2003. Fecal indicator bacteria are abundant in wet sand at freshwater beaches. Water Res. 37:3978-3982. [DOI] [PubMed] [Google Scholar]
- 33.Whitman, R. L., D. A. Shively, H. Pawlik, M. B. Nevers, and M. N. Byappanahalli. 2003. Occurrence of Escherichia coli and enterococci in Cladophora (Chlorophyta) in nearshore water and beach sand of Lake Michigan. Appl. Environ. Microbiol. 69:4714-4719. [DOI] [PMC free article] [PubMed] [Google Scholar]