Abstract
The Bcl-2/CED-9 family of proteins, which includes both antiapoptotic and proapoptotic members, plays key regulating roles in programmed cell death. We report here the identification and characterization of Drob-1, the first Drosophila member of the Bcl-2/CED-9 family to be isolated. Drob-1 contains four conserved Bcl-2 homology domains (BH1, BH2, BH3, and BH4) and a C-terminal hydrophobic domain. Ectopic expression of Drob-1 in the developing Drosophila eye resulted in a rough-eye phenotype. Furthermore, when overexpressed in Drosophila S2 cells, Drob-1 induced apoptosis accompanied by elevated caspase activity. This Drob-1-induced cell death, however, could not be antagonized by baculovirus p35, a broad-spectrum caspase inhibitor. Drob-1 was localized to the intracytoplasmic membranes, predominantly to the mitochondrial membranes, and a mutant Drob-1 lacking the hydrophobic C terminus lost both its mitochondrial localization and its proapoptotic activity. These results suggest that Drob-1 promotes cell death by inducing both caspase-dependent and -independent pathways at the mitochondria. Our identification of Drob-1 and further genetic analysis should provide increased understanding of the universal mechanisms by which the Bcl-2/CED-9 family members and other related proteins regulate apoptosis.
Programmed cell death or apoptosis, an evolutionarily conserved process of cell suicide, plays an important role in normal development, tissue homeostasis, and many pathological conditions (1, 2). Genetic studies in the nematode Caenorhabditis elegans have identified three essential components of a central cell death pathway, CED-3, CED-4, and CED-9 (3, 4). These proteins are highly structurally and functionally related to the mammalian proteins, caspases, Apaf-1, and the Bcl-2 family of proteins, respectively (5). The caspases comprise a family of cysteine proteases that participate in a proteolytic cascade, cleaving downstream caspases and a number of cellular proteins that ultimately execute apoptotic biochemical events such as DNA fragmentation and chromatin condensation (6). Apaf-1 functions at the initial step in this cascade to activate the initiator caspase, caspase-9, in the presence of cytochrome c and ATP/dATP (7, 8). The Bcl-2 family of proteins consists of both antiapoptotic and proapoptotic members, which control the cell-death decision by regulating such processes as mitochondrial cytochrome c release and caspase activation through adapter protein Apaf-1, and/or by neutralizing the effects of opposing Bcl-2 family members (9, 10). To date, at least 16 Bcl-2 family members have been identified in mammalian cells; however, their precise mechanisms and the functional interactions of each family member under physiological conditions remain to be elucidated.
In Drosophila, widespread programmed cell death occurs during embryonic development and metamorphosis and also can be induced by various stimuli, such as DNA damage (11, 12). A genetic screen has led to the identification of three genes required for Drosophila cell death (13): reaper (rpr) (13, 14), head involution defective (hid) (15), and grim (16). Products of all three genes induce apoptosis via a pathway that requires caspase action (14–16). These cell death activators have been shown to bind to the Drosophila inhibitor-of-apoptosis proteins, DIAP1 and DIAP2 (17, 18), and a recent study has shown that one mechanism by which Rpr, Hid, and Grim promote apoptosis is by disrupting inhibitor of apoptosis-caspase interactions (19). Furthermore, four Drosophila caspases, DCP-1 (20), DrICE (21), DCP-2/Dredd (22), and DRONC (23), have been characterized and implicated in the regulation of apoptosis. In addition, more recently, a Drosophila homolog of Apaf-1/CED4, Dark/Dapaf-1/HAC-1 (24, 25, 47), has been identified. Thus, a requirement for caspase function and the presence of molecules involved in the activation of a caspase cascade during Drosophila apoptotic cell death have been well documented, although no Bcl-2/CED-9 family member has been identified in flies.
In this paper, we describe the identification and characterization of Drob-1, the first Drosophila member of the Bcl-2/CED-9 family. Our observations suggest that Drob-1 is a proapoptotic member of the Drosophila Bcl-2 family that positively regulates apoptosis, through both caspase-dependent and -independent pathways.
Materials and Methods
Identification of the drob-1 cDNA.
Partial nucleotide sequences of Drosophila cDNAs homologous to Bcl-xL (26) were identified in the expressed sequence tag (EST) database of GenBank by using tblastn (National Center for Biotechnology Information). The nucleotide sequences of EST clones GH01265 (GenBank accession no. AI513093), GH01665 (GenBank accession no. AI062455), and LD12719 (GenBank accession no. AA440915) were determined by dideoxy sequencing. Clones GH01265 and GH01665 both had sequences that were identical to LD12719 except for a 2-bp deletion between 1248 and 1249. Clone LD12719 and the genomic sequences of Drob-1 (GenBank accession nos. AC007624 and AC007593) were identical. A DNA fragment containing the ORF of Drob-1 was obtained from Drosophila S2 cells and Drosophila embryos by reverse transcription–PCR (RT-PCR) using the following 5′ and 3′ untranslated region primers: 5′-ACGCTCCCTTCACGCCGGCGGACCCATGAC-3′ and 5′-CCCTCGACAGCTGTCGCGATATGTTTGTG-3′.
Generation of GMRdrob-1 Transgenic Flies.
An EcoRI fragment containing drob-1 cDNA was inserted into the pGMR (glass multimer reporter) vector (27) to generate the pGMRdrob-1 transgene. This plasmid was injected into w1118;Dr/TMS,Sb P[ry+,Δ2–3] embryos to produce transgenic flies as described (28). Several independent transformant lines were obtained. Canton-S or white1118 was used as a wild-type strain. Scanning electron microscopy and the preparation of semithin sections of adult heads were performed as described (29).
Northern Blotting, RT-PCR, and in Situ Hybridization.
Total RNA was prepared by using Trizol (GIBCO/BRL) according to the manufacturer's instructions, then fractionated by electrophoresis, blotted, and hybridized as described (25). A 32P-labeled DNA fragment containing the first 363 bases of drob-1 cDNA was used as a probe. For RT-PCR analysis, drob-1 cDNA was amplified by using the following primer set: 5′-GAACAGCATGGGCAAGGAACTG-3′ and 5′-CAATCAGGCACTGTAGGTAGTCG-3′. The primer sequences used for detection of glyceraldehyde-phosphate dehydrogenase cDNA have been described (25). Whole-mount in situ hybridization was performed as described (14).
Expression Constructs.
All expression plasmids, except for a reporter plasmid, were constructed with the pUAST expression vector, which bears five GAL4 binding sites as an upstream activation sequence (UAS) (30). The full-length Drob-1 cDNA was cloned into pUAST with an N-terminal hemagglutinin (HA) tag epitope sequence, to generate pUAS-HA-drob-1. An EcoRI fragment of HA-drob-1 was subcloned into pBluescriptII SK(+) (Stratagene), then a fragment was deleted with SmaI, and an EcoRI–XbaI fragment with a TAG termination sequence released from the subclone was inserted into pUAST, creating a C-terminal deletion mutant-expressing vector, pUAS-HA-drob-1ΔTM. cDNA fragments encoding Rpr, p35, and DIAP2, which were released from pCaspeR-hs-reaper, -p35, or -diap2 (29), were inserted into pUAST to generate pUAS-rpr, -p35, and -diap2. A cDNA fragment encoding hid was obtained by RT-PCR from Drosophila embryos, and the fragment was inserted into pUAST to generate pUAS-hid. pUAS-mDRONC was constructed with a cDNA fragment encoding the active-site mutant (C318G) of DRONC that was excised from pcDNA3-dronc (C318G) (23). An expression vector bearing GAL4 binding regulatory sequences (UAS), G4E/hGH (31), was used to construct the β-galactosidase-expressing plasmid, pUAS-LacZ (named pM91). pUAS-GFP (green fluorescent protein) was a kind gift from R. Niwa, Kyoto Univ., Kyoto, Japan (32, 33). A driver plasmid that expresses GAL4 under the control of the actin5C promoter, pWAGAL4 (Y. Hiromi, personal communication), was a kind gift from Y. Hiromi, National Institute of Genetics, Mishima, Japan. GAL4/236 gene was released from pMMTV-GAL4/236-SV40 (31) and cloned into the HindIII site of pcDNA3 (Invitrogen) to generate pcDNA3-GAL4 (named pM92). The construction of pCaspeR-hs-lacZ has been described (34).
Cell Culture, Transfection, Immunostaining, Subcellular Fractionation, and Western Blotting.
S2 cells and COS7 cells were cultured and transfected by using Cellfectin or Lipofectamine (GIBCO/BRL), as described (34). For immunofluorescence, S2 cells or COS7 cells were transfected, fixed, and immunostained with a monoclonal anti-HA antibody (12CA5, Boehringer Mannheim) and FITC-conjugated secondary antibody as described (34). For mitochondrial labeling, MitoTracker red CMXRos (Molecular Probes) was added to the cells at a concentration of 200 nM for 30 min before fixation. Images were obtained by using an Axioplan2 fluorescence microscope (Zeiss). For subcellular fractionation, lysates were made with a Dounce homogenizer from 4 × 107 S2 cells in extraction buffer: 50 mM Pipes, pH 7.9/50 mM KCl/5 mM EDTA/2 mM MgCl2/1 mM DTT/1 mM PMSF. Nuclei and unbroken cells were separated at 700 g for 10 min as the low-speed pellet (P1). This supernatant was centrifuged at 24,000 g for 10 min to collect the heavy membrane (HM) pellet. This supernatant was centrifuged at 54,000 g for 1 hr to yield the light membrane (LM) pellet and final soluble (S) fraction. The HM fraction is enriched for mitochondria. The LM fraction contains the endoplasmic reticulum and plasma membrane, and the S fraction represents the cytosol. Immunoblotting was performed as described (29) using the same antibody as above.
Cell Death Assay and Caspase Assay.
S2 cells were cultured in 24-well plates (1 × 105 cells/well) and transfected with various amounts of the indicated plasmids plus 2–2.5 ng of driver plasmid, pWAGAL4. Each transfection mixture contained 20 ng of pCaspeR-hs-lacZ, which encodes β-galactosidase under the control of the hsp70 promoter, as a reporter plasmid. Twenty four hours after transfection, cells were heat-shocked at 37°C, as described (34), and cultured at 27°C for another 24 hr. At 48 hr, cells were lysed in 300 μl of 1× Reporter lysis buffer (Promega), and each lysate was assayed for β-galactosidase activity in a reaction mixture containing 1 mg/ml o-nitrophenyl-β-d-galactopyranoside, as described (35). Caspase activity was determined at 24 hr after transfection with fluorescent enzyme substrates, DEVD-4-methyl-coumaryl-7-amide (MCA) and YVAD-MCA (Peptide Institute, Osaka), as described (29).
Results
Identification of Drob-1, a Drosophila Bcl-2/CED-9 Family Member.
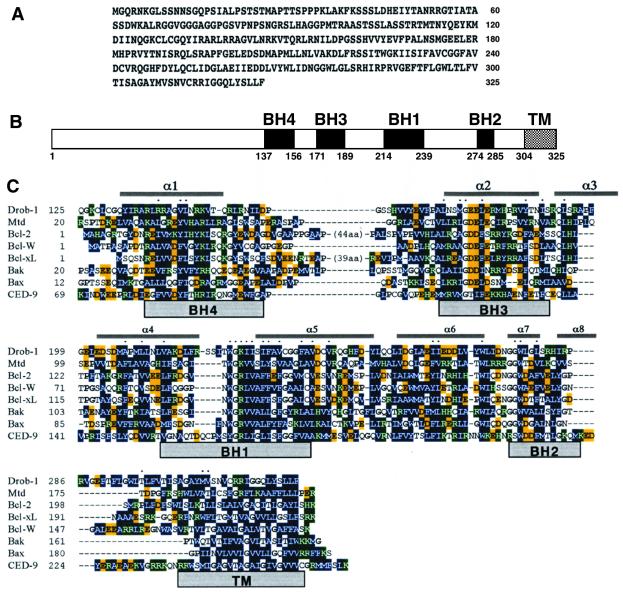
To identify Drosophila Bcl-2/CED-9 family members, we screened the GenBank database for cDNAs encoding proteins with homology to Bcl-xL (26). Using the tblastn program, we identified a Drosophila expressed sequence tag containing a nucleotide sequence with statistically significant amino acid homology to all known Bcl-2 family members. Analysis of its nucleotide sequence revealed an ORF of 325 aa with a predicted relative molecular mass of 35.4 kDa (Fig. 1A). We designated this protein as Drob-1 (Drosophila ortholog of the Bcl-2 family-1). Alignment analysis revealed that Drob-1 was a Bcl-2-related protein with eight predicted α-helices, four Bcl-2 homology (BH) domains (BH1, BH2, BH3, and BH4), and a C-terminal hydrophobic tail (Fig. 1 B and C). Drob-1 showed significant structural and amino acid homology with all known Bcl-2 family members including Bcl-2, Bcl-xL, Bcl-w, Bax, Bak, Mtd/Bok, and C. elegans CED-9 (Fig. 1C). Drob-1 was most homologous to Mtd/Bok (36, 37), a proapoptotic member of the mammalian Bcl-2 family, and also showed similar functions to this killer protein (see below).
Figure 1.
Sequence and structure of Drob-1. (A) Deduced amino acid sequence of Drob-1. (B) Schematic structure of Drob-1. The conserved BH1, BH2, BH3, and BH4 regions and the putative transmembrane (TM) region are shown by closed boxes. (C) The amino acid sequence of Drob-1 is aligned with those of mouse Mtd, human Bcl-2, Bcl-xL, Bcl-w, Bak, Bax, and C. elegans CED-9. Hydrophobic residues are shown in dark blue. Negatively and positively charged residues are highlighted in yellow and green, respectively. Proline and glycine residues (α/β breakers) are in bold. Conserved residues are indicated by dots. Residues that form the flexible loop in Bcl-2 (44 aa) and Bcl-xL (39 aa) are not included for simplicity. A transmembrane domain predicted by the Kyte and Doolittle method is indicated as TM.
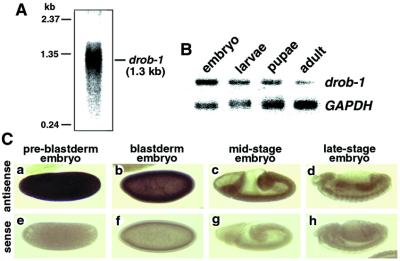
Drob-1 Is Expressed at All Stages of Development.
Northern blot analysis of drob-1 detected a 1.3-kb transcript, consistent with the size of the cDNA (Fig. 2A). RT-PCR analysis demonstrated that drob-1 mRNA was expressed at all stages of development (Fig. 2B). In addition, in situ hybridization with digoxigenin-labeled drob-1 RNA probes revealed that drob-1 was highly expressed in early-stage embryos (Fig. 2C a and b). As development proceeded, expression levels of drob-1 mRNA were reduced (Fig. 2C c and d). In the third-instar larval stages, drob-1 was expressed in leg discs, eye discs, wing discs, and midgut, but we failed to detect the signal in salivary glands and the brain (data not shown).
Figure 2.
drob-1 mRNA is detectable at all developmental stages in Drosophila. (A) The Northern blot of total RNA from adult flies showed a single 1.3-kb drob-1 transcript. (B) Total RNA from various developmental stages of Drosophila was reverse-transcribed, and PCR amplification was performed for 28 cycles with specific primers corresponding to the sequences for drob-1 or glyceraldehyde-phosphate dehydrogenase (GAPDH). (C) Whole-mount in situ hybridization to wild-type embryos was performed at various stages of development using drob-1 antisense (a–d) or sense (e–h) RNA probes.
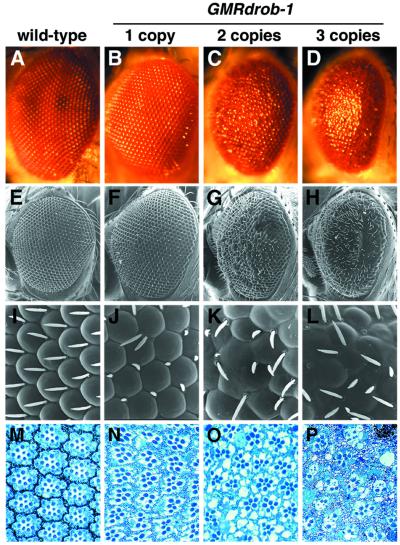
Transgenic Flies Misexpressing Drob-1 in the Eyes Exhibit a Rough-Eye Phenotype.
To assess the role of Drob-1 in regulating apoptosis in Drosophila, drob-1 cDNA was placed under the control of an eye-specific regulatory element in the pGMR transformation vector (27). Using the resulting construct, pGMRdrob-1, we generated transgenic flies that misexpress Drob-1 in the developing retina. Whereas transformants carrying a single copy of GMRdrob-1 had eyes that were slightly abnormal (Fig. 3 B, F, and J), compared with those of wild-type flies (Fig. 3 A, E, and I), severely rough eyes resulted from the increased copies of the transgene (Fig. 3 C, D, G, H, K, and L). The Drosophila compound eye is composed of an array of cell clusters, called ommatidia, each containing eight photoreceptor neurons. Tangential sections of the eyes of flies carrying the transgene showed a loss of photoreceptor morphology as well as reduced number or complete loss of the photoreceptor neurons in ommatidia in a gene dosage-dependent manner (Fig. 3 N–P). These results suggest that Drob-1 functions as a proapoptotic factor in these conditions.
Figure 3.
Ectopic expression of Drob-1 in the developing Drosophila eye causes a rough-eye phenotype. Light level (A–D), scanning electron microscopic (E–H), and its high magnification (I–L) images of the compound eyes of wild-type (A, E, and I), GMRdrob-1/Y; +/CyO (1 copy: B, F, and J), GMRdrob-1/Y; GMRdrob-1/CyO (2 copies: C, G, and K), and GMRdrob-1/Y; GMRdrob-1/GMRdrob-1 (3 copies: D, H, and L) flies are shown. Toluidine blue-stained semithin sections of wild-type (M) and transgenic fly (N-P) eyes also are shown.
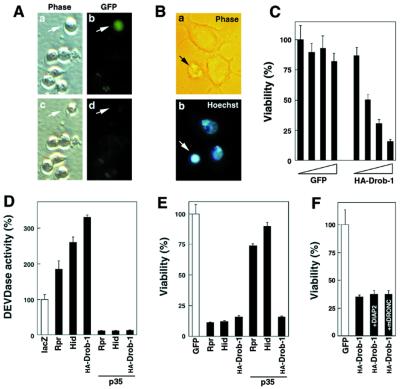
Overexpression of Drob-1 Promotes Apoptotic Cell Death and Caspase Activation in Drosophila S2 Cells.
The rough-eye phenotype seen in the GMRdrob-1 transgenic flies may have resulted from excess cell death induced by a proapoptotic function of Drob-1. To examine this possibility, HA-tagged Drob-1 was transiently overexpressed in Drosophila S2 cells. When the HA-Drob-1 expression plasmid was cotransfected with a GFP expression vector, the majority of GFP-positive cells, which were assumed to be overexpressing HA-Drob-1, died as early as 24 hr posttransfection, as defined by disappearance of the fluorescence (Fig. 4A a–d). Those dying cells displayed morphological features of apoptosis, such as cell-body shrinkage (Fig. 4Ac), membrane blebbing, and nuclear condensation (Fig. 4B), and detached from the dish. For the cell death assay, a lacZ expression vector was cotransfected and the viability of the transfected cells was determined by measuring β-galactosidase activity. Whereas almost all cells transfected with the GFP control vector remained viable, those transfected with the HA-Drob-1 plasmid showed a significant decrease in cell viability in a dose-dependent manner (Fig. 4C). In addition, expression of Drob-1 strongly stimulated a caspase activity that cleaved the caspase-3 substrate, DEVD, but not the caspase-1-like protease substrate, YVAD (data not shown), similar to other Drosophila cell death proteins, Rpr and Hid (Fig. 4D).
Figure 4.
Drob-1 induces apoptotic cell death and caspase activation. (A) Drosophila S2 cells were cotransfected with 5 ng of pUAS-HA-drob-1 and 1 ng of pUAS-GFP, together with 2 ng of driver plasmid pWAGAL4. Twenty two hours after the transfection, cells bearing expression plasmids could be identified by the GFP fluorescence (a and b, arrows). At about 24 hr posttransfection, the GFP-positive cells underwent apoptosis (c and d). (B) Cells transfected with the HA-Drob-1 expression plasmid displayed cell-body shrinkage (a) and condensed nuclei (b) when stained with Hoechst 33342. (C) Cell viability was determined by cell death assay 48 hr after transfection with 0.1, 0.5, 1, or 5 ng of HA-Drob-1 or GFP expression plasmid, together with 2 ng of pWAGAL4. (D) Cells were cultured in 12-well plates and transfected with 70 ng of pUAS-lacZ, -rpr, -hid, or HA-drob-1 with or without 140 ng of pUAS-p35, together with 40 ng of pWAGAL4. The total amount of plasmid DNA was adjusted to 250 ng/well by adding pUAS-lacZ. Twenty four hours after transfection, caspase activity was measured by using the fluorescent enzyme substrate, DEVD-4-methyl-coumaryl-7-amide. (E) Cells were transfected with 2.5 ng of pUAS-rpr, -hid, or -HA-drob-1 together with 5 ng of the GFP (the three filled bars on the left) or p35 (the three filled bars on the right) expression plasmid, and viability was assessed as in A. Expression of the transfected genes was driven by cotransfection of 2 ng of pWAGAL4. (F) Cells were transfected with pWAGAL4 (2 ng) plus pUAS-HA-drob-1 (1 ng) together with the DIAP2 (2 ng) or mDRONC (2 ng) expression plasmid. pUAS-GFP was added to adjust the total amount of expression plasmids to 5 ng/well, and viability was determined as in A.
Drob-1 Induces Cell Death Through a Caspase-Independent Pathway.
As reported previously, both Rpr- and Hid-induced cell death were blocked by coexpression of baculovirus p35 (Fig. 4E). In contrast, overexpression of p35 showed no effect on Drob-1-induced apoptosis (Fig. 4E), although its intracellular DEVDase activity was completely abolished (Fig. 4D). Higher levels of p35 expression exhibited only a slight protective effect against Drob-1-induced cell killing (data not shown). Similarly, DIAP2, which inhibited both Rpr- and Hid-induced apoptosis (data not shown), or an active-site mutant of the CARD (caspase recruitment domain)-containing Drosophila caspase, DRONC, showed little effect on Drob-1-induced apoptosis (Fig. 4F), suggesting that the Drob-1-stimulated cell-death pathway is downstream or independent of DIAP2 and DRONC. Thus, overexpression of Drob-1 induces apoptosis, probably through a caspase-independent pathway partly distinct from that used by other known Drosophila killer proteins, Rpr, Hid, and Grim. Drob-1 overexpressed in human embryonic kidney 293T cells also exhibited a proapoptotic activity that was only slightly inhibited by p35 (N.I. and G.N., unpublished data).
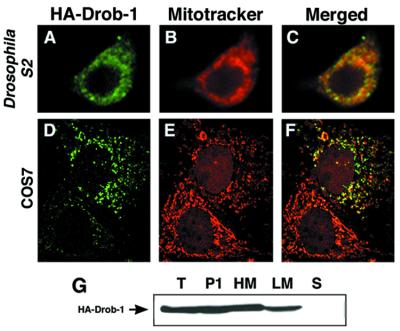
Drob-1 Localizes to the Mitochondrial Membranes in Drosophila Cells.
To determine subcellular localization, HA-tagged Drob-1 was overexpressed in S2 cells, and the cells were stained with an anti-HA antibody. Confocal microscopy revealed that Drob-1 had a granular cytoplasmic distribution, consistent with an association with intracellular organelles (Fig. 5A). To better identify the localization of Drob-1, transfected cells were colabeled with a mitochondria-specific fluorescent dye, MitoTracker. The pattern of mitochondrial staining was similar to that of Drob-1 (Fig. 5B). Merged images (Fig. 5C) of Drob-1- and MitoTracker-stained cells showed that the two proteins colocalized. This colocalization also was observed when HA-Drob-1 was overexpressed in mammalian COS7 cells (Fig. 5 D–F).
Figure 5.
Drob-1 localizes to the intracytoplasmic membranes. Drosophila S2 cells (A–C) or COS7 cells (D–F) were transiently transfected with pUAS-HA-drob-1 plus pWAGAL4, or pUAS-HA-drob-1 plus pM92, respectively. Twenty four hours after transfection, cells were stained with MitoTracker, fixed, and immunostained with an anti-HA mAb and FITC-conjugated secondary antibody. Confocal images visualized by FITC (A and D), MitoTracker (B and E), or both (C and F) are shown. Magnifications: ×1,000. (G) Subcellular fractionation of lysates from S2 cells expressing HA-Drob-1. S2 cells were transfected with pUAS-HA-drob-1 and pWAGAL4. Twenty four hours after transfection, cells were homogenized and fractionated into low-speed pellet (P1), HM fraction, LM fraction, and soluble (S) fraction by differential centrifugation. P1 (29 μg), HM (13 μg), LM (35 μg), and soluble (S; 25 μg) fractions as well as total cell lysate (T; 37 μg) were analyzed by Western blot with anti-HA mAb.
Subcellular fractionation studies in S2 cells revealed that HA-Drob-1 was enriched in the HM fraction. HA-Drob-1 was also present in P1 and LM fractions with a smaller amount but not in the cytosolic (S) fraction (Fig. 5G). Together with the microscopic data, these results suggest that Drob-1 localizes to the intracytoplasmic membranes, predominantly to the mitochondrial membranes.
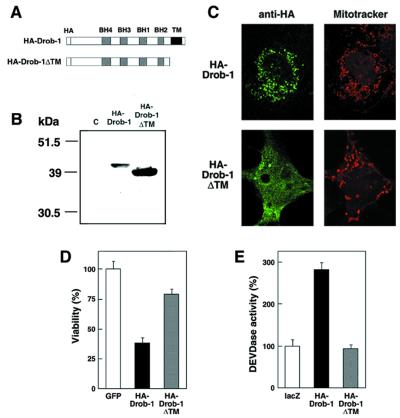
Intracytoplasmic Membrane Localization of Drob-1 Is Critical for Cell Killing and Caspase Activation.
To determine whether the C-terminal hydrophobic region of Drob-1 is necessary for its intracytoplasmic membrane docking or proapoptotic function, an expression plasmid producing a truncated form of Drob-1 (Drob-1ΔTM) lacking the last 39-aa residues, which contain the putative transmembrane domain, was constructed (Fig. 6 A and B). In contrast to the membrane localization of full-length Drob-1, the Drob-1ΔTM mutant displayed a diffuse distribution throughout the cytosol, when overexpressed in either COS7 cells (Fig. 6C) or S2 cells (data not shown). A cell death assay revealed that this mutation dramatically reduced the ability of Drob-1 to kill S2 cells (Fig. 6D). Furthermore, the caspase-stimulating activity of the Drob-1ΔTM mutant was completely eliminated (Fig. 6E). These results suggest that the cell killing and caspase-stimulating activities of Drob-1 both require that it be targeted to subcellular components, most likely to the mitochondria, through its C-terminal transmembrane domain.
Figure 6.
The C-terminal hydrophobic region of Drob-1 is required for its intracytoplasmic membrane distribution and proapoptotic activity. (A) Schematic structures of wild-type (HA-Drob-1) and mutant (HA-Drob-1ΔTM) proteins. (B) Drosophila S2 cells were cultured in 6-well plates and transfected with 0.8 μg of pUAS-HA-drob-1 or pUAS-HA-drob-1ΔTM, together with 0.2 μg of pWAGAL4. The lysates were prepared at 24 hr after transfection and analyzed by Western blotting with an anti-HA mAb. Lane C, the lysate from cells transfected with pWAGAL4 only. (C) pUAS-HA-drob-1 or pUAS-HA-drob-1ΔTM was transfected together with pM92 into COS7 cells, and their subcellular distributions were assessed by anti-HA immunostaining and mitochondrial staining as in Fig. 5. (D and E) pUAS-HA-drob-1 (1 ng or 200 ng) or pUAS-HA-drob-1ΔTM (1 ng or 200 ng) was transfected together with pWAGAL4 (2 ng or 50 ng) into S2 cells, and their abilities to kill cells and stimulate caspase activity were assessed as in Fig. 4 C and E, respectively.
Discussion
Previous and recent studies demonstrating the presence of at least four caspases and an Apaf-1 homolog in flies strongly argue the existence of a Drosophila caspase cascade similar to the mammalian cell death machinery. These findings have also predicted a role for the Bcl-2/CED-9 family in Drosophila apoptotic cell death. In this study, we identified the first Drosophila ortholog of the Bcl-2/CED-9 family, Drob-1. Drob-1 has the evolutionarily conserved BH domains and showed significant structural and sequence homology with all known Bcl-2 family members. Its identification supports the idea that a system for cell-death decisions regulated by Bcl-2 family members also may exist in Drosophila.
Functional analysis using transgenic flies and cultured cells revealed that Drob-1 acts as a proapoptotic component in Drosophila. In addition, GMRdrob-1 transgenic flies did not suppress the eye-ablation phenotype caused by overexpression of Rpr or Hid (T.I., H.K., and M.M., unpublished data). Furthermore, at any ratio examined, the transfection of a Drob-1 expression plasmid did not protect against the apoptosis induced by the transfection of a Rpr- or Hid-expressing vector in S2 cells (data not shown). These results suggest that Drob-1 is a proapoptotic member of the Drosophila Bcl-2 family. A mutant form of Drob-1 lacking the C-terminal hydrophobic region lost both its proapoptotic activity and mitochondrial distribution, similar to the truncated form of mammalian Bax (38). Many proapoptotic and antiapoptotic members of the mammalian Bcl-2 family such as Bcl-2, Bcl-xL, Bax, and Bid have been shown to function at the mitochondria to regulate its membrane permeability and/or the release of apoptogenic factors from the mitochondria to the cytosol (10). Recent studies demonstrating the appearance of an altered epitope of cytochrome c before apoptotic cell death (39) and a requirement for both Dapaf-1 and cytochrome c for caspase activation in flies (25) imply a central role for the mitochondria in regulating Drosophila programmed cell death.
Although Drob-1 strongly stimulated caspase activation when overexpressed in S2 cells, its ability to kill cells was barely antagonized by the caspase-related apoptosis inhibitors, p35 and DIAP2, or by an active-site mutant of a Drosophila apical caspase, DRONC. Furthermore, in transgenic flies, rough-eye phenotype caused by overexpression of Drob-1 was not completely suppressed by coexpression of p35 (T.I., H.K., and M.M., unpublished data). Similarly, previous studies on the proapoptotic Bax, Bak, and Mtd suggest that they all induce apoptosis in the presence of broad caspase inhibitors (37, 40, 41). Moreover, both Bax and Bak can induce mitochondrial dysfunction and also kill yeast, which lacks endogenous caspases (42, 43). Drob-1, like mammalian Bax, Bak, and Mtd, may activate cell death by inducing both caspase-dependent and -independent pathways. The latter pathway probably can be antagonized by an as-yet-unidentified antiapoptotic member of the Drosophila Bcl-2 family. In the nematode C. elegans, in which two Bcl-2/CED-9 family members, CED-9 and EGL-1 (44), have been identified, all cell deaths occur in a caspase-dependent manner. The presence of a Bax-like protein, Drob-1, in Drosophila might indicate the acquisition of a caspase-independent cell death pathway through evolution.
Drosophila is an appropriate animal model for the genetic analysis of apoptosis and also has emerged as a model for neurodegenerative diseases such as Huntington's disease and Machado-Joseph disease. In these transgenic disease models, expression of p35 shows little or partial protective effect on the neuronal degeneration caused by expanded polyglutamine proteins in Drosophila eyes (45, 46). The Drosophila cell death proteins so far identified, Rpr, Hid, and Grim, all require caspase action for their function (14–16). Our identification of Drob-1 and further genetic screening should help elucidate the evolutionarily conserved mechanisms of physiological cell death regulation by Bcl-2 family members, as well as the caspase-independent cell death pathways that exist in some neurodegenerative disorders.
Acknowledgments
We are grateful to R. Shimamura for making the fly medium, S. Kumar for the mutant DRONC cDNA, Y. Hiromi for pWAGAL4 plasmid, J. B. Gurdon and R. Niwa for GFP plasmid, P. Leder for UAS and GAL4 genes, and G. M. Rubin for pGMR vector. This work was supported in part by grants from the Japanese Ministry of Education, Science, Sports and Culture to H.O. and M.M., the Human Frontier Science Program to H.O., and the SPSBS, Science and Technology Agency of Japan to K.S. This work also was supported in part to H.O. by Core Research for Evolutional Science and Technology (CREST), Japan Science and Technology Corporation. H.K. is a research fellow of the Japan Society for the Promotion of Science.
Abbreviations
- BH
Bcl-2 homology
- DIAP
Drosophila inhibitor-of-apoptosis protein
- GMR
glass multimer reporter
- UAS
upstream activation sequence
- GFP
green fluorescent protein
- CED
cell death abnormal
- RT-PCR
reverse transcription–PCR
- HA
hemagglutinin
- HM
heavy membrane
- LM
light membrane
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AB032430).
References
- 1.Raff M C. Nature (London) 1992;356:397–400. doi: 10.1038/356397a0. [DOI] [PubMed] [Google Scholar]
- 2.Thompson C B. Science. 1995;267:1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- 3.Ellis H M, Horvitz H R. Cell. 1986;44:817–829. doi: 10.1016/0092-8674(86)90004-8. [DOI] [PubMed] [Google Scholar]
- 4.Hengartner M O, Ellis R E, Horvitz H R. Nature (London) 1992;356:494–499. doi: 10.1038/356494a0. [DOI] [PubMed] [Google Scholar]
- 5.Vaux D L, Korsmeyer S J. Cell. 1999;96:245–254. doi: 10.1016/s0092-8674(00)80564-4. [DOI] [PubMed] [Google Scholar]
- 6.Thornberry N A, Lazebnik Y. Science. 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 7.Zou H, Henzel W J, Liu X, Lutschg A, Wang X. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- 8.Li P, Nijhawan D, Budihardjo I, Srinivasula S M, Ahmad M, Alnemri E S, Wang X. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 9.Adams J M, Cory S. Science. 1998;281:1322–1326. doi: 10.1126/science.281.5381.1322. [DOI] [PubMed] [Google Scholar]
- 10.Gross A, McDonnell J M, Korsmeyer S J. Genes Dev. 1999;13:1899–1911. doi: 10.1101/gad.13.15.1899. [DOI] [PubMed] [Google Scholar]
- 11.Abrams J M, White K, Fessler L I, Steller H. Development (Cambridge, UK) 1993;117:29–43. doi: 10.1242/dev.117.1.29. [DOI] [PubMed] [Google Scholar]
- 12.Jiang C, Baehrecke E H, Thummel C S. Development (Cambridge, UK) 1997;124:4673–4683. doi: 10.1242/dev.124.22.4673. [DOI] [PubMed] [Google Scholar]
- 13.White K, Grether M E, Abrams J M, Young L, Farrell K, Steller H. Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- 14.White K, Tahaoglu E, Steller H. Science. 1996;271:805–807. doi: 10.1126/science.271.5250.805. [DOI] [PubMed] [Google Scholar]
- 15.Grether M E, Abrams J M, Agapite J, White K, Steller H. Genes Dev. 1995;9:1694–1708. doi: 10.1101/gad.9.14.1694. [DOI] [PubMed] [Google Scholar]
- 16.Chen P, Nordstrom W, Gish B, Abrams J M. Genes Dev. 1996;10:1773–1782. doi: 10.1101/gad.10.14.1773. [DOI] [PubMed] [Google Scholar]
- 17.Vucic D, Kaiser W J, Harvey A J, Miller L K. Proc Natl Acad Sci USA. 1997;94:10183–10188. doi: 10.1073/pnas.94.19.10183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vucic D, Kaiser W J, Miller L K. Mol Cell Biol. 1998;18:3300–3309. doi: 10.1128/mcb.18.6.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang S L, Hawkins C J, Yoo S J, Muller H A, Hay B A. Cell. 1999;98:453–463. doi: 10.1016/s0092-8674(00)81974-1. [DOI] [PubMed] [Google Scholar]
- 20.Song Z, McCall K, Steller H. Science. 1997;275:536–540. doi: 10.1126/science.275.5299.536. [DOI] [PubMed] [Google Scholar]
- 21.Fraser A G, Evan G I. EMBO J. 1997;16:2805–2813. doi: 10.1093/emboj/16.10.2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen P, Rodriguez A, Erskine R, Thach T, Abrams J M. Dev Biol. 1998;201:202–216. doi: 10.1006/dbio.1998.9000. [DOI] [PubMed] [Google Scholar]
- 23.Dorstyn L, Colussi P A, Quinn L M, Richardson H, Kumar S. Proc Natl Acad Sci USA. 1999;96:4307–4312. doi: 10.1073/pnas.96.8.4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodriguez A, Oliver H, Zou H, Chen P, Wang X, Abrams J M. Nat Cell Biol. 1999;1:272–279. doi: 10.1038/12984. [DOI] [PubMed] [Google Scholar]
- 25.Kanuka H, Sawamoto K, Inohara N, Matsuno K, Okano H, Miura M. Mol Cell. 1999;4:757–769. doi: 10.1016/s1097-2765(00)80386-x. [DOI] [PubMed] [Google Scholar]
- 26.Boise L H, Gonzalez G M, Postema C E, Ding L, Lindsten T, Turka L A, Mao X, Nunez G, Thompson C B. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-n. [DOI] [PubMed] [Google Scholar]
- 27.Hay B A, Wolff T, Rubin G M. Development (Cambridge, UK) 1994;120:2121–2129. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- 28.Sawamoto K, Taguchi A, Hirota Y, Yamada C, Jin M-H, Okano H. Cell Death Differ. 1998;5:262–270. doi: 10.1038/sj.cdd.4400342. [DOI] [PubMed] [Google Scholar]
- 29.Kanuka H, Hisahara S, Sawamoto K, Shoji S, Okano H, Miura M. Proc Natl Acad Sci USA. 1999;96:145–150. doi: 10.1073/pnas.96.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brand A H, Perrimon N. Development (Cambridge, UK) 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 31.Ornitz D M, Moreadith R W, Leder P. Proc Natl Acad Sci USA. 1991;88:698–702. doi: 10.1073/pnas.88.3.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zernicka-Goetz M, Pines J, Ryan K, Siemering K R, Haseloff J, Evans M J, Gurdon J B. Development (Cambridge, UK) 1993;118:401–415. doi: 10.1242/dev.122.12.3719. [DOI] [PubMed] [Google Scholar]
- 33.Usui T, Shima Y, Shimada Y, Hirano S, Burgess R W, Schwarz T L, Takeichi M, Uemura T. Cell. 1999;98:585–595. doi: 10.1016/s0092-8674(00)80046-x. [DOI] [PubMed] [Google Scholar]
- 34.Hisahara S, Kanuka H, Shoji S I, Yoshikawa S, Okano H, Miura M. J Cell Sci. 1998;111:667–673. doi: 10.1242/jcs.111.6.667. [DOI] [PubMed] [Google Scholar]
- 35.del Peso L, Gonzalez V M, Nunez G. J Biol Chem. 1998;273:33495–33500. doi: 10.1074/jbc.273.50.33495. [DOI] [PubMed] [Google Scholar]
- 36.Hsu S Y, Kaipia A, McGee E, Lomeli M, Hsueh A J. Proc Natl Acad Sci USA. 1997;94:12401–12406. doi: 10.1073/pnas.94.23.12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inohara N, Ekhterae D, Garcia I, Carrio R, Merino J, Merry A, Chen S, Nunez G. J Biol Chem. 1998;273:8705–8710. doi: 10.1074/jbc.273.15.8705. [DOI] [PubMed] [Google Scholar]
- 38.Wolter K G, Hsu Y T, Smith C L, Nechushtan A, Xi X G, Youle R J. J Cell Biol. 1997;139:1281–1292. doi: 10.1083/jcb.139.5.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varkey J, Chen P, Jemmerson R, Abrams J M. J Cell Biol. 1999;144:701–710. doi: 10.1083/jcb.144.4.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiang J, Chao D T, Korsmeyer S J. Proc Natl Acad Sci USA. 1996;93:14559–14563. doi: 10.1073/pnas.93.25.14559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCarthy N J, Whyte M K, Gilbert C S, Evan G I. J Cell Biol. 1997;136:215–227. doi: 10.1083/jcb.136.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zha H, Fisk H A, Yaffe M P, Mahajan N, Herman B, Reed J C. Mol Cell Biol. 1996;16:6494–6508. doi: 10.1128/mcb.16.11.6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ink B, Zornig M, Baum B, Hajibagheri N, James C, Chittenden T, Evan G. Mol Cell Biol. 1997;17:2468–2474. doi: 10.1128/mcb.17.5.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Conradt B, Horvitz H R. Cell. 1998;93:519–529. doi: 10.1016/s0092-8674(00)81182-4. [DOI] [PubMed] [Google Scholar]
- 45.Warrick J M, Paulson H L, Gray B G, Bui Q T, Fischbeck K H, Pittman R N, Bonini N M. Cell. 1998;93:939–949. doi: 10.1016/s0092-8674(00)81200-3. [DOI] [PubMed] [Google Scholar]
- 46.Jackson G R, Salecker I, Dong X, Yao X, Arnheim N, Faber P W, MacDonald M E, Zipursky S L. Neuron. 1998;21:633–642. doi: 10.1016/s0896-6273(00)80573-5. [DOI] [PubMed] [Google Scholar]
- 47.Zhou L, Song Z, Tittel J, Steller H. Mol Cell. 1999;4:745–755. doi: 10.1016/s1097-2765(00)80385-8. [DOI] [PubMed] [Google Scholar]