Abstract
External carbon sources can enhance denitrification rates and thus improve nitrogen removal in wastewater treatment plants. The effects of adding methanol and ethanol on the genetic and metabolic diversity of denitrifying communities in activated sludge were compared using a pilot-scale plant with two parallel lines. A full-scale plant receiving the same municipal wastewater, but without external carbon source addition, was the reference. Metabolic profiles obtained from potential denitrification rates with 10 electron donors showed that the denitrifying communities altered their preferences for certain compounds after supplementation with methanol or ethanol and that methanol had the greater impact. Clone libraries of nirK and nirS genes, encoding the two different nitrite reductases in denitrifiers, revealed that methanol also increased the diversity of denitrifiers of the nirS type, which indicates that denitrifiers favored by methanol were on the rise in the community. This suggests that there might be a niche differentiation between nirS and nirK genotypes during activated sludge processes. The composition of nirS genotypes also varied greatly among all samples, whereas the nirK communities were more stable. The latter was confirmed by denaturing gradient gel electrophoresis of nirK communities on all sampling occasions. Our results support earlier hypotheses that the compositions of denitrifier communities change during predenitrification processes when external carbon sources are added, although no severe effect could be observed from an operational point of view.
Considerable efforts have been made to improve the technology for efficient and economic removal of nitrogen by denitrification in municipal activated sludge. Denitrification is an anaerobic microbial respiration process with a stepwise reduction of nitrate or nitrite via nitric oxide to nitrous oxide or dinitrogen. In the ideal activated sludge process, bacteria denitrify by using carbon compounds in the influent wastewater as electron donors. As a means to improve control strategies for nitrogen removal, external carbon compounds can be added to enhance denitrification rates (10, 14, 18, 19, 34, 35). Higher rates allow reductions in the hydraulic retention time in the anoxic zones, making it possible to minimize basin volumes (23). For this purpose, methanol and ethanol are most commonly used in practice, although acetate has been reported to give the highest rates in most cases (12, 24, 28-30).
Although much is known about process performance after the addition of external carbon sources, little effort has been made to explain the specific effects on the denitrifying communities in activated sludge. The selective pressure from the carbon compound added might affect the composition of the denitrifying community, which would influence metabolic diversity and functional properties of the denitrifiers. For example, it was shown that acetate as the major carbon source in a laboratory-scale reactor selected for denitrifiers with nonflocculating properties, which would adversely affect sedimentation if they became abundant (8). Methanol addition to activated sludge often requires an adaptation period of up to several months before its effect on denitrification rates is fully recognized, but the response to ethanol or acetate seems to be faster (12, 13, 19, 22, 25, 29). Recent studies have shown that ethanol and acetate select for the Azoarcus, Dechloromonas, Thauera, and Acidovorax-like denitrifiers commonly found in treatment processes, which could explain the rapid adaptation to these external carbon sources (8, 17). The lag period observed before the denitrification rate increases after methanol addition has been assumed to be an effect of population shifts within the denitrifying community, and this supposition is strongly supported by the results of Ginige et al. (7). By employing stable isotope probing, they identified members of the family Methylophilales that were enriched in a laboratory-scale reactor operated under denitrifying conditions with methanol as the sole carbon source. Older studies using culture-dependent techniques reported that methanol selects for Hyphomicrobium spp. and Paracoccus spp. in postdenitrification processes (1, 3, 24, 26, 37), i.e., with anoxic zones for denitrification placed before the aerobic ones. Predenitrification processes, with denitrification as the first step, are more complex, and a large variety of carbon compounds is present in the influent wastewater. These systems should therefore allow for more diverse denitrifying communities to thrive than those in the cases mentioned above, where fewer easily degradable carbon sources other than the organic compound added were available.
The objective of this work was to compare the effects of methanol and ethanol as external carbon sources on the genetic and metabolic diversity of denitrifying communities in predenitrifying activated sludge. The genes nirK and nirS, which encode the two structurally different but functionally equal nitrite reductases in the denitrification pathway, were exploited as markers to target the dominant nir genotypes in two parallel pilot-scale plants fed with methanol or ethanol and a full-scale plant without external carbon sources. To compare all samples, we analyzed the nirK communities by denaturing gradient gel electrophoresis (DGGE), whereas clone libraries of nirK and nirS genes were compared in samples from processes completely adapted to methanol or ethanol and from full-scale and pilot-scale processes without external carbon addition. In addition, the community function and metabolic diversity were assessed by non-growth-dependent electron donor use profiling.
MATERIALS AND METHODS
Wastewater treatment plant and sampling.
Activated sludge samples were collected from an experimental line (1,700 m3) in the full-scale plant and from a 2.4-m3 pilot-scale plant with two parallel lines (2) at the Kungsängen municipal wastewater treatment plant in Uppsala, Sweden. Both the full-scale plant and the pilot-scale plant were operated as single-sludge systems with a predenitrification design, i.e., the anoxic zone for denitrification was located before the aerobic zone. The nitrate produced by the nitrifying bacteria was therefore recirculated to the anaerobic denitrification zone to meet the influent flow of wastewater. The total sludge retention time was 26 days in the full-scale plant and 11 days in the pilot plant. All lines received the same preprecipitated sewage with an average chemical oxygen demand (COD) of 80 mg filtered COD liter−1. Both lines in the pilot-scale plant were initially operated without the addition of an external carbon source under stable conditions for 2 months. At the start of the experiment, methanol or ethanol was added to the influent in amounts equivalent to 100 mg COD liter−1, and this concentration was kept throughout the entire experiment. The full-scale line received nothing but preprecipitated sewage. The nitrogen removal was 80% for all three treatment plants during the experimental period.
From the methanol line in the pilot plant, samples were taken 6 days prior to methanol addition (RP2) and after 21 (MP1) and 65 days (MP2) of continuous methanol dosage. To compare the effects of methanol and ethanol addition, the parallel ethanol line was also sampled 6 days prior to ethanol addition (RP1) and on day 28 (EP1) and day 65 (EP2). The full-scale plant without an external carbon source was used as a reference and was sampled twice during the experimental period. The first sample (RF1) was taken on the same day as the RP1 and RP2 samples to compare processes without external carbon addition, and the second (RF2) was taken on day 30 to determine the stability and reproducibility of community patterns and metabolic profiles in the reference plant. On each sampling occasion, a 1-liter sample was withdrawn from the return-sludge flow from the last anaerobic zone in each line and transferred to three 250-ml flasks under anaerobic conditions, and the activity was measured within 1 h of sampling. In addition, 10 1-ml aliquots of each sludge sample were centrifuged for 10 min at 14,000 rpm, and the pellets were frozen at −20°C prior to DNA extraction.
Potential denitrification activity assay for metabolic profiles.
The potential denitrification activities with 10 different electron donors (formate, acetate, propionate, butyrate, ethanol, methanol, glycerol, glucose, glutamine, and benzoate) were measured in triplicate using the acetylene inhibition technique (42) to generate electron donor use profiles for samples from the processes without external carbon addition (RP1, RP2, RF1, and RF2), with methanol addition (MP2), and with ethanol addition (EP2). The electron donors were selected to cover a diversity of metabolic pathways. Rates obtained with only nitrate added (NA) in the assay were the controls. Activated sludge (10 ml) was added to 39 ml 10 mM sodium phosphate buffer (pH 7.2) in glass flasks (118 ml) with an anaerobic atmosphere but was otherwise treated as described by Hallin and Pell (11). The substrate was added by injecting 1 ml of a stock solution to reach an initial concentration of 2 mM KNO3-N and 100 mg C liter−1. Samples were incubated at 15°C for 1 h, and 0.5-ml gas samples were withdrawn every 12 min.
Nitrous oxide was analyzed on a gas chromatograph (Chrompack 9000; Chrompack, Rotterdam, The Netherlands) equipped with a 63Ni electron capture detector. Denitrification rates were calculated from linear regression of the N2O produced during incubation and expressed on a mixed-liquor volatile suspended solids (MLVSS) basis (11). MLVSS were measured according to Swedish standards (SS-EN 872, Swedish Standards Institute [http://www.sis.se]). In short, the amount of suspended solids was first determined by vacuum filtration of the sludge sample through a preignited 0.7-μm fiberglass filter and drying at 105°C. The amount of MLVSS was then calculated from the weight loss after incineration at 550°C.
Comparative study.
The data from the electron donor use profiles were merged with those from previously published profiles obtained from other experiments at the same treatment plant (12). Variables from the data in the previous study that had no equivalent in the new data were excluded and vice versa. The data were normalized by dividing each individual variable by the total sum of rates for each sample profile. The total data set was then subjected to principal component analysis in order to elucidate patterns of electron donor use profiles in the differently operated processes and over time, using the Unscrambler software package (Camos AS, Oslo, Norway).
DNA extraction and PCR amplification of nirK and nirS.
Genomic DNAs were extracted in triplicate from all samples, except for the RP1 sample, which was lost, by using a FastDNA Spin kit for soil (Qbiogene Inc., Carlsbad, CA) according to the manufacturer's protocol, with a few modifications. The sludge pellets were first dissolved in 978 μl phosphate buffer supplied with the kit and then transferred to multimix tubes to which 122 μl MT buffer from the kit was added. The isolated DNAs were stored at −20°C.
Amplification of a 473-bp nirK fragment for DGGE analysis and cloning and of a 425-bp nirS fragment for cloning was performed in triplicate reactions for each DNA extract, using the F1aCu/R3Cu primer set, with and without a GC clamp, and the cd3aF/R3cd primer set (36), respectively. The oligonucleotides were purchased from Invitrogen (Carlsbad, CA). The gene fragments were amplified with initial denaturation of the DNA at 94°C for 2 min, followed by 28 cycles of 30 s at 94°C, 1 min at 57°C, and 1 min at 72°C. The reactions were completed after 10 min at 72°C. PCRs were carried out in a minicycler (MJ Research, Waltham, MA), and each 25-μl reaction mix contained 2.5 μl of 10× PCR buffer (500 mM KCl, 15 mM MgCl2, and 100 mM Tris-HCl, pH 9.0, at room temperature), a 200 μM concentration of each deoxynucleoside triphosphate, 1.25 U of Taq polymerase (GE Healthcare, Giles, United Kingdom), 1.0 μM of each primer, and approximately 20 ng DNA. For nirK and nirS reactions, bovine serum albumin (GE Healthcare) was added to reach final concentrations of 400 and 1,000 ng μl−1, respectively.
DGGE of nirK.
The nirK amplicons with a GC clamp were resolved by DGGE analysis. Prior to DGGE, the three PCR products from the same DNA extract were pooled, but the triplicate DNA extracts from each sample were treated separately throughout the subsequent analysis. The nirK PCR products were separated using a Dcode system (Bio-Rad Laboratories Inc., Hercules, CA) according to the method of Throbäck et al. (36). Gels composed of 37.5:1 acrylamide:bisacrylamide (7% [vol/vol]) with a gradient of 50 to 70% denaturant (formamide and urea) were cast with a gradient maker (Bio-Rad Laboratories Inc.). After electrophoresis at 130 V for 13 h at 60°C in TAE buffer (40 mM Tris-acetate, 1 mM EDTA, pH 7.5), the migration patterns were visualized by staining with 1:10,000 SYBR green (Molecular Probes, Eugene, Oreg.) for 30 min followed by UV transillumination. Images were documented with a video camera (Ikegami Tsushinki Co., Tokyo, Japan), and the pictures were scanned to create digital images.
Cloning of nirK and nirS and RFLP analysis.
Four clone libraries for each of the amplified nirK and nirS genes were constructed from samples taken from the pilot-scale plant before methanol addition (RP2) and after 65 days of continuous methanol dosage (MP2), from the parallel plant fed with ethanol on day 65 (EP2), and from the full-scale plant at the start of the experimental period (RF1). Prior to cloning, the nirK or nirS PCR products from the same DNA extract were pooled, separated by agarose gel (1%) electrophoresis, and purified with a JETquick gel extraction spin kit (Genomed, Löhne, Germany) to avoid insertions of incorrect amplicons. The PCR products were then cloned by using a TOPO TA cloning kit for sequencing (Invitrogen, Carlsbad, CA). Small amounts of cell material from randomly picked clones were collected with toothpicks and resuspended in 25-μl preprepared PCR mixtures, and the inserts were amplified as described above. In total, 70 nirK and 60 nirS PCR products of the correct size from each library were digested overnight at 37°C, using HaeIII for nirK and HhaI for nirS, and then screened by restriction fragment length polymorphism (RFLP). The digested products were separated by 2% DNA agarose gel electrophoresis at 80 V for 2.0 h.
The RFLP patterns were compared by using Quantity One 1-D analysis software (Bio-Rad Laboratories Inc.) and were grouped into pattern types. Library coverage (C) was estimated as follows: C = 1 − nN−1, where n is the number of different RFLP pattern types from a clone library that were encountered only once and N is the total number of clones analyzed. The diversity of RFLP pattern types for nirK and nirS was analyzed using Analytic Rarefaction software (http://www.uga.edu/∼strata/software/Software.html).
Sequencing and phylogenetic analysis.
A selection of nirK and nirS clones from all RFLP pattern types represented by at least two clones were grown at 37°C overnight in LB medium, and the plasmids were isolated for sequencing using the QIAprep Spin miniprep kit protocol (QIAGEN, Valencia, CA). The nirK and nirS inserts were sequenced on one strand by Macrogen Inc. (Seoul, Korea), using an ABI3730 XL automatic DNA sequencer and the vector primer M13F. For sequencing of nirK bands from DGGE gels, the middle sections of the visible bands were excised from the denaturing gels, placed in 160 μl ultrapure water, and stored at −70°C until used for sequencing. To elute the DNA from the polyacrylamide gels, the samples were thawed for 1 h at room temperature, frozen at −70°C for 1 h, and then thawed again at 8°C for 12 h. The eluted fragments were PCR amplified with the nirK primers F1aCu and R3Cu without a GC clamp, using 4 μl template DNA in a total reaction volume of 50 μl, but otherwise following the method described above. Prior to sequencing, 45 μl of each PCR product was purified with a MicroSpin S-400 HR column (GE Healthcare). Both strands were sequenced in 10-μl reaction mixtures with a DYEnamic ET Terminator cycle sequencing kit (GE Healthcare), using an ABI PRISM 377 (Perkin-Elmer, Wellesley, MA) automated DNA sequencer with F1aCu and R3Cu as sequencing primers. The DGGE bands harboring more than one nirK sequence were further cloned by the procedure described above. From each mixed DGGE band, 10 clones with the correct insert were chosen for sequencing. Both strands of the insert were sequenced as described above, using the vector primers M13F and M13R.
The nirK and nirS sequences obtained in this study were aligned with nirK and nirS gene sequences from cultivated strains and a selection of sequences from environmental nir clones obtained in other studies available from the NCBI database, using CLUSTAL W software (http://www.ebi.ac.uk/clustalw/). Tree analysis was performed with TREECON software (39). Distance matrix analyses were performed with the Jukes and Cantor correction (20). The trees were reconstructed by using the neighbor-joining method of Saitou and Nei (31), and tree topology was evaluated by bootstrap analysis using 100 replicates. The resulting phylograms are given in the supplemental material.
Nucleotide sequence accession numbers.
The partial nirK and nirS gene sequences from this study have been deposited in the GenBank database under accession numbers DQ182155 to DQ182227 and DQ182111 to DQ182154, respectively.
RESULTS
Denitrification potentials and metabolic profiles.
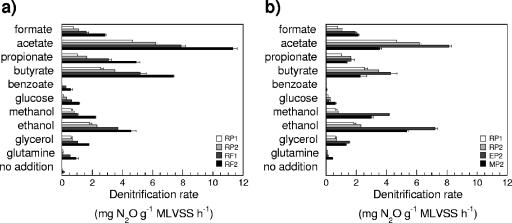
The metabolic profiles demonstrated that the full-scale reference plant had an overall higher potential to denitrify with all 10 different electron donors than the nonamended pilot-scale plant samples (RP1 and RP2), and between the last two samples, higher rates were determined throughout for the RP2 sample (Fig. 1a). There was also a difference between the two sampling occasions in the full-scale plant, showing that the total activity could vary from time to time. Nevertheless, even though the absolute values differed, the four profiles were similar. After continuous methanol dosage for 65 days, the denitrifiers in the activated sludge demonstrated an increased potential activity with methanol (Fig. 1b). In addition, the potential to denitrify with formate, ethanol, glucose, glutamine, and glycerol was enhanced. The capacity to denitrify with acetate, butyrate, and to some extent, propionate as electron donors was instead lower after 65 days of methanol dosage. The denitrifiers in the activated sludge in the ethanol-fed line (EP2) had a high potential to use nearly all 10 electron donors, except for glucose and glutamine, in comparison to the pilot-scale plant with or without methanol addition (Fig. 1b, MP2, RP1, and RP2). The denitrification potentials were especially pronounced for ethanol, methanol, acetate, and butyrate.
FIG. 1.
Potential denitrification rates with 10 organic compounds supplied in excess (means ± standard deviations; n = 3) to generate metabolic profiles for samples from (a) the pilot-scale plants prior to ethanol (RP1) and methanol (RP2) addition and the full-scale plant without an external carbon source on two occasions (RF1 and RF2) and (b) the pilot-scale plants with ethanol (EP2) and methanol (MP2) addition on day 65 in comparison to the pilot-scale plants prior to the addition of ethanol (RP1) and methanol (RP2).
Comparative study of metabolic profiles.
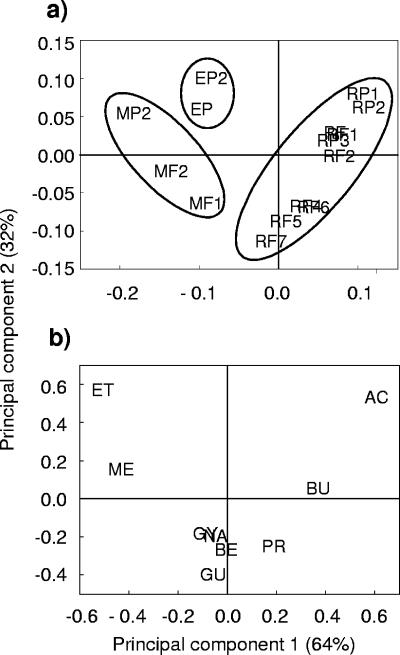
In the comparative study, principal components 1 and 2 together explained 96% of the total variation in the metabolic profiles (Fig. 2). The samples were divided into three clusters described by the influent carbon source (Fig. 2a). The observed clustering of the samples from processes without external carbon source addition demonstrates the similarity in electron donor preferences, although the full-scale samples (all denoted RF) showed greater variation than those from the pilot-scale plants (all denoted RP). The discriminating variables of the three clusters turned out to be the alcohols methanol and ethanol and the fatty acids acetate and butyrate (Fig. 2b). Methanol-fed sludge had increased activity with methanol and ethanol and decreased activity with fatty acids, as indicated by the locations in the score plot. The ethanol-fed sludge had an overall higher capacity to use even-numbered fatty acids and primary alcohols, thus shifting the activity up and to the right in the score plot. The covariance values between butyrate and acetate, between methanol and ethanol, and between benzoate, glucose, glycerol, and the no-addition control (NA) are illustrated in the loading plot.
FIG. 2.
Score plot (a) and loading plot (b) obtained by principal component analysis of potential denitrification rates with different electron donors to compare the metabolic profiles reported by Hallin and Pell (12) to those from this study, including those for the pilot-scale plants prior to ethanol (RP1) and methanol (RP2) addition, the full-scale plant without an external carbon source on two occasions (RF1 and RF2), and the pilot-scale plants with methanol (MP2) and ethanol (EP2) addition on day 65. Samples in score plot: P, pilot-scale plants; F, full-scale plants; E, addition of ethanol; M, addition of methanol; R, no external carbon source. Variables in loading plot: ME, methanol; ET, ethanol; AC, acetate; PR, propionate; BU, butyrate; GU, glucose; GY, glycerol; BE, benzoate; NA, no additional electron donor. Ellipses indicate processes with similar modes of operation.
DGGE analysis of nirK.
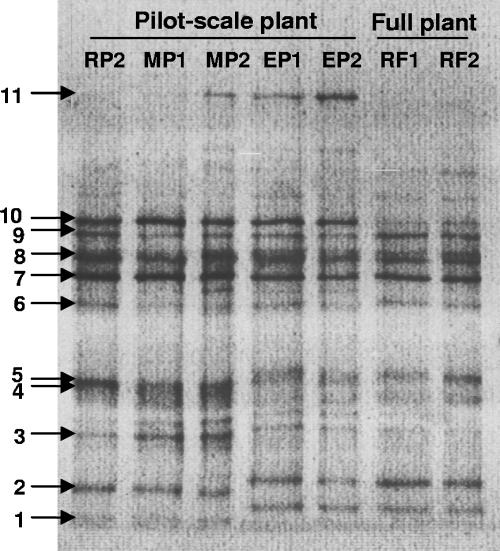
The GC-clamped nirK primers amplified a single band of the expected size (approximately 470 bp) from all samples. With the DGGE fingerprints, we detected at least 15 bands for each sludge sample (Fig. 3). The patterns were reproducible and identical for the three replicate DNA extracts for all samples (not shown). Ten bands dominated the pattern for all of the samples. Nevertheless, band 11 (denoted Kasp11) only appeared in the MP2 sample from the methanol-fed plant and in the EP1 and EP2 samples from the parallel plant receiving ethanol as an external carbon source.
FIG. 3.
DGGE analysis with SYBR green staining of PCR-amplified partial nirK genes from the pilot-scale plant prior to methanol addition (RP2), the pilot-scale plant with methanol addition on day 21 (MP1) and day 65 (MP2), the pilot-scale plant with ethanol addition on day 28 (EP1) and day 65 (EP2), and the full-scale plant without external carbon addition at the start of the experiment (RF1) and on day 30 (RF2).
The identities of the most dominant bands were confirmed by direct sequencing of the excised and reamplified fragments in bands 1 to 11 in Fig. 3. These bands were excised from all gels, and at least five randomly picked ones that had migrated to the same level were sequenced. In all cases, sequences retrieved from the same level were identical. Bands Kasp1 and Kasp6 harbored three different sequences each, and band Kasp5 held two, while all other bands each contained a single sequence. The 16 sequences derived from the DGGE analysis were related to other nirK sequences. Compared to environmental nirK clones, preferably from activated sludge, as well as to pure cultures by neighbor joining, 13 of the sequences showed <85% similarity to any known nirK sequence. The remaining three nirK sequences from the DGGE analysis were distributed all over the phylogenetic tree (see the supplemental material). The nirK sequence from band number 10 (Kasp10) was located within the Rhodobacter sphaeroides cluster, while sequences from band 8 (Kasp8) and clone Kasp6a from band number 6 clustered in the middle of the tree, where nirK sequences from many denitrifying strains and from different environments were located. The latter of the two was related to the nirK genes from Bradyrhizobium japonicum and Blastobacter denitrificans.
nirK and nirS clone libraries.
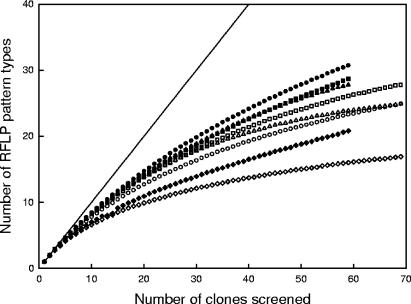
A total of 280 nirK- and 240 nirS-positive clones were screened with RFLP. The coverage for the eight clone libraries, based on RFLP pattern types, varied from 69 to 91% (Table 1). The rarefaction curves for each library did not reach a plateau, indicating that both nirK and nirS genotype diversity was high in all samples (Fig. 4). Nevertheless, the diversity of nirS genotypes was higher than the nirK diversity within each plant operation mode. The lowest level of nir diversity was found in the clone libraries from the full-scale plant without an external carbon source. The rarefaction curves indicated less nirK diversity in the processes with methanol or ethanol addition than in the pilot-scale plant without supplemental carbon (RP2), but there was more nirS diversity in the methanol sample. Of the 280 nirK clones screened, 79 different RFLP pattern types were found, and 41 of these patterns were represented by more than one clone. For nirS, 96 different RFLP patterns were detected, and 36 of these were represented by at least two clones among the 240 screened.
TABLE 1.
Coverage of nirK and nirS clone libraries based on RFLP pattern typesa
| Sample | Library coverage (%)
|
|
|---|---|---|
| nirK | nirS | |
| RP2 | 81 | 70 |
| MP2 | 84 | 69 |
| EP2 | 88 | 75 |
| RF1 | 91 | 78 |
RP2, sample from the pilot-scale plant prior to methanol addition; MP2, sample from the pilot-scale plant with methanol addition; EP2, sample from the pilot-scale plant with ethanol addition; RF1, sample from the full-scale plant without an external carbon source.
FIG. 4.
Rarefaction curves showing the numbers of RFLP pattern types obtained from the numbers of nirK (white) and nirS (black) clones screened from the pilot-scale plant sludge prior to methanol addition (RP2; squares), after methanol adaptation on day 65 (MP2; circles), and after ethanol adaptation on day 65 (EP2; triangles) and the sludge from the full-scale plant without external carbon addition at the start of the experiment (RF1; diamonds). The solid line shows the maximum possible level of diversity.
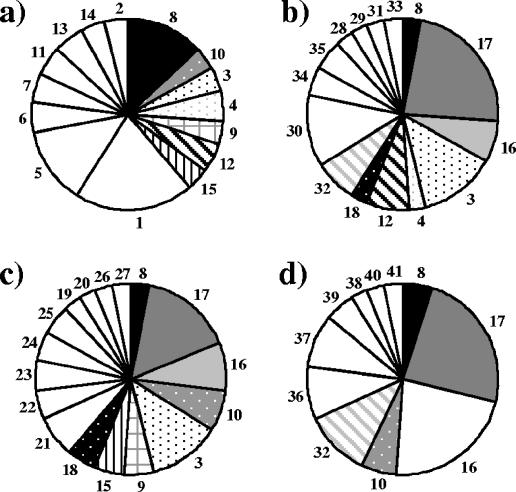
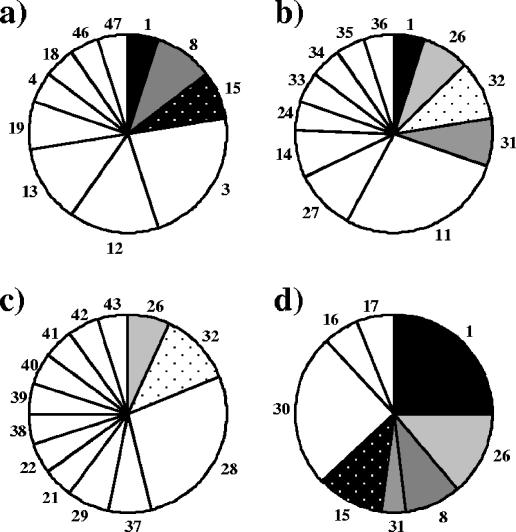
On the basis of the distribution of RFLP patterns represented by at least two clones in a library, it was shown that the four nirK libraries were more similar to each other than were the nirS libraries (Fig. 5 and 6). However, all eight libraries contained many RFLP pattern types that were specific for the samples, especially the nirS libraries. For the nirK libraries, the methanol and ethanol samples shared five RFLP pattern types (patterns 3, 8, 16, 17, and 18), and these constituted about 50% of each library (Fig. 5). Pattern types 8, 16, and 17 made up half of the library for the sample from the full-scale plant, while the pilot-scale plant sample taken prior to the addition of methanol was dominated by sample-specific pattern types. The distribution of dominant RFLP pattern types for the nirS clones illustrates that all samples were almost unique, although the ethanol plant sample differed the most (Fig. 6).
FIG. 5.
Distributions of RFLP pattern types represented by at least two nirK clones in (a) the pilot-scale plant prior to methanol addition (RP2), (b) the methanol-fed pilot-scale plant on day 65 (MP2), (c) the ethanol-fed plant on day 65 (EP2), and (d) the full-scale plant without an external carbon source at the start of the experiment (RF1). White pie slices represent RFLP pattern types specific for the sample, and the numbers refer to the RFLP pattern types.
FIG. 6.
Distributions of RFLP pattern types represented by at least two nirS clones in (a) the pilot-scale plant prior to methanol addition (RP2), (b) the methanol-fed pilot-scale plant on day 65 (MP2), (c) the ethanol-fed plant on day 65 (EP2), and (d) the full-scale plant without an external carbon source at the start of the experiment (RF1). White pie slices represent RFLP pattern types specific for the sample, and the numbers refer to the RFLP pattern types.
In total, 57 nirK and 44 nirS clones from RFLP pattern types represented by at least two clones were sequenced. One to four clones representing each RFLP pattern per library were sequenced, and all showed homology to other nirK or nirS sequences. With a few exceptions, clones sequenced from one pattern type had similar sequences, indicating that nir diversity could be resolved by RFLP analysis. The nir sequences from the activated sludge samples in this study were scattered all over the neighbor-joining phylograms, with no specific clustering due to treatment (see the supplemental material). A few nirK sequences clustered with nirK genes from Rhodobacter sphaeroides, Alcaligenes spp., Mesorhizobium sp., Blastobacter denitrificans, or Bradyrhizobium japonicum, although none of these were represented in the most dominant RFLP types. Several of the nirS sequences were found in the same cluster as Paracoccus sp. sequences. Those from RFLP pattern type 30 were related to genes form Alcaligenes faecalis, Pseudomonas stutzeri, Thauera terpenica, and Azoarcus spp., while the others were related to different environmental clones. Sequences from nirK pattern types 9 and 12 as well as from nirS pattern type 16 were only very distantly related to any deposited nir gene sequences.
DISCUSSION
Electron donor use profiles showed that functional alterations in the denitrifying community occurred in predenitrification systems supplemented with an external carbon source, which supports previous findings (12). Despite the observed differences in absolute values of potential denitrification rates over time in the reference sludge and the difference between the two pilot-scale plants prior to the addition of the external carbon sources, the patterns in the four profiles were similar and indicative of a typical electron donor use preference for nonamended sludge. This demonstrated treatment effect supports the interpretation of the altered profiles for the sludge with methanol or ethanol addition. The profile from the pilot-scale plant with methanol addition differed the most, while that from the ethanol plant was more similar to profiles from the plants without an external carbon source. Denitrification rates with acetate and butyrate showed a decrease in sludge from the process receiving methanol, even though a variety of electron donors were available in the influent wastewater. This confirms previous results obtained when methanol was added to a full-scale plant within the same municipal wastewater treatment facility (12). A reduced capacity to denitrify with fatty acids can be a negative development in predenitrification processes, since it reflects a lower potential to exploit the substances naturally occurring in wastewater. Volatile fatty acids represent the major fraction of easily degradable COD in wastewater, and acetate probably accounts for 5 to 10% of the total COD (15). The addition of either ethanol or methanol as an external carbon source enhanced the denitrification capacity with primary alcohols. This is in agreement with the results from postdenitrification systems reported by Nyberg et al. (28). In addition, they demonstrated that ethanol could be used in the start-up phase of a nitrogen removal process before switching over to methanol dosage to avoid the long lag period that is common for methanol addition.
The comparative analysis showed that treatment plants within a certain operational mode did not differ substantially from each other. The metabolic pattern stability observed was most likely due to stable operation of the treatment plant and similarity of the influent wastewater over time at the treatment facility. The pattern from principal component analysis was related to that obtained in the study by Hallin and Pell (12), even though the analysis in the present study included not only electron donor use profiles with fewer variables but also profiles from both full-scale and pilot-scale plant experiments conducted in different years. This strengthens the proposed preference for certain electron donors in activated sludge from plants operated either without an external carbon source or with methanol or ethanol addition, as discussed above. The profiles obtained from the pilot-scale plants treated with ethanol in the two experiments run in different years were more similar to each other than were those from full-scale and pilot-scale plants with methanol addition. This is likely explained by more controlled operation of the pilot-scale plants giving more reproducible results but also shows that there are differences that might depend on the size of the treatment plant. Nevertheless, the similarity in the metabolic profiles among the methanol-amended plants indicates that the pilot-scale plant was sufficiently large to mimic a full-scale plant.
Not only was activity affected differently by the external carbon sources, but the diversity in terms of richness in the denitrifying communities was also affected. In comparison to that in the sample obtained prior to commencement of methanol addition, the diversity of nirK genotypes decreased, while the nirS diversity increased when methanol was added to the process. The amounts of nirK diversity were similar in the methanol and ethanol plants, but the amount of nirS diversity was higher in the methanol plant than in the plant with ethanol addition. This indicates that methanol caused the growth of additional denitrifier populations that mainly use methanol even in predenitrification processes, as hypothesized from activity measurements in a previous study by Hallin et al. (10). The level of diversity of nirS genotypes was always higher than the level of nirK diversity within each operational mode. This is in agreement with the nirK and nirS distributions in cultured denitrifiers from a municipal wastewater treatment plant (43) and in clone libraries from a saline metallurgic wastewater treatment plant (41). Results from a biofilm reactor treating wastewater suggest that the concentration of organic compounds is a major factor in the competition between nirK and nirS genotypes, with lower concentrations favoring nirK (4). Yan et al. (40) speculated that nitrate levels might alter the proportions of nirK and nirS denitrifiers in groundwater and that the community dynamics of nirK genotypes affect the nirS community and vice versa.
The composition of nirK and nirS genotypes in the different samples implies that the denitrifier community structure was affected by external carbon sources. The effect was most pronounced on the nirS denitrifiers, while the nirK communities appeared to be more stable. All four nirS libraries differed from each other, showing distinctly different RFLP pattern types. The sample from the ethanol-fed plant differed the most, and the clone library was dominated by treatment-specific nirS RFLP types. The similarity among the nirK libraries fits with the DGGE analysis of nirK genes, which demonstrated that the same dominant denitrifiers of the nirK type were present on all sampling occasions. The resolution is lower for DGGE analysis than for clone library analysis, and therefore minor differences among the treatments could be detected by DGGE. The addition of methanol to the plant resulted in the appearance of an additional band, suggesting that a new nirK population was on the rise. This band was also detected in samples from the ethanol-fed plant. The sequence from this band clustered with other nirK sequences from this study, but nirK sequences from cultivated strains were not found within this cluster. The sequence was identical to sequences of RFLP type 10 in the nirK libraries, but this RFLP type did not dominate any of the clone libraries. Surprisingly, it was found in all libraries except that from the methanol plants, although the coverage of the nirK clone library from this sample was 84%.
As reported in other environmental studies of the functional genes in the denitrification pathway (e.g., see references 5 and 33), most of the dominant nir types in our study clustered with other environmental clones. This could indicate a large diverse population of unknown denitrifying bacteria in activated sludge. Since methanol added to the process increased the capacity of the denitrifiers in the sludge to utilize both methanol and formate for denitrification, methylotrophic denitrifiers would certainly be present. We speculate that some of these unknown nir genotypes are affiliated with nir genes from as yet uncultivated denitrifying methylotrophs. Recent results have shown that many bacteria involved in C1 cycling remain uncultivated and uncharacterized (27), and methanol was shown to select for methylotrophs not previously known as denitrifiers in activated sludge (7). Nevertheless, some of the dominant nirS types in our study were related to nirS genes from Paracoccus, Thauera, Azoarcus, and Alcaligenes faecalis, although none of them could be assigned to any specific operational mode. Accordingly, both cultivation and 16S rRNA-based studies have suggested that the families Comamonadaceae and Rhodocyclaceae, dominated by Azoarcus, Paracoccus, Hydrogenophaga, and Acidovorax species, are key players in denitrification in municipal and industrial activated sludge processes (6, 16, 21, 32). However, the identification of denitrifiers from the retrieved 16S rRNA sequences is difficult. Surprisingly, only a few functional gene approaches to target denitrifiers in municipal activated sludge have been published. Sequencing of the dominant nirK, nirS, and nosZ genes in activated sludge samples from two different full-scale treatment plants revealed that some genotypes were distantly related to those from Rhizobium spp., Azospirillum spp., Rhodobacter sphaeroides, and Paracoccus denitrificans, although the majority clustered with other environmental clones (36). In contrast, most nosZ clones clustered with nosZ sequences from well-known denitrifiers within the α (Azospirillum)- and γ-proteobacteria (Pseudomonas) in a biofilm pilot-scale reactor treating urban wastewater (9). Nearly 70% of the nirS clones analyzed in a sequencing batch reactor process with combined nitrogen and phosphorus removal were similar to nirS genes in Azoarcus and Thauera, within the Rhodocyclus group (38).
To conclude, we used a community-level approach to study the effects of methanol or ethanol as an external carbon source on both the structure and function of denitrifying bacterial communities in municipal activated sludge. Our results support an earlier hypothesis that the denitrifier community composition changes not only in postdenitrification processes, but also in predenitrification systems, when external carbon sources are added to speed up denitrification rates. Methanol had a greater impact than ethanol on the metabolic profiles of the communities, and this was linked to an increased level of diversity of denitrifiers of the nirS type. This indicates that denitrifying bacteria favored by methanol were on the rise in the community. The composition of nirS genotypes in the communities varied greatly among the samples, whereas the nirK communities were more stable, suggesting that there might be a niche differentiation between nirS and nirK genotypes in activated sludge processes. The results do not indicate any severe negative effect of either methanol or ethanol on denitrification and the denitrifying communities. Based on this study, we cannot conclude which carbon source to prefer. In the operation of sewage treatment plants, other aspects, such as sludge production, sludge settling properties, economy, etc., are equally important to consider when making a choice.
Supplementary Material
Acknowledgments
We thank the staff at the Kungsängen wastewater treatment plant in Uppsala, Sweden, for operating the pilot-scale plant during the experimental period.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Blaszczyk, M., E. Galka, E. Sakowicz, and R. Mycielski. 1985. Denitrification of high concentrations of nitrites and nitrates in synthetic medium with different sources of organic carbon. III. Methanol. Acta Microbiol. Pol. 34:195-206. [PubMed] [Google Scholar]
- 2.Carlsson, B., S. Hasselblad, E. Plaza, S. Mårtensson, and C.-F. Lindberg. 1997. Design and operation of a pilot-scale activated sludge plant. Vatten 53:27-32. [Google Scholar]
- 3.Claus, G., and H. J. Kutzner. 1985. Denitrification of nitrate and nitric acid with methanol as carbon source. Appl. Microbiol. Biotechnol. 22:378-381. [Google Scholar]
- 4.Cole, A. C., M. J. Semmens, and T. M. LaPara. 2004. Stratification of activity and bacterial community structure in biofilms grown on membranes transferring oxygen. Appl. Environ. Microbiol. 70:1982-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enwall, K., L. Philippot, and S. Hallin. 2005. Activity and composition of the denitrifying bacterial community respond differently to long-term fertilization. Appl. Environ. Microbiol. 71:8335-8343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Etchebehere, C., I. Errazquin, E. Barradeguy, P. Dabert, R. Moletta, and L. Muxí. 2001. Evaluation of the denitrifying microbiota of anoxic reactors. FEMS Microbiol. Ecol. 35:259-265. [DOI] [PubMed] [Google Scholar]
- 7.Ginige, M. P., P. Hugenholtz, H. Daims, M. Wagner, J. Keller, and L. L. Blackall. 2004. Use of stable-isotope probing, full-cycle rRNA analysis, and fluorescence in situ hybridization-microautoradiography to study a methanol-fed denitrifying microbial community. Appl. Environ. Microbiol. 70:588-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ginige, M. P., J. Keller, and L. L. Blackall. 2005. Investigation of an acetate-fed denitrifying microbial community by stable isotope probing, full-cycle rRNA analysis, and fluorescent in situ hybridization-microautoradiography. Appl. Environ. Microbiol. 71:8683-8691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gómez-Villalba, B., C. Calvo, R. Vilchez, J. González-López, and B. Rodelas. 4. January 2006, posting date. TGGE analysis of the diversity of ammonia-oxidizing and denitrifying bacteria in submerged filter biofilms for the treatment of urban wastewater. Appl. Microbiol. Biotechnol. [Online.] doi: 10.1007/s00253-005-0272-7. [DOI] [PubMed]
- 10.Hallin, S., C.-F. Lindberg, M. Pell, E. Plaza, and B. Carlsson. 1996. Microbial adaptation, process performance and a suggested control strategy in a pre-denitrifying system with ethanol dosage. Water Sci. Technol. 34:91-99. [Google Scholar]
- 11.Hallin, S., and M. Pell. 1994. Acetylene inhibition for measuring denitrification rates in activated sludge. Water Sci. Technol. 30:161-167. [Google Scholar]
- 12.Hallin, S., and M. Pell. 1998. Metabolic properties of denitrifying bacteria adapting to methanol and ethanol in activated sludge. Water Res. 32:13-18. [Google Scholar]
- 13.Hallin, S., M. Rothman, and M. Pell. 1996. Adaptation of denitrifying bacteria to acetate and methanol in activated sludge. Water Res. 30:1445-1450. [Google Scholar]
- 14.Hasselblad, S., and S. Hallin. 1998. Intermittent addition of external carbon to enhance denitrification in activated sludge. Water Sci. Technol. 37:227-233. [Google Scholar]
- 15.Henze, M., G. H. Kristensen, and R. Strube. 1994. Rate-capacity characterization of wastewater for nutrient removal processes. Water Sci. Technol. 29:101-107. [Google Scholar]
- 16.Hoshino, T., T. Terahara, S. Tsuneda, A. Hirata, and Y. Inamori. 2005. Molecular analysis of microbial population transition associated with the start of denitrification in a wastewater treatment process. J. Appl. Microbiol. 99:1165-1175. [DOI] [PubMed] [Google Scholar]
- 17.Hwang, C., W.-M. Wu, T. Gentry, J. Carley, S. Carroll, C. Schadt, D. Watson, P. Jardine, J. Zhou, R. Hickey, C. Criddle, and M. Fields. Posting date, 15 November 2005. Changes in bacterial community structure correlate with initial operating conditions of a field-scale denitrifying fluidized bed reactor. Appl. Microbiol. Biotechnol. [Online.] doi: 10.1007/s00253-005-0189-1. [DOI] [PubMed]
- 18.Isaacs, S. H., and M. Henze. 1995. Controlled carbon source addition to an alternating nitrification-denitrification wastewater treatment process including biological P removal. Water Res. 29:77-89. [Google Scholar]
- 19.Isaacs, S. H., M. Henze, H. Søeberg, and M. Kümmel. 1994. External carbon source addition as a means to control an activated sludge nutrient removal process. Water Res. 28:511-520. [Google Scholar]
- 20.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism, vol. III. Academic Press, New York, N.Y. [Google Scholar]
- 21.Juretschko, S., A. Loy, A. Lehner, and M. Wagner. 2002. The microbial community composition of a nitrifying-denitrifying activated sludge from an industrial sewage treatment plant analyzed by the full-cycle rRNA approach. Syst. Appl. Microbiol. 25:84-99. [DOI] [PubMed] [Google Scholar]
- 22.Koch, G., and H. Siegrist. 1997. Denitrification with methanol in tertiary filtration at wastewater treatment plant Zürich-Werdhölzi. Water Sci. Technol. 36:165-172. [Google Scholar]
- 23.Lee, H., J. A. Brereton, D. S. Mavnic, R. A. Fiorante, W. K. Oldham, and J. K. Paisley. 2001. Nutrient removal with methanol as a carbon source full-scale continuous inflow SBR application. Environ. Technol. 10:1223-1235. [DOI] [PubMed] [Google Scholar]
- 24.Lee, N. M., and T. Welander. 1996. The effect of different carbon sources on respiratory denitrification in biological wastewater treatment. J. Ferment. Bioeng. 82:277-285. [Google Scholar]
- 25.McCarty, P. L., L. Beck, and P. S. Amant. 1969. Biological denitrification of wastewaters by addition of organic materials, p. 1271-1285. Proceedings of the 24th Purdue Industrial Waste Conference. Purdue Industrial Waste Conference, Lafayette, Ind.
- 26.Neef, A., A. Zaglauer, H. Meier, R. Amann, H. Lemmer, and K.-H. Schleifer. 1996. Population analysis in a denitrifying sand filter: conventional and in situ identification of Paracoccus spp. in methanol-fed biofilms. Appl. Environ. Microbiol. 62:4329-4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nercessian, O., E. Noyes, M. G. Kalyuzhnaya, M. E. Lidstrom, and L. Chistoserdova. 2005. Bacterial populations active in metabolism of C1 compounds in the sediment of Lake Washington, a freshwater lake. Appl. Environ. Microbiol. 71:6885-6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nyberg, U., B. Andersson, and H. Aspegren. 1996. Long-term experiences with external carbon sources for nitrogen removal. Water Sci. Technol. 33:109-116. [Google Scholar]
- 29.Nyberg, U., H. Aspegren, B. Andersson, and J. L. C. Jansen. 1992. Full-scale application of nitrogen removal with methanol as carbon source. Water Sci. Technol. 26:1077-1086. [Google Scholar]
- 30.Purtschert, I., H. Siegrist, and W. Gujer. 1996. Enhanced denitrification with methanol at WWTP Zürich-Werdhölzli. Water Sci. Technol. 33:117-126. [Google Scholar]
- 31.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 32.Snaidr, J., R. Amann, I. Huber, W. Ludwig, and K.-H. Schleifer. 1997. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl. Environ. Microbiol. 63:2884-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stres, B., I. Mahne, G. Augustin, and J. M. Tiedje. 2004. Nitrous oxide reductase (nosZ) gene fragments differ between native and cultivated Michigan soils. Appl. Environ. Microbiol. 70:301-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tam, N. F. Y., G. L. W. Leung, and Y. S. Wong. 1994. The effects of external carbon loading on nitrogen removal in sequencing batch reactors. Water Sci. Technol. 30:73-81. [Google Scholar]
- 35.Tam, N. F. Y., Y. S. Wong, and G. Leung. 1992. Effect of exogenous carbon sources on removal of inorganic nutrient by the nitrification-denitrification process. Water Res. 26:1229-1236. [Google Scholar]
- 36.Throbäck, I., K. Enwall, Å. Jarvis, and S. Hallin. 2004. Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiol. Ecol. 49:401-417. [DOI] [PubMed] [Google Scholar]
- 37.Timmermans, P., and A. Van Haute. 1983. Denitrification with methanol. Fundamental study of the growth and denitrification capacity of Hyphomicrobium sp. Water Res. 17:1249-1255. [Google Scholar]
- 38.Tsuneda, S., R. Miyauchi, T. Ohno, and A. Hirata. 2005. Characterization of denitrifying polyphosphate-accumulating organisms in activated sludge based on nitrite reductase gene. J. Biosci. Bioeng. 99:403-407. [DOI] [PubMed] [Google Scholar]
- 39.Van de Peer, Y., and R. De Wachter. 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 40.Yan, T., M. Fields, L. Wu, Y. Zu, J. M. Tiedje, and J. Zhou. 2003. Molecular diversity and characterization of nitrite reductase gene fragments (nirK and nirS) from nitrate- and uranium-contaminated groundwater. Environ. Microbiol. 5:13-24. [DOI] [PubMed] [Google Scholar]
- 41.Yoshie, S., N. Noda, S. Tsuneda, A. Hirata, and Y. Inamori. 2004. Salinity decreases nitrite reductase gene diversity in denitrifying bacteria of wastewater treatment systems. Appl. Environ. Microbiol. 70:3152-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshinari, T., and R. Knowles. 1976. Acetylene inhibition of nitrous oxide reduction by denitrifying bacteria. Biochem. Biophys. Res. Commun. 69:705-710. [DOI] [PubMed] [Google Scholar]
- 43.You, S. J. 2005. Identification of denitrifying bacteria diversity in an activated sludge system by using nitrite reductase genes. Biotechnol. Lett. 27:1477-1482. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.