Abstract
The influence of altitude and salinity on bacterioplankton community composition (BCC) in 16 high-mountain lakes located at altitudes of 2,817 to 5,134 m on the Eastern Qinghai-Xizang (Tibetan) Plateau, China, spanning a salinity gradient from 0.02% (freshwater) to 22.3% (hypersaline), was investigated. Three different methods, fluorescent in situ hybridization, denaturing gradient gel electrophoresis (DGGE) with subsequent band sequencing, and reverse line blot hybridization (RLB) with probes targeting 17 freshwater bacterial groups, were used for analysis of BCC. Furthermore, the salt tolerances of 47 strains affiliated with groups detected in or isolated from the Tibetan habitats were investigated. Altitude was not found to influence BCC significantly within the investigated range. Several groups of typical freshwater bacteria, e.g., the ACK-M1 cluster and the Polynucleobacter group, were detected in habitats located above 4,400 m. Salinity was found to be the dominating environmental factor controlling BCC in the investigated lakes, resulting in only small overlaps in the BCCs of freshwater and hypersaline lakes. The relative abundances of different classes of Proteobacteria showed a sharp succession along the salinity gradient. Both DGGE and RLB demonstrated that a few freshwater bacterial groups, e.g., GKS98 and LD2, appeared over wide salinity ranges. Six freshwater isolates affiliated with the GKS98 cluster grew in ecophysiological experiments at maximum salinities of 0.3% to 0.7% (oligosaline), while this group was detected in habitats with salinities up to 6.7% (hypersaline). This observation indicated ecologically significant differences in ecophysiological adaptations among members of this narrow phylogenetic group and suggested ecological significance of microdiversity.
Saline lakes constitute 45 percent of total inland water (running and stagnant waters) (47). Despite the quantitative importance of saline lakes, only a few studies have investigated the diversity of bacterioplankton in such habitats (13, 15, 23). By contrast, the influence of salinity on bacterioplankton community composition (BCC) in dynamic saline systems, such as estuaries (6, 11, 12, 25, 26, 37) and coastal solar salterns (3, 7, 8), has been well investigated. Therefore, the current knowledge on the influence of salinity on BCC is almost completely based on investigations of systems characterized by rapid changes in salinity. These systems are too dynamic to allow inhabitants to evolutionarily adapt to the changing environment. All studies on such systems indicate that salinity strongly controls BCC, i.e., changes in salinity are assumed to result in replacement of suboptimally adapted taxa by taxa better adapted to the current salinity conditions. By contrast, the slow evolution of large saline lakes from freshwater lakes may have allowed bacterial taxa originally adapted to freshwater conditions to adapt to saline conditions.
In order to reveal the influence of salinity on the BCC of stagnant inland waters, 16 lakes located at the Qinghai-Xizang (Tibetan) Plateau were investigated. The Qinghai-Xizang Plateau, located in the western part of China, is an endorheic area of ca. 2 × 106 km2, with an average altitude of 4,500 m above sea level (53). This region comprises several thousand lakes, which cover a total area of 4.5 × 104 km2 (44). The lakes are characterized by a geographic salinity gradient, which results from a gradual decrease of annual precipitation from south to north (51). The ages of lakes located at the Tibetan Plateau were estimated to be 2 to 8 million years (53).
The investigated lakes are characterized by a broad salinity gradient from freshwater to hypersaline, as well as by an altitude gradient ranging from 2,817 to 5,134 m. The set of investigated lakes includes Lake Qinghai, which is the seventh largest saline lake in the world, as well as five lakes located at altitudes above 4,400 m, which are among the most elevated lakes in the world.
The goals of this study were (i) to reveal the influence of salinity on BCC in habitats with stable salinity conditions, (ii) to investigate the salt tolerance of strains representing phylogenetic groups of bacteria detected in the investigated habitats or isolated from these habitats, and (iii) to explore whether typical groups of freshwater bacteria, which have frequently been detected in lowland habitats, are also present in high-mountain lakes located at extreme altitudes.
MATERIALS AND METHODS
Study sites and sampling.
Sixteen lakes located in the eastern part of the Qinghai-Xizang (Tibetan) Plateau at altitudes ranging from 2,817 to 5,134 m above sea level were investigated (see Table S1 in the supplemental material). The lakes were chosen in order to cover a salinity gradient from 0.2 to 222.6 g liter−1 (51) as well as an altitude gradient. Water samples were collected from surface waters (top 50 cm) with a 5-liter Schindler sampler. In the case of large, deep lakes, water samples were also taken from different water depths (see Table S1 in the supplemental material). Water samples for determination of numbers of prokaryotes and heterotrophic nanoflagellates (HNF) were fixed with 2% formaldehyde (final concentration) on location and analyzed within 3 months. Water samples for fluorescent in situ hybridization (FISH) were fixed with paraformaldehyde solution (2% final concentration) on location for 2 to 4 h, filtered onto 0.2-μm-pore-size Isopore membrane filters (Millipore), rinsed with phosphate-buffered saline buffer and Milli-Q water, and dried at room temperature. Bacterioplankton samples (250 to 500 ml water) for denaturing gradient gel electrophoresis (DGGE) and reverse line blot hybridization (RLB) analyses were collected on 0.2-μm-pore-size Isopore filters. Filters for both FISH and DNA extraction were stored in liquid nitrogen during the field campaign as well as during transport to the laboratory. Untreated water samples (2 to 3 liters) were transported to the laboratory for immediate chemical analysis.
Measurement of physical and chemical parameters.
Water temperature, pH, conductivity, and Secchi depth were measured on location. Concentrations of the eight major ions potassium, sodium, calcium, magnesium, chloride, sulfate, carbonate, and bicarbonate, as well as the concentration of total nitrogen (TN) and total phosphorus (TP), were measured according to standard methods (18) after transportation of samples to the laboratory. The salinity (salt concentration) of the investigated habitats was determined by summing up the concentrations of the eight major ions (47).
Microbial counts and FISH.
Total bacterial numbers and numbers of HNF were determined by epifluorescence microscopy after DAPI (4′,6′-diamidino-2-phenylindole) staining as described previously (50). FISH on filters was performed according to the protocol of Alfreider et al. (1) and as previously described (48). The probes used in this study were ARCH915 (39), EUB338 (2), ALF968 (32), BET42a (30), GAM42a (30), CFB319a (29), HGC69a (34), and NON338 (43).
DNA extraction and purification.
DNA was extracted from biomass collected on filters by a standard phenol-chloroform extraction, precipitation with ethanol, and purification with the Wizard DNA clean-up kit (Promega) and concentrated to a volume of 50 μl (36).
PCR and DGGE.
Two nanograms of microbial DNA was used as template for PCR amplification of partial 16S rRNA genes by using Bacteria-specific primers 358f (5′ CCT ACG GGA GGC AGC AG 3′, with a 40-bp GC clamp) and 907rM (5′ CCG TCA ATT CMT TTG AGT TT 3′) (31, 36). DGGE was carried out with a DCode system (Bio-Rad) by using a 6% polyacrylamide gel with a 30% to 70% gradient of a DNA denaturant agent as described previously (36). Eight hundred nanograms of PCR products was loaded, and the gel was run at 100 V for 16 h at 60°C in 1× Tris-acetate-EDTA buffer. The gel was stained with the nucleic acid stain SybrGold (Molecular Probes) for 45 min, rinsed with 1× Tris-acetate-EDTA buffer, and visualized under UV light.
Analysis of DGGE bands by sequencing.
DGGE bands were excised, transferred into 20 μl MilliQ water, and incubated overnight for elution of DNA. Two microliters of the supernatant was used for reamplification with the original primer pair (without a GC clamp). PCR products were purified with a QIAquick PCR purification kit (QIAGEN) and sequenced with primer 358f without a GC clamp. All long unambiguous sequences (ca. 500 bp) were phylogenetically analyzed using the ARB software package (http://www.arb-home.de), and phylogenetic trees were constructed as described previously (19). In the case of DGGE sequences containing ambiguities, the affiliations at the phylum or class level were determined by BLAST (http://www.ncbi.nlm.nih.gov/BLAST/) but were omitted from further phylogenetic analyses.
RLB.
RLB with antisense oligonucleotide probes targeting 16S rRNA sequences was performed as described previously (55). Eighteen probes, including 10 previously published (55) and 8 newly designed probes, were applied. These probes target bacterial 16S rRNA genes of 17 phylogenetic clusters of typical freshwater bacteria (54). The specificity and hybridization conditions of the applied probes are presented in Table S2 in the supplemental material. All probes were tested and have shown positive hybridization signals against natural samples or cultivated controls.
Salt tolerance of bacterial strains.
The salt tolerances of 47 cultivated strains of aquatic bacteria (see Table S3 in the supplemental material) were determined. Thirty-seven out of the 47 strains have previously been isolated from different freshwater habitats. These strains are affiliated with five phylogenetic groups of typical freshwater bacteria, i.e., GKS98 (54), Polynucleobacter subclusters PnecC and PnecD (19, 49), Luna-1 (acII A to C) (21, 45), and Luna-2 (acII D) (21, 45). Furthermore, 10 additional strains, representing four taxa affiliated with Alphaproteobacteria and Gammaproteobacteria, isolated from the studied lakes were investigated (see Table S3 in the supplemental material). The tested strains were precultured at room temperature in NSY medium (22). Ten microliters of the cultures growing in the exponential phase was dropped on NSY agar plates with salinities of 0.6 to 50 g liter−1, or 1-ml samples of precultures were inoculated into liquid NSY medium for testing salinities above 50 g liter−1. Twenty-one different salinities ranging from 0.06% to ∼40% (0.6 to ∼400 g liter−1 [satiated solution]) were used in these ecophysiological experiments. The salinity was adjusted by the addition of sodium chloride to NSY medium, which has a salinity equivalent to 0.6 g liter−1. The agar plates or liquid cultures were incubated at room temperature for 4 weeks and were inspected for bacterial growth weekly.
Statistics.
Canonical correspondence analysis (CCA) was used for revealing relationships between BCC, as determined by FISH or DGGE, and environmental variables (40). CCA was performed with the software CANOCO 4.53 (Scientia Software). CCA requires a unimodal species-environment relationship (41), which can be assumed in cases where the length of the first detrended correspondence analysis axis run on species data is >2. This requirement was fulfilled by our data set. The environmental factors best describing the most influential gradients were identified by forward selection. Explanatory variables were added until the addition of further variables did not result in significant (P < 0.05) improvements to the model's explanatory power. This was assessed in permutation tests with 499 unrestricted Monte Carlo permutations. The relative abundances of major bacterial groups determined by FISH were transformed as log(x + 1). For analysis of DGGE fingerprints by CCA, a binary matrix was constructed by scoring the presence (1) and absence (0) of DGGE bands. This matrix was used for CCA without transformation. The environmental variables tested were salinity, TP, TN, altitude, lake area, temperature, pH, and abundance of HNF. Missing data of TP, TN, and water temperature of Lake Yanghu (see Table S1 in the supplemental material) were replaced by data measured in previous investigations during the same season (44). All environmental data, except pH, were transformed for CCA as log(x + 1). CCA was performed separately for FISH and DGGE data sets. Principle components analysis was applied to illustrate the main gradients in ion concentrations and their relationship with salinity.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences obtained from DGGE bands and isolates have been deposited under the accession numbers AM182266 to AM182325.
RESULTS
Environmental characteristics of the investigated lakes.
The major geographical and physicochemical characteristics of the investigated lakes are summarized in Table S1 in the supplemental material. The salinity ranged from 0.2 g liter−1 (0.02%; freshwater) to 222.6 g liter−1 (22.3%, hypersaline). All investigated saline habitats were characterized by high concentrations of sodium, chloride, and sulfate, and principal components analysis run on the data set of concentrations of the eight major ions resulted in clustering of all investigated habitats in one group. Furthermore, a positive relationship between salinity and the concentration of total nitrogen (r2 = 0.74, P < 0.001, n = 23) was found.
Microbial numbers and FISH.
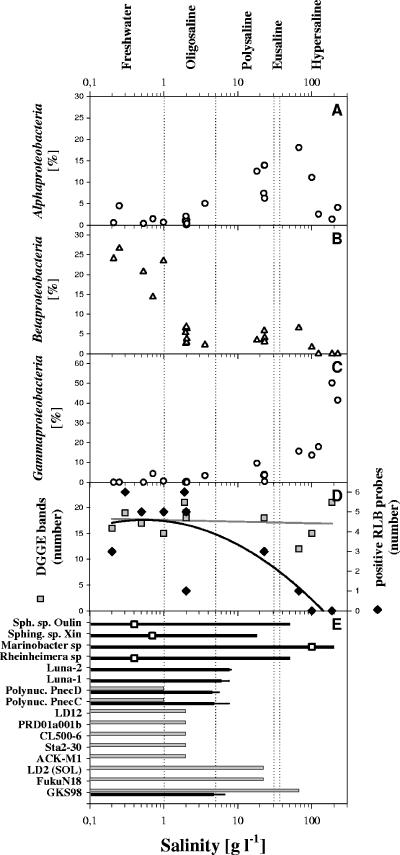
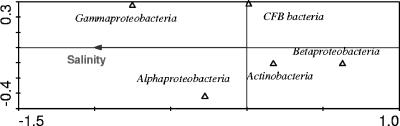
The total prokaryotic abundances varied from 0.4 × 106 to 13.9 × 106 cells ml−1 (Table 1). In general, higher cell numbers were observed in lakes with a higher concentration of total phosphorus and higher salinity. Analysis by FISH revealed that no single bacterial group dominated across all investigated lakes (Table 1). The relative abundances of Alphaproteobacteria, Betaproteobacteria, and Gammaproteobacteria varied systematically along the salinity gradient (Fig. 1A to C). The relative abundance of Alphaproteobacteria exhibited a peaked distribution with higher relative abundances in the range of 20 to 100 g liter−1 (poly- to hypersaline) and peaked at a salinity of approximately 70 g liter−1. Betaproteobacteria were abundant in all investigated freshwater lakes (14 to 27% of DAPI-stained cells), but in saline lakes, relative abundances never exceeded 7%, even though Betaproteobacteria could be detected in habitats with salinities up to 100 g liter−1 (Fig. 1B). The presence of Betaproteobacteria in saline lakes was confirmed by DGGE and RLB (Fig. 1E). By contrast, the relative abundance of Gammaproteobacteria increased with increasing salinity (Fig. 1C). In the CCA model, salinity was found to be the dominating environmental factor statistically explaining variations in composition of major bacterial groups in the investigated lakes (Fig. 2).
TABLE 1.
Results obtained by FISH, DGGE, and RLB
| Lake | Lake type | Salinity (g liter−1)c | Sampling depth (m) | Prokaryotic abundance (106 cells ml−1) | % of DAPI-stained cells ina:
|
No. of DGGE bands | No. of RLB signalsb | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ALF968 | BET42a | GAM42a | CF319a | HGC69a | EUB338 | ARCH915 | |||||||
| High lake 1 | Freshwater | 0.2 | 0.5 | 1.7 | 0.5 | 24.0 | 0 | 5.7 | 5.2 | 66.4 | 1.2 | 16 | 3 |
| High lake 2 | Freshwater | 0.3 | 0.5 | 5.6 | 4.5 | 26.6 | 0 | 10.0 | 11.5 | 55.3 | 0.6 | 19 | 6 |
| High lake 3 | Freshwater | 0.5 | 0.5 | 2.5 | 0.3 | 20.7 | 0 | 5.7 | 35.1 | 60.0 | 1.2 | 17 | 5 |
| Kelike | Freshwater | 0.7 | 0.5 | 2.3 | 1.4 | 14.3 | 4.6 | 1.6 | 27.2 | 51.9 | 1.2 | NDd | ND |
| Pond | Freshwater | 1.0 | 0.5 | 2.6 | 0.6 | 23.5 | 0.5 | 20.6 | 9.4 | 66.5 | 0 | 15 | 5 |
| Yanghu | Oligosaline | 1.9 | 0.5 | ND | ND | ND | ND | ND | ND | ND | ND | 21 | 6 |
| Narnocuo | Oligosaline | 2.0 | 0.5 | 1.2 | 0.9 | 5.4 | 0 | 1.0 | 9.1 | 65.6 | 0.4 | 19 | 5 |
| Narnocuo | Oligosaline | 2.0 | 5 | 1.4 | 1.2 | 6.6 | 0.1 | 2.4 | 9.1 | 69.3 | 1.6 | 19 | 5 |
| Narnocuo | Oligosaline | 2.0 | 10 | 1.7 | 0.8 | 6.8 | 0.2 | 1.1 | 11.5 | 69.6 | 0.4 | 19 | 5 |
| Narnocuo | Oligosaline | 2.0 | 20 | 1.6 | 2.0 | 2.7 | 0.1 | 0.6 | 9.7 | 60.9 | 1.0 | 19 | 5 |
| Narnocuo | Oligosaline | 2.0 | 30 | 2.0 | 0 | 2.8 | 0.4 | 1.0 | 9.6 | 44.0 | 0.6 | 19 | 5 |
| Narnocuo | Oligosaline | 2.0 | 67 | 0.9 | 0.3 | 3.8 | 0 | 0.7 | 8.9 | 43.6 | 1.4 | 19 | 5 |
| Erhai | Oligosaline | 2.0 | 0.5 | 13.9 | 1.2 | 6.3 | 0 | 1.0 | 12.2 | 38.4 | 0 | 18 | 1 |
| Agecuo | Oligosaline | 3.6 | 0.5 | 4.5 | 5.0 | 2.2 | 3.3 | 2.0 | 6.6 | 39.0 | 0.2 | ND | ND |
| Kuhai | Polysaline | 18.1 | 0.5 | 6.3 | 12.5 | 3.1 | 9.5 | 4.4 | 19.3 | 55.5 | 0 | ND | ND |
| Qinghai | Polysaline | 22.6 | 0.5 | 0.4 | 13.9 | 5.8 | 4.0 | 1.8 | 3.4 | 35.2 | 0 | 18 | 3 |
| Qinghai | Polysaline | 22.4 | 5 | 0.5 | 7.4 | 3.5 | 3.4 | 4.4 | 5.2 | 31.3 | 0 | 18 | 3 |
| Qinghai | Polysaline | 23.0 | 10 | 0.6 | 6.2 | 4.1 | 3.6 | 4.1 | 4.3 | 25.5 | 0 | 18 | 3 |
| Qinghai | Polysaline | 22.9 | 17 | 0.8 | 13.9 | 3.0 | 0.3 | 2.6 | 6.2 | 29.8 | 0 | 18 | 3 |
| Gahai 1 | Hypersaline | 66.7 | 0.5 | 0.6 | 18.1 | 6.5 | 15.6 | 5.6 | 9.8 | 51.9 | 1.5 | 12 | 1 |
| Gahai 2 | Hypersaline | 99.8 | 0.5 | 3.5 | 11.0 | 1.6 | 13.6 | 3.5 | 9.1 | 71.5 | 0 | 15 | 0 |
| High lake 4 | Hypersaline | 122.9 | 0.5 | 3.7 | 2.5 | 0 | 17.9 | 10.5 | 42.0 | 77.5 | 0 | ND | ND |
| Xiaochaidan | Hypersaline | 187.1 | 0.5 | 2.3 | 1.3 | 0 | 50.1 | 6.3 | 1.7 | 83.5 | 0 | 21 | 0 |
| Dachaidan | Hypersaline | 222.6 | 0.5 | 2.1 | 4.1 | 0 | 41.4 | 9.6 | 2.0 | 72.6 | 0 | ND | ND |
Alphaproteobacteria, probe ALF968; Betaproteobacteria, probe BET42a; Gammaproteobacteria, probe GAM42a; many Bacteroidetes, probe CF319a; Actinobacteria, probe HGC69a; Bacteria, probe EUB338; Archaea, probe ARCH915. Counts have been corrected by subtracting NON338 counts. Almost all NON338 counts had the typical characteristics of picocyanobacterial autofluorescence signals.
Number of probes which gave a signal.
Salinity equals the sum of the concentrations of the ions potassium, sodium, calcium, magnesium, chloride, sulfate, carbonate, and bicarbonate.
ND, not done.
FIG. 1.
(A to C) Relative abundances of Alphaproteobacteria (A), Betaproteobacteria (B), and Gammaproteobacteria (C) in Tibetan lakes along a salinity gradient as determined by FISH. (D) Number of DGGE bands (gray squares) and number of RLB probes with positive hybridization signals (black diamonds). Trends in changes of numbers with salinity are indicated by first (gray line) and second (black line) order regression lines. (E) Maximum salinity at which the growth of cultivated representatives of nine bacterial taxa was observed in laboratory experiments (black bars) and maximum salinity at which the respective bacterial groups were detected in the environment by RLB (gray bars). The results shown for the nine different taxa represent results from 2 to 15 investigated strains (see Table S3 in the supplemental material). The black bars indicate the average maximum salinities at which growth of strains was observed, and the narrow extension indicates the highest salinity at which a member of the group grew. The open squares overlaying the respective bars indicate the salinity of the habitats from which the Tibetan strains were obtained. RLB probes were available only for the lower 10 bacterial groups. Sph. sp. Oulin, Sphingomonas sp. “Oulin”; Sphing. sp. Xin, Sphingomonas sp. “Xin”; Polynuc., Polynucleobacter.
FIG. 2.
CCA biplot based on FISH and environmental data of 15 investigated lakes (see Table S1 in the supplemental material). For Lake Qinghai and Namocuo, only data from the epilimnion were used. The analyzed bacterial groups were Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, Actinobacteria, and Bacteroidetes (CFB bacteria). Salinity was the dominant factor significantly (P < 0.05) explaining the differences in composition of major bacterial groups. The x axis explained 37% of the observed variation.
DGGE analyses.
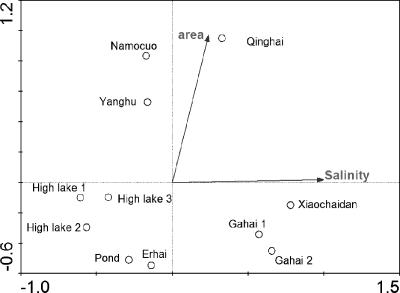
Nineteen samples from 11 lakes were analyzed by DGGE (see Table S1 and Fig. S1 in the supplemental material). Between 12 and 21 bands were observed per sample (see Fig. S1 in the supplemental material). Analysis of DGGE fingerprints by CCA resulted in four clusters (Fig. 3) separated by salinity and lake area. The first cluster contained hypersaline lakes (Gahai 1, Gahai 2, and Xiaochaidan), the second contained polysaline (Qinghai), the third contained large oligosaline lakes (Namocuo and Yanghu), and the fourth cluster contained all freshwater lakes (High lake 1, High lake 2, High lake 3, and Pond) as well as a small oligosaline lake (Erhai).
FIG. 3.
CCA biplot based on DGGE data and environmental factors of 11 investigated lakes (see Table S1 in the supplemental material). The plot illustrates the differences in BCC in relation to the two environmental factors significantly (P < 0.05) influencing BCC. For Lake Qinghai and Namocuo, only data from the epilimnion were used. The eigenvalues of the x and y axes were 0.97 and 0.63, respectively. The two axes explained 45% of the observed variation in BCC. Salinity was strongly correlated with the x axis.
Sequencing of 92 DGGE bands (see Fig. S1 in the supplemental material) resulted in 55 sequences of acceptable quality over lengths of about 500 bp (see Table S4 in the supplemental material) and 33 sequences with many ambiguous positions (only fragments of 110 to 450 bp could be used for analysis), and sequencing of four bands failed completely. Several DGGE band sequences obtained from freshwater habitats clustered within well-characterized groups of freshwater bacteria, while only a few of the sequences obtained from saline or hypersaline lakes were affiliated with typical freshwater bacteria (see Table S4 in the supplemental material). For example, band Gahai1-8 retrieved from hypersaline Lake Gahai 1 (salinity, 66 g liter−1) clustered (see Fig. S2 in the supplemental material) within the freshwater bacterial group GKS98 (54) and shared a similarity of 98.6% with strain QLW-P2DMWB-4, which has been isolated from an acidic freshwater pond (Q. L. Wu and M. W. Hahn, unpublished data). Band Qinghai-14 obtained from polysaline Lake Qinghai, clustered within the picocyanobacterial Synechococcus group B (10) and was found to be closely related (sequence similarity, 99.6%) to Synechococcus sp. strain MW76B2, which has been isolated from freshwater Lake Mondsee (10).
Members of some other bacterial groups appeared only in saline and hypersaline lakes over wide salinity ranges. For instance, sequences of bands Gahai2-1 and Gahai2-2 (both from salinities of 100 g liter−1) as well as Xiaochadan-1 to Xiaochadan-7 (salinity, 187 g liter−1) were closely related to Psychroflexus tropicus (see Tables S2 and S4 in the supplemental material), which is an obligately halophilic bacterium isolated from a Hawaiian hypersaline lake (14). Sequences of bands Qinghai-11 to -13 and Gahai1-7 were affiliated with Loktanella vestfoldensis, a bacterium isolated from Antarctic lakes (42). These sequences were obtained from salinities of 22 g liter−1 and 66 g liter−1, respectively (see Tables S2 and S4 in the supplemental material).
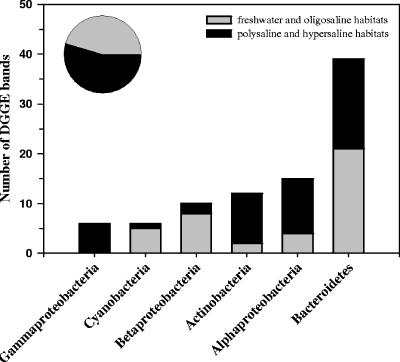
Analysis of the 88 sequences obtained from DGGE bands revealed that they were affiliated with six major phylogenetic groups (Fig. 4). The majority of analyzed bands represented bacteria affiliated with Bacteroidetes, followed by Alphaproteobacteria and Actinobacteria. In five of these groups, >73% of sequences originated either from freshwater and oligosaline (salinity, ≤2 g liter−1) or from polysaline and hypersaline (salinity, >22 g liter−1) habitats. Only the sequences affiliated with Bacteroidetes almost equally originated from these two groups of habitats differing in salinity.
FIG. 4.
Phylogenetic affiliation of the 88 sequences obtained from DGGE bands. The origins of sequences from habitats with salinities of ≤2 g liter−1 (freshwater and oligosaline) and >22 g liter−1 (polysaline and hypersaline) were distinguished. The pie chart shows the contribution of the two habitat types to the total number of analyzed sequences.
RLB.
The BCCs in 11 habitats were investigated by RLB. In total, 10 of the 15 targeted groups were detected in the investigated high-mountain lakes (Fig. 1E). The number of detected groups decreased with increasing salinity (Fig. 1D). While all 10 detected groups were found in at least one of the seven investigated freshwater and oligosaline habitats, only 3 groups were detected in the four investigated habitats with salinities higher than 2 g liter−1 (Fig. 1E). Thus, only a few of the investigated groups of typical freshwater bacteria occurred over broad salinity ranges. These groups were GKS98 (Betaproteobacteria), which was detected at salinities of 22 g liter−1 and 66 g liter−1, and LD2 (SOL cluster, Bacteroidetes) and FukuN18 (Verrucomicrobia), both of which were detected at a salinity of 22 g liter−1. In contrast, the three probes targeting the Polynucleobacter group (Betaproteobacteria) were positive only in freshwater habitats, with salinities of <1 g liter−1. Groups ACK-M1, CL500-6, and PRD01a001b were detected across all freshwater and oligosaline lakes, while groups Sta2-30 and LD12 were found just in oligosaline lakes (Lake Yanghu and Lake Namocuo). In hypersaline Lakes Gahai 2 and Xiaochaidan, no bacterial groups were detected by the applied RLB probes.
Groups of typical freshwater bacteria detected in lakes above 4,400 m elevation.
Eight phylogenetic groups of bacteria, previously detected frequently in lowland freshwater habitats, were detected by RLB or DGGE/band sequencing in the five lakes located at altitudes of >4,400 m (see Table S1 in the supplemental material). These five lakes included freshwater and oligosaline lakes. The groups ACK-M1 (Actinobacteria), CL500-6 (Bacteroidetes), and PRD01a001b (Bacteroidetes) were detected in all five lakes. Three other groups, i.e., Polynucleobacter (Betaproteobacteria), LD12 (Alphaproteobacteria), and Sta2-30 (Actinobacteria), were each detected in two out of these five lakes, and two groups, i.e., Rhodoferax sp. BAL47 (Betaproteobacteria) and Synechococcus cluster 6b (Cyanobacteria), were each detected in one out of these five lakes.
Salt tolerance of cultivated strains.
The maximum salinities at which growth of tested bacterial strains belonging to Luna-1, Luna-2, Polynucleobacter subclusters PnecC and PnecD, and GKS98 was observed were 7.6, 8.1, 7.6, 5.6, and 6.6 g liter−1, respectively (Fig. 1E). In contrast, the 10 strains, representing four taxa, isolated from the studied Tibetan lakes demonstrated a much higher salinity tolerance (Fig. 1E). Representatives of three of these four taxa were obtained from freshwater habitats, but despite this origin, the strains were able to grow under polysaline (all taxa) or even hypersaline conditions (two taxa).
DISCUSSION
Currently, the factors controlling the distribution of bacterial groups known to inhabit many freshwater lakes are not well established. In order to learn more about these controlling factors, bacterioplankton in several high-mountain lakes covering two major gradients was investigated by cultivation-independent methods. The first major gradient was altitude, ranging from 2,817 m to 5,134 m, and the second was salinity, ranging from 0.2 g liter−1 (0.02%, freshwater) to 222.6 g liter−1 (22.3%, hypersaline).
Influence of altitude on bacterioplankton community composition.
Solar UV radiation (UVR) is a crucial environmental factor in high-mountain lakes because of the natural increase of the UVR flux with elevation and the usually higher water transparency of high-mountain lakes (38). The increase of UVR with altitude is influenced by several factors, including atmospheric turbidity and the solar zenith angle. Most previous estimations of the UV altitude effect were in the range of 10% to 20% UVR increase per 1,000 m altitude (4, 38). According to these previous estimates, the potential UVR impacts on the investigated Tibetan lakes differed due to altitude by 20% to 40%. The UVR impact differences between the investigated Tibetan lakes and lowland lakes greatly exceeded this range.
Despite the expected strong altitude-dependent differences in UVR intensity, CCA did not indicate an influence of altitude on BCC (Fig. 2 and 3). Warnecke et al. (46) observed a positive correlation between the relative abundance of Actinobacteria affiliated with the acI lineage and ambient levels of solar UV radiation in eight lakes located above the timberline at altitudes of 1,650 to 2,799 m and hypothesized a high UV resistance by bacteria affiliated with this linage. In that study, Actinobacteria, which were >90% affiliated with the acI lineage, contributed up to 70% of the total bacterial abundances (46). By contrast, our investigation did not reveal a prevalence of Actinobacteria or of the acI linage in the studied Tibetan habitats. In fact, the ACK-M1 group was detected in all five of the studied freshwater lakes, and the Sta2-30 group was detected in two out of the five studied freshwater lakes located at >4,400 m. These groups are major subgroups of the acI linage. Previous investigations of lowland freshwater lakes also detected the ACK-M1 group in all of the 81 (55) and 15 (28) investigated lakes. The other acI subgroup, i.e., Sta2-30, was previously detected in >85% of the habitats investigated by RLB (28, 55). Thus, the presence of acI Actinobacteria (i.e., ACK-M1 and/or Sta2-30) in all investigated Tibetan freshwater habitats cannot be interpreted as a result of a high UVR resistance of acI bacteria. Interestingly, the vertical profiles of BCC in the thermally stratified Lake Namocuo (Table 1) revealed by DGGE and FISH did not indicate a pronounced change of BCC with depths (see Fig. S1 in the supplemental material). Variation of BCC with depths could have been a second possible evidence of a structuring influence of UVR. Thus, we did not obtain any hints on a structuring influence of UVR on BCC. However, since these results might be due to other effects overlapping with a potential impact of UVR, ecophysiological experiments with isolates representing several groups of typical freshwater bacteria could be attempted to reveal potential differences in UVR sensitivity.
Two-thirds of the groups of typical freshwater bacteria targeted by RLB probes were detected in the investigated high-mountain habitats, which indicates that the distribution of these groups was not limited by UVR or altitude within the investigated ranges. The absence of the other five groups targeted by RLB probes could be a result of the small number of investigated freshwater habitats as well as of their relatively narrow ecological variation among the investigated freshwater habitats. Due to similar geological settings, the investigated Tibetan freshwater habitats had similar ion compositions and displayed a narrow pH range (see Table S1 in the supplemental material). Restrictions in distribution of freshwater bacteria due to water chemistry have previously been shown (35). The distribution of group CL0-14 (Verrucomicrobia), which was not detected in Tibetan habitats, was found to be negatively correlated with pH (28); therefore, the lack of detection of this group could have been a result of the high pH in the Tibetan lakes. In previous studies, group Urk-014 (Actinobacteria) was detected only in 9 out of 81 lakes representing a broad ecological diversity (55) and in 2 out of 15 diverse lakes (28). Groups Sta2-35 (Verrucomicrobia) and FukuN47 (Bacteroidetes) were not previously investigated by RLB. Both phylogenetic groups are represented only by a few 16S rRNA sequences and are known only from a few habitats (54). The probe targeting Sta2-35 and probe FukuN47 match only two and six ribosomal sequences deposited in GenBank, respectively. Therefore, the lack of detection of these five groups of freshwater bacteria in the Tibetan habitats cannot be interpreted as an inability by these bacteria to inhabit high-mountain lakes.
Altogether, our data clearly demonstrated that some groups of bacteria, which are well known from lowland freshwater habitats (54), were also present in the investigated high-mountain freshwater lakes. Thus, altitude and/or intensity of UVR did not appear to limit the distribution of these phylogenetic groups of bacteria. On the other hand, we cannot exclude that the distribution of groups not detected in our investigation was limited by these factors.
Influence of salinity on bacterioplankton community composition.
Salinity was clearly the major factor responsible for differences in BCC in the investigated habitats. A clear succession of proteobacterial groups along the salinity gradient was observed (Fig. 1A to C), and 70% of the typical freshwater bacteria groups were restricted to salinities of <0.2% (Fig. 1D). Only a few groups of bacteria, which were detected previously or in the current study in habitats with salinities of <0.2%, were also detected in Tibetan habitats with salinities of >0.2%. These groups were GKS98, LD2, FukuN18, acIII, and Synechococcus group B (Fig. 1E and 4). The betaproteobacterial GKS98 group (54) was detected by both RLB and DGGE/sequencing in hypersaline Lake Gahai 1, and the detection of LD2 bacteria by RLB in the large, polysaline Lake Qinghai confirmed a previous report on the detection of this group in the same habitat by an LD2-specific FISH probe (35).
The observed succession of proteobacterial groups along the salinity gradient is consistent with previous observations from dynamic saline systems but was previously never observed as clearly as in our investigation on stable freshwater and saline habitats. The high relative abundance of Betaproteobacteria in freshwater habitats and their decrease with increasing salinity was observed previously in studies on inland waters (13, 17) or estuaries and other habitats influenced by seawater (3, 6, 9, 25). A positive but weak correlation of the relative abundance of Alphaproteobacteria with salinity was observed in the Delaware Estuary (25), and investigations on habitats exceeding the salinity of seawater also demonstrated high relative abundances of this bacterial group (3, 23). High percentages of Gammaproteobacteria under hypersaline conditions were also observed in a solar saltern (3) and in Mono Lake (23) but not under the eusaline or polysaline conditions present in estuaries (6, 25). The observed relationship between the relative abundance of these three proteobacterial groups and salinity seems to be independent of the stability of the habitats. This relationship was consistently found in dynamic habitats, such as solar salterns (3) and estuaries (6), as well as in the Tibetan lakes characterized by stable salinity. Thus, relatively rapid increases of salinity to certain salinity levels (as in solar salterns) and stable salinity conditions at the same level of salinity select for taxa within the same proteobacterial classes but not for the same taxa.
According to general ecological principles, more-extreme environments are expected to be inhabited by less diverse communities (16). The performed DGGE analysis of BCC along the investigated salinity gradient seems to be in contradiction with this principle (Fig. 1D), since the number of DGGE bands did not decrease with increasing salinity. The same phenomenon was observed in investigations of BCC along a salinity gradient in a solar saltern (8). In both investigations, the decrease of bacterial diversity with increasing salinity was superimposed by a trend of increasing microdiversity (i.e., the appearance of closely related ribotypes slightly differing in 16S rRNA sequences) within the remaining groups of bacteria. Closely related sequences affiliated with such microdiverse groups appeared in the DGGE analysis as separate bands and compensated the disappearance of other bands. We observed this phenomenon in three independent groups, i.e., Psychroflexus spp., acIII lineage, and Loktanella spp. (see Fig. S2 in the supplemental material). Part of this microdiversity might result from heteroduplex formation; however, a substantial part of the microdiversity observed under hypersaline conditions might be real (8).
Ecophysiological adaptation to salinity.
Most of the groups of typical freshwater bacteria detected in the Tibetan habitats exclusively appeared at salinities of ≤2 g liter−1. The 37 strains representing five groups of typical freshwater bacteria (three of them were detected in Tibetan habitats) demonstrated in ecophysiological tests rather uniform salt tolerances, with the highest tolerated salinities, 5.6 to 8.1 g liter−1. These results may indicate the possible width of the fundamental niches of the strains; however, it is expected that the realized niches are significantly narrower. In agreement with this assumption, the two investigated Polynucleobacter subclusters appeared only in the Tibetan habitats at much lower salinities than those tolerated in the experiments (Fig. 1E). In contrast, the maximal salinity at which the six freshwater isolates (obtained from non-Tibetan habitats) affiliated with the GKS98 cluster grew was 10-fold lower than the maximal salinity at which this group was detected in the Tibetan habitats. This indicated significant differences in ecophysiological adaptations among members of the phylogenetically narrow GKS98 cluster (54). Obviously, these differences in ecophysiological adaptations are of importance in the colonization of ecologically diverse habitats. Members of the GKS98 cluster have been detected in soft water (33) in 13 out of 15 diverse freshwater lakes in Scandinavia (28) as well as in an acidic and humic freshwater pond (see Table S3 in the supplemental material).
In contrast to the strains affiliated with typical groups of freshwater bacteria, the seven strains isolated from Tibetan freshwater habitats demonstrated pronounced halotolerances in the ecophysiological experiment. This may indicate a larger niche width in terms of salinity. Interestingly, the three taxa represented by these seven isolates have never been reported to be abundant in freshwater or saline habitats. This may indicate a trade-off between adaptations providing a broad halotolerance and adaptations providing a high competitiveness.
Conclusions.
Salinity was the dominating environmental factor controlling BCC in the investigated lakes. Especially, the observed systematic succession of proteobacterial groups along the investigated salinity gradient demonstrated the structuring influence of salinity. No other previously revealed factor influencing BCC in lacustrine inland waters (27, 28, 35, 46, 52) has been found to be as powerful as salinity. In contrast to salinity, altitude and/or UVR impact did not limit the distribution of several groups of typical freshwater bacteria. Only minor compositional overlaps between bacterioplankton communities of freshwater and hypersaline lakes were observed, while in all previous comparisons of BCC between ecologically diverse freshwater habitats (differing in pH, trophic status, dissolved organic carbon, etc.), much larger overlaps have been observed (28, 52, 55). These observed overlaps of BCCs of different freshwater systems, as well as the rare overlaps between freshwater and saline systems, were attributed to the presence of phylogenetic groups with high ecological plasticity. The most pronounced ecological plasticity was observed for the GKS98 group (54), which inhabits freshwater habitats with very low salt concentrations (e.g., Lake Gossenköllesee [33]) as well as hypersaline lakes with a twofold-higher salinity than seawater (Fig. 1E). In the case of this group, the ecological plasticity was not an intrinsic trait of all strains affiliated with the group. In fact, the ecological plasticity seemed to result from a diversity of specific ecophysiological adaptations among members of this group. This conclusion is in accordance with previously demonstrated ecophysiological diversity within microdiverse groups of freshwater bacteria (5, 20, 24, 50) and highlights the significance of microdiversity in the ecology of at least some freshwater bacteria.
. . . .
Supplementary Material
Acknowledgments
We would like to thank Xiangdong Yang, Xingqi Liu, and Weilang Xia for their assistance in sampling of the lakes and Hongxi Pang for water chemistry analysis.
The China National Key Basic Research Project (G2005CB422000) supported the field sampling, and the National Natural Science Foundation of China (grant 30370278) and the Austrian Science Foundation (grant P15655) funded the research.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alfreider, A., J. Pernthaler, R. Amann, B. Sattler, F. Glöckner, A. Wille, and R. Psenner. 1996. Community analysis of the bacterial assemblages in the winter cover and pelagic layers of a high mountain lake by in situ hybridization. Appl. Environ. Microbiol. 62:2138-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benlloch, S., A. López-López, E. O. Casamayor, L. Øvreås, V. Goddard, F. L. Daae, G. Smerdon, R. Massana, I. Joint, F. Thingstad, C. Pedrós-Alió, and F. Rodríguez-Valera. 2002. Prokaryotic genetic diversity throughout the salinity gradient of a coastal solar saltern. Environ. Microbiol. 4:349-360. [DOI] [PubMed] [Google Scholar]
- 4.Blumthaler, M., W. Ambach, and R. Ellinger. 1997. Increase in solar UV radiation with altitude. J. Photochem. Photobiol. B 39:130-134. [Google Scholar]
- 5.Boenigk, J., P. Stadler, A. Wiedlroither, and M. W. Hahn. 2004. Strain-specific differences in the grazing sensitivity of closely related ultramicrobacteria affiliated with the Polynucleobacter cluster. Appl. Environ. Microbiol. 70:5787-5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouvier, T. C., and P. A. del Giorgio. 2002. Compositional changes in free living bacterial communities along a salinity gradient in two temperate estuaries. Limnol. Oceanogr. 47:453-470. [Google Scholar]
- 7.Casamayor, E. O., J. I. Calderón-Paz, and C. Pedrós-Alió. 2000. 5S rRNA fingerprints of marine bacteria, halophilic archaea and natural prokaryotic assemblages along a salinity gradient. FEMS Microbiol. Ecol. 34:113-119. [DOI] [PubMed] [Google Scholar]
- 8.Casamayor, E. O., R. Massana, S. Benlloch, L. Øvreas, B. Diez, V. J. Goddard, J. M. Gasol, I. Joint, F. Rodriguez-Valera, and C. Pedros-Alio. 2002. Changes in archaeal, bacterial and eukaryal assemblages along a salinity gradient by comparison of genetic fingerprinting methods in a multi-pond solar saltern. Environ. Microbiol. 4:338-348. [DOI] [PubMed] [Google Scholar]
- 9.Cottrell, M. T., and D. L. Kirchman. 2003. Contribution of major bacterial groups to bacterial biomass production (thymidine and leucine incorporation) in the Delaware Estuary. Limnol. Oceanogr. 48:168-178. [Google Scholar]
- 10.Crosbie, N. D., M. Pöckl, and T. Weisse. 2003. Dispersal and phylogenetic diversity of nonmarine picocyanobacteria, inferred from 16S rRNA gene and cpcBA-intergenic spacer sequence analyses. Appl. Environ. Microbiol. 69:5716-5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crump, B. C., C. S. Hopkinson, M. L. Sogin, and J. E. Hobbie. 2004. Microbial biogeography along an estuarine salinity gradient: combined influences of bacterial growth and residence time. Appl. Environ. Microbiol. 70:1494-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crump, B. C., E. V. Armbrust, and J. A. Baross. 1999. Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia River, its estuary, and the adjacent coastal ocean. Appl. Environ. Microbiol. 65:3192-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Demergasso, C., E. O. Casamayor, G. Chong, P. Galleguillos, L. Escudero, and C. Pedrós-Alió. 2004. Distribution of prokaryotic genetic diversity in athalassohaline lakes of the Atacama Desert, northern Chile. FEMS Microbiol. Ecol. 48:57-69. [DOI] [PubMed] [Google Scholar]
- 14.Donachie, S. P., J. P. Bowman, and M. Alam. 2004. Psychroflexus tropicus sp. nov., an obligately halophilic Cytophaga-Flavobacterium-Bacteroides group bacterium from an Hawaiian hypersaline lake. Int. J. Syst. Evol. Microbiol. 54:935-940. [DOI] [PubMed] [Google Scholar]
- 15.Donachie, S. P., S. Hou, K.-S. Lee, C. W. Riley, A. Pikina, C. Belisle, S. Kempe, T. S. Gregory, A. Bossuyt, J. Boerema, J. Liu, T. A. Freitas, A. Malahoff, and M. Alam. 2004. The Hawaiian archipelago: a microbial diversity hotspot. Microb. Ecol. 48:509-520. [DOI] [PubMed] [Google Scholar]
- 16.Frontier, S. 1985. Diversity and structure in aquatic ecosystems. Oceanogr. Mar. Biol. Annu. Rev. 23:253-312. [Google Scholar]
- 17.Glöckner, F. O., B. M. Fuchs, and R. Amann. 1999. Bacterioplankton composition of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl. Environ. Microbiol. 65:3721-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenberg, A. E., L. S. Clesceri, and A. D. Eaton. 1992. Standard methods for the examination of water and wastewater. American Public Health Association, Washington, D.C.
- 19.Hahn, M. W. 2003. Isolation of strains belonging to the cosmopolitan Polynucleobacter necessarius cluster from freshwater habitats located in three climatic zones. Appl. Environ. Microbiol. 69:5248-5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hahn, M. W., and M. Pöckl. 2005. Ecotypes of planktonic Actinobacteria with identical 16S rRNA genes adapted to thermal niches in temperate, subtropical, and tropical freshwater habitats. Appl. Environ. Microbiol. 71:766-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hahn, M. W., H. Lünsdorf, Q. L. Wu, M. Schauer, M. G. Höfle, J. Boenigk, and P. Stadler. 2003. Isolation of novel ultramicrobacteria classified as Actinobacteria from five freshwater habitats in Europe and Asia. Appl. Environ. Microbiol. 69:1442-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hahn, M. W., P. Stadler, Q. L. Wu, and M. Pöckl. 2004. The filtration-acclimatization method for isolation of an important fraction of the not readily cultivable bacteria. J. Microbiol. Methods 57:379-390. [DOI] [PubMed] [Google Scholar]
- 23.Humayoun, S. B., N. Bano, and J. T. Hollibaugh. 2003. Depth distribution of microbial diversity in Mono Lake, a meromictic soda lake in California. Appl. Environ. Microbiol. 69:1030-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaspers, E., and J. Overmann. 2004. Ecological significance of microdiversity: identical 16S rRNA gene sequences can be found in bacteria with highly divergent genomes and ecophysiologies. Appl. Environ. Microbiol. 70:4831-4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirchman, D. L., A. I. Dittel, R. R. Malmstrom, and M. T. Cottrell. 2005. Biogeography of major bacterial groups in the Delaware Estuary. Limnol. Oceanogr. 50:1697-1706. [Google Scholar]
- 26.Langenheder, S., V. Kisand, J. Wikner, and L. J. Tranvik. 2003. Salinity as a structuring factor for the composition and performance of bacterioplankton degrading riverine DOC. FEMS Microbiol. Ecol. 45:189-202. [DOI] [PubMed] [Google Scholar]
- 27.Lindström, E. 2001. Investigating influential factors on bacterioplankton community composition: results from a field study of five mesotrophic lakes. Microb. Ecol. 42:598-605. [DOI] [PubMed] [Google Scholar]
- 28.Lindström, E. S., M. P. Kamst-Van Agterveld, and G. Zwart. 2005. Distribution of typical freshwater bacterial groups is associated with pH, temperature, and lake water retention time. Appl. Environ. Microbiol. 71:8201-8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manz, W., R. Amann, W. Ludwig, M. Vancanneyt, and K.-H. Schleifer. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology 142:1097-1106. [DOI] [PubMed] [Google Scholar]
- 30.Manz, W., R. Amann, W. Ludwig, M. Wagner, and K.-H. Schleifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 31.Muyzer, G., S. Hottenträger, A. Teske, and C. Waver. 1995. Denaturing gradient gel electrophoresis of PCR-amplified 16S rDNA—a new molecular approach to analyse the genetic diversity of mixed microbial communities, p. 3.4.4.1-3.4.4.22. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual, 2nd ed. Kluwer, Dordrecht, The Netherlands.
- 32.Neef, A. 1997. Anwendung der in situ-Einzelzell-Identifizierung von Bakterien zur Populationsanalyse in komplexen mikrobiellen Biozönosen. Ph.D. thesis. Technische Universität München, Munich, Germany.
- 33.Pernthaler, J., F.-O. Glöckner, S. Unterholzner, A. Alfreider, R. Psenner, and R. Amann. 1998. Seasonal community and population dynamics of pelagic bacteria and archaea in a high mountain lake. Appl. Environ. Microbiol. 64:4299-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roller, C., M. Wagner, R. Amann, W. Ludwig, and K.-H. Schleifer. 1994. In situ probing of gram-positive bacteria with high DNA G + C content using 23S rRNA-targeted oligonucleotides. Microbiology 140:2849-2858. [DOI] [PubMed] [Google Scholar]
- 35.Schauer, M., C. Kamenik, and M. W. Hahn. 2005. Ecological differentiation within a cosmopolitan group of planktonic freshwater bacteria (SOL cluster, Saprospiraceae, Bacteroidetes). Appl. Environ. Microbiol. 71:5900-5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schauer, M., V. Balagué, C. Pedrós-Alió, and R. Massana. 2003. Seasonal changes in the taxonomic composition of bacterioplankton in a coastal oligotrophic system. Aquat. Microb. Ecol. 31:163-174. [Google Scholar]
- 37.Selje, N., and M. Simon. 2003. Composition and dynamics of particle-associated and free-living bacterial communities in the Weser Estuary, Germany. Aquat. Microb. Ecol. 30:221-237. [Google Scholar]
- 38.Sommaruga, R. 2001. The role of solar UV radiation in the ecology of alpine lakes. J. Photochem. Photobiol. B 62:35-42. [DOI] [PubMed] [Google Scholar]
- 39.Stahl, D. A., and R. Amann. 1991. Development and application of nucleic acid probes, p. 205-248. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons Ltd., Chichester, United Kingdom.
- 40.ter Braak, C. J. F., and P. F. M. Verdonschot. 1995. Canonical correspondence analysis and related multivariate methods in aquatic ecology. Aquat. Sci. 57:255-289. [Google Scholar]
- 41.ter Braak, C. J. F. 1987. The analysis of vegetation-environment relationships by canonical correspondence analysis. Vegetatio 69:69-77. [Google Scholar]
- 42.Van Trappen, S., J. Mergaert, and J. Swings. 2004. Loktanella salsilacus gen. nov., sp. nov., Loktanella fryxellensis sp. nov. and Loktanella vestfoldensis sp. nov., new members of the Rhodobacter group, isolated from microbial mats in Antarctic lakes. Int. J. Syst. Evol. Microbiol. 54:1263-1269. [DOI] [PubMed] [Google Scholar]
- 43.Wallner, G., R. Amann, and W. Beisker. 1993. Optimizing fluorescent in situ-hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14:136-143. [DOI] [PubMed] [Google Scholar]
- 44.Wang, S., and H. Dou. 1998. Lakes in China. Science Press, Beijing, China. (In Chinese.)
- 45.Warnecke, F., R. Amann, and J. Pernthaler. 2004. Actinobacterial 16S rRNA genes from freshwater habitats cluster in four distinct lineages. Environ. Microbiol. 6:242-253. [DOI] [PubMed] [Google Scholar]
- 46.Warnecke, F., R. Sommaruga, R. Sekar, J. S. Hofer, and J. Pernthaler. 2005. Abundances, identity, and growth state of Actinobacteria in mountain lakes of different UV transparency. Appl. Environ. Microbiol. 71:5551-5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wetzel, R. G. 2001. Limnology. Academic Press, San Diego, Calif.
- 48.Wu, Q. L., and M. W. Hahn. 2006. Differences in structure and dynamics of Polynucleobacter communities in a temperate and a subtropical lake revealed at three phylogenetic levels. FEMS Microbiol. Ecol. 57:67-79. [DOI] [PubMed] [Google Scholar]
- 49.Wu, Q. L., and M. W. Hahn. High predictability of the seasonal dynamics of a species-like Polynucleobacter population in a freshwater lake. Environ. Microbiol., in press. [DOI] [PubMed]
- 50.Wu, Q. L., J. Boenigk, and M. W. Hahn. 2004. Successful predation of filamentous bacteria by a nanoflagellate challenges current models of flagellate bacterivory. Appl. Environ. Microbiol. 70:332-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang, X. D., C. Kamenik, R. Schimidt, and S. M. Wang. 2003. Diatom-based conductivity and water-level inference models from eastern Tibetan (Qinghai-Xizang) Plateau lakes. J. Paleolimnol. 30:1-19. [Google Scholar]
- 52.Yannarell, A. C., and E. W. Triplett. 2004. Within- and between-lake variability in the composition of bacterioplankton communities: investigations using multiple spatial scales. Appl. Environ. Microbiol. 70:214-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng, D., and T. D. Yao. 2004. Uplifting of Tibetan Plateau with its environmental effects. Science Press, Beijing, China. (In Chinese.)
- 54.Zwart, G., B. C. Crump, M. P. Kamst-van Agterveld, F. Hagen, and S.-K. Han. 2002. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat. Microb. Ecol. 28:141-155. [Google Scholar]
- 55.Zwart, G., E. J. van Hannen, M. P. Kamst-van Agterveld, K. van der Gucht, E. S. Lindström, J. Van Wichelen, T. Lauridsen, B. C. Crump, S.-K. Han, and S. Declerck. 2003. Rapid screening for freshwater bacterial groups by using reverse line blot hybridization. Appl. Environ. Microbiol. 69:5875-5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.