Abstract
The understanding of microbial interactions and trophic networks is a prerequisite for the elucidation of the turnover and transformation of organic materials in soils. To elucidate the incorporation of biomass carbon into a soil microbial food web, we added 13C-labeled Escherichia coli biomass to an agricultural soil and identified those indigenous microbes that were specifically active in its mineralization and carbon sequestration. rRNA stable isotope probing (SIP) revealed that uncultivated relatives of distinct groups of gliding bacterial micropredators (Lysobacter spp., Myxococcales, and the Bacteroidetes) lead carbon sequestration and mineralization from the added biomass. In addition, fungal populations within the Microascaceae were shown to respond to the added biomass after only 1 h of incubation and were thus surprisingly reactive to degradable labile carbon. This RNA-SIP study identifies indigenous microbes specifically active in the transformation of a nondefined complex carbon source, bacterial biomass, directly in a soil ecosystem.
The flux of organic carbon into and within soil carbon pools is controlled by complex biotic interactions and food webs. Energy flow and nutrient cycling at the ecosystem level are controlled by trophic interactions between the saprophytic soil microbiota (e.g., bacteria and fungi), various predatory groups of the micro- and mesofauna, and plants (29). Trophic levels can be coupled by both bottom-up and top-down effects which control soil biological communities and nutrient availability (5, 9), and the role of eukaryotic grazers and their importance for system productivity are fairly well understood (3, 30). However, the net activity of soil microbial communities is essential for ecosystem processes, and the specific contribution and the identity of key prokaryotes in terrestrial carbon transformations are largely unknown. Up to now, heterotrophic soil microbiota have mostly been regarded as a “black-box” system (28), and detailed information on the specific activity and function of individual community members is scarce.
New developments in stable-isotope approaches, particularly nucleic acid-based stable isotope probing (SIP) (for a recent review, see reference 4) have facilitated the targeted detection and identification of microbes metabolizing defined, mostly low-molecular-weight carbon sources in soil (6, 14, 17, 19). However, carbon sources in a soil environment usually are mixtures of different biomacromolecules, typical of biomass itself. The fate of biomacromolecule mixtures may differ considerably from that of individual components, because (i) a substrate mixture may result in competition, inhibition, and priming effects that change microbial degradation kinetics and (ii) some of the substrates may not be easily accessible to microbes due to chemical or physical protection mechanisms within the soil structure. Here, we followed the fate of fully 13C-labeled Escherichia coli cells as a proxy for labile biomass introduced into the soil (e.g., by manure or feces). The sequestration of this biomass-derived carbon pool was followed into the soil microbial food web, and microbial populations specifically active in its initial mineralization and assimilation were identified by SIP of rRNA.
MATERIALS AND METHODS
Biomass labeling and microcosm incubations.
E. coli strain RFM443 (25) was cultivated in mineral medium containing 2 g liter−1 13C6-labeled glucose (99 atom%) as described elsewhere (7). Early-stationary-phase cells were harvested, washed, and determined to be more than 98 atom% 13C labeled by elementary analysis-mass spectrometry (EuroEA 3000 elementary analyzer [Eurovector, Milan, Italy] and a Balzers QMG 422 quadrupole mass spectrometer [Balzers Instruments, Balzers, Liechtenstein]).
13C-labeled biomass was added to microcosms at ∼1.3 × 108 cells per g dry weight (gdw) of soil as determined by microscopic cell counting and most-probable-number analysis in Luria-Bertani medium to check viability (7). The soil material used, the Ah horizon of a haplic phaeozem, was sampled at the long-term agricultural research site “Ewiger Roggenbau” (Halle, Germany). A detailed description of soil characteristics, incubation setup, experimental conditions, analytical procedures, and sampling is provided elsewhere (7). The microcosm bioreactor system was modified from that described in reference 31 and comprised circulating humidified airflow, CO2 traps, and temperature control (20°C). Pressure loss caused by CO2 removal was compensated for by addition of oxygen from a flexible gas bag acting as a self-dosing system. In order to obtain representative soil samples and to average microscale heterogeneities of the soil, multiple independent vertical soil cores were sampled at different locations in each reactor at every sampling time. The cores were pooled, homogenized, and subsampled for analysis. Soil samples were taken after 1 h and after 1, 2, 4, and 8 weeks of incubation. Unlabeled E. coli cells were used for a 12C control series. Samples for molecular analyses were frozen immediately (−20°C) and stored until analysis. The reactor content was remixed after sampling to ensure homogeneous distribution of the remaining material. Trapped CO2 was analyzed for quantity and isotopic composition as published (18).
RNA extraction and centrifugation.
RNA was extracted from the soil samples using a previously described lysis protocol (13). Five hundred nanograms of RiboGreen-quantified rRNA extracts was density resolved by equilibrium density gradient centrifugation in cesium trifluoroacetate (CsTFA) (16). After centrifugation, the density of collected gradient fractions was determined refractometrically, and rRNA was precipitated for subsequent community analyses (13). Control gradients were conducted with rRNA from unamended soil, from soil incubated with unlabeled E. coli cells, and from unlabeled and 13C-labeled E. coli cells to calibrate the centrifugation system for the range of expected buoyant densities.
Quantitative and qualitative community analyses.
Bacterial and fungal rRNA templates precipitated and reeluted from gradient fractions were quantified by quantitative reverse transcription-PCR using assays previously described (13, 15) and standardized to relative ng μl−1 units using a dilution series of pure-culture rRNA extracts of E. coli and Fusarium oxysporum. Terminal restriction fragment length polymorphism (T-RFLP) fingerprinting of bacterial communities was conducted as published elsewhere (14, 15). Additional fingerprints were generated as follows. Digested amplicons (∼50 ng in 10 μl) were desalted by using DyeEx spin columns (QIAGEN). Desalted digests (1 μl) were mixed with 13 μl of Hi-Di formamide (Applied Biosystems) containing a 400-fold dilution of a 6-carboxy-X-rhodamine-labeled MapMarker 1000 ladder (Bio-Ventures, Murfreesboro, Tenn.), denatured (3 min at 95°C), cooled on ice, and size separated on a 3730 DNA analyzer (Applied Biosystems). Electrophoresis was performed with POP-7 polymer in a 50-cm capillary array under the following conditions: 10-s injection time, 2-kV injection voltage, 7-kV run voltage, 66°C run temperature, and 63-min analysis time. Electropherograms were analyzed using the GeneMapper 3.5 software package (Applied Biosystems). Cloning and sequencing of bacterial and fungal communities in resolved gradient fractions were done as described previously (15). Cloned reverse transcription-PCR amplicons were fully sequenced, resulting in ∼900 bp for bacteria and ∼600 bp for fungi. Phylogenetic trees were reconstructed using the ARB software package (11) and maximum-likelihood algorithms as described previously (12).
Nucleotide sequence accession numbers.
Sequence data were deposited with GenBank under accession numbers DQ643629 to DQ643792.
RESULTS
13C-labeled biomass was added to an agricultural soil at ∼1.3 × 108 cells per gdw final concentration. The amount of carbon added corresponded to ∼36 μg 13C per gdw of soil, to ∼0.2% of the indigenous soil organic carbon (∼19 mg Corg per gdw), to ∼26% of the soil bacterial biomass, and to ∼18% of the natural 13C content. Within 2 and 4 weeks of incubation, more than 30 and 40% of the added carbon was oxidized to 13CO2, respectively. Over the entire incubation period of 32 weeks, around 55% of the label was recovered as 13CO2, indicating that one-half of the added biomass was not oxidized but rather was sequestered into the indigenous soil microbiota and nonliving soil organic matter (7).
Identification of 13C-assimilating indigenous bacteria.
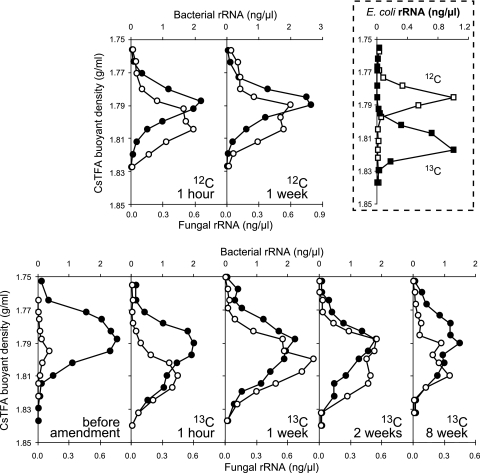
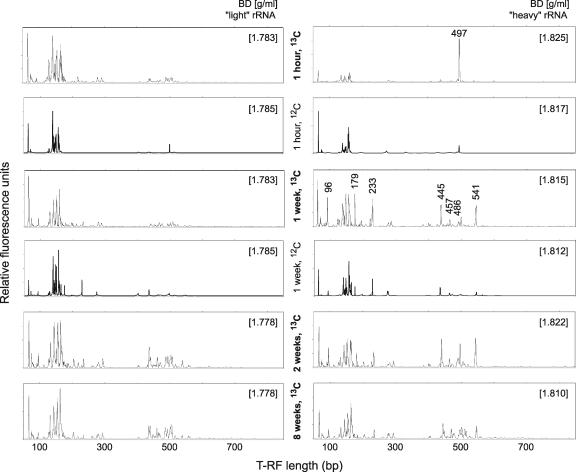
In order to identify distinct members of the soil microbiota that had been predominantly active in incorporating the added 13C label, 13C-enriched (or “heavy”) rRNA molecules were separated from unlabeled bulk rRNA by density gradient centrifugation. RNA for gradients was extracted after 1 h and after 1, 2, and 8 weeks of incubation. At 1 h, highly 13C-labeled bacterial rRNA was present (Fig. 1) at a buoyant density (BD) matching that of fully labeled E. coli rRNA (Fig. 1, dashed box). In the controls, such “heavy” rRNA was not found for bacteria. Just 1 h after addition, the “heavy” rRNA detected was likely to originate directly from intact or lysed E. coli cells, as the amendment had certainly not been completely degraded already at this early time point. T-RFLP fingerprinting of bacterial populations in gradient fractions ranging from “light” to “heavy” confirmed that, 1 h after amendment, the 497-bp terminal restriction fragment (T-RF) characteristic of E. coli rRNA dominated the “heavy” fractions (Fig. 2). In the unlabeled E. coli control gradient for this time point, fingerprints of both “light” rRNA and unlabeled rRNA tailing into the “heavy” fractions were highly alike, and the 497-bp T-RF was not allocated to specific fractions.
FIG. 1.
Quantitative profiles of RNA-SIP centrifugation gradients. Shown are the distributions of bacterial (solid circles) and fungal (open circles) small-subunit (SSU) rRNA in density gradient fractions of rRNA extracted from soil microcosms incubated with unlabeled (12C, top row) and 13C-labeled E. coli biomass (bottom row) at successive times after amendment as specified. Domain-specific template distribution within gradient fractions was quantified by quantitative reverse-transcription-PCR (13). Dashed box, banding of E. coli pure-culture rRNA in density centrifugation gradients, showing the quantitative distribution of fully 13C-labeled (solid squares) and unlabeled (open squares) E. coli SSU rRNA in separate CsTFA density gradients.
FIG. 2.
T-RFLP fingerprints of density-resolved bacterial communities. Profiles were generated from representative “light” and “heavy” rRNA gradient fractions of the labeled (13C) and unlabeled (12C) soil incubations as specified. CsTFA BDs (g ml−1) of fractions are given in brackets. T-RFs in “heavy” rRNA (after 1 week) characteristic for “heavy” fractions are specified, along with their respective fragment lengths, and were subsequently identified by cloning and sequencing.
After 1 week of incubation, substantial amounts of “heavy” bacterial rRNA were still present in the 13C series (Fig. 1) but, in comparison to the controls and 1-h gradients, T-RFLP fingerprints of “heavy” gradient fractions changed dramatically (Fig. 2). Several T-RFs specifically predominating the “heavy” rRNA appeared. The fact that these T-RFs were hardly detected in “light” fractions after 1 h and also after 1 week substantiated that certain members of the indigenous soil microbial community were more or less specifically utilizing the added 13C-labeled biomass. Also the fingerprints from the unlabeled E. coli control gradient after 1 week showed an increase of certain characteristic T-RFs, such as the 233- and 445-bp T-RFs (Fig. 2). Here again, however, the fingerprints of both “light” rRNA and unlabeled rRNA tailing into the “heavy” fractions were very similar, and a specific enrichment of certain T-RFs within the “heavy” fractions was not evident. This was in clear contrast to the 13C time series.
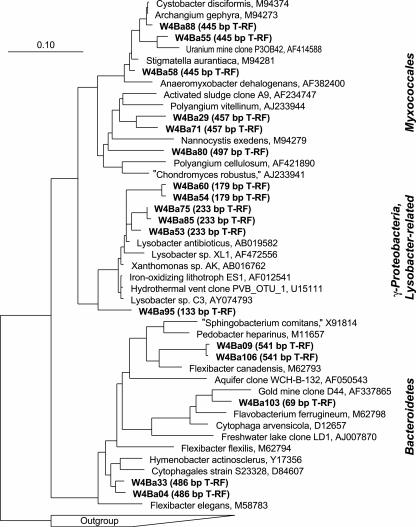
After 1 week of incubation, rRNA from a “heavy” gradient fraction (BD of 1.815 g ml−1) was cloned in order to identify the putative key players in biomass degradation represented by the “heavy” T-RFs. A total number of 99 clones were randomly selected, sequenced, and phylogenetically analyzed. As indicated already by the T-RFLP fingerprint of the corresponding gradient fraction (Fig. 2), the composition of phylotypes in the clone library was rather diverse (Table 1). However, a number of clones (n = 23) within distinct phylogenetic lineages were identified that were specifically represented within the T-RFs characteristic for “heavy” rRNA (Fig. 3; Table 1): a cluster of sequences of uncultivated γ-Proteobacteria within the Xanthomonadaceae, most closely related to Lysobacter spp. (179- and 233-bp T-RFs), diverse sequence types within the Myxococcales (445- and 457-bp T-RFs), and some uncultivated members of the phylum Bacteroidetes (451- and 486-bp T-RFs). Furthermore, an abundant 96-bp T-RF was observed in “heavy” rRNA after 1 week of incubation, but only one Bacteroidetes clone was found to display such a fragment. Since bias by undersampling in clone libraries must be taken into account, it is also possible that this peak represents Bacteroidetes members specifically allocated to the “heavy” rRNA. Remarkably, all three lineages identified by T-RFLP to be selectively enriched in “heavy” rRNA (Lysobacter spp., Myxococcales, and Bacteroidetes) are well known to comprise unicellular gliding bacterial micropredators (22-24).
TABLE 1.
Composition of the “heavy” bacterial clone librarya
| Phylogenetic lineage | No. of clones | Characteristic T-RF lengths (bp)b |
|---|---|---|
| α-Proteobacteria | 9 | |
| β-Proteobacteria | 8 | |
| γ-Proteobacteria | 2 | |
| Lysobacter related | 10 | 179, 233 |
| δ-Proteobacteria, Myxococcales | 7 | 445, 457 |
| Bacteroidetes | 6 | 486, 541 |
| Planctomycetes | 5 | |
| Acidobacteria | 10 | 201, 293 |
| Cyanobacteria | 2 | |
| Firmicutes, Bacilli | 8 | 148, 154 |
| Actinobacteria | 30 | 140, 143, 146, 162, 276 |
| Other uncultured | 2 |
Phylogenetic affiliation, characteristic T-RFs, and number of bacterial 16S rRNA clones retrieved from a library generated from “heavy” (BD = 1.815 g ml−1 in CsTFA) rRNA 1 week after 13C-biomass amendment. Lineages comprising unicellular gliders and related data are in boldface.
T-RFs detected for a major group of clones within a lineage.
FIG. 3.
Phylogenetic affiliation of representative bacterial 16S rRNA clones found to be specific for “heavy” rRNA fractions. Clones are marked in boldface, and their predicted T-RFs given in parentheses. Clones were generated from “heavy” rRNA (1.815 g ml−1) after 1 week of degradation of added 13C-E. coli biomass in soil. The scale bar represents 10% sequence divergence. GenBank accession numbers of reference sequences are given.
However, not only these glider-related clones were found in the “heavy” clone library. Almost one-third of the clones were related to different genera within the Actinobacteria, and sequence data revealed that they, together with members of the Bacilli, were mostly represented within the T-RFs between 135 and 165 bp (Fig. 2; Table 1). These T-RFs, however, were not specifically detected in “heavy” rRNA but rather were found to predominate all “light” gradient fractions. Hence, a specialized role of these bacteria in the degradation of the added biomass cannot be inferred. Rather, they seem to represent the numerically most abundant microbes constituting the bulk of unlabeled rRNA or partially labeled rRNA at later time points. As observed also for the 12C controls, a certain amount of such “light” templates is known to contaminate “heavy” gradient fractions due to the limited capacity of CsTFA gradients to focus mixed rRNA species into precisely defined bands (13, 16).
Over time, differences between the fingerprints of “light” and “heavy” rRNA became less pronounced (Fig. 2). This indicates that the 13C label was no longer specifically retained by certain bacterial populations but rather was spreading, via trophic networks, within the entire soil microbiota. Highly labeled bacterial rRNA remained detectable even after 8 weeks of incubation (Fig. 1). Of the ∼45% of added labeled carbon remaining in the soil after this time (7), around half was still present within the soil microbial community itself and persisted within the living fraction of the soil carbon pool. The other half of the remaining carbon appears to be present in nonliving soil organic matter, as suggested by phospholipid fatty acid analysis (7).
Dynamics of fungal rRNA within gradient fractions.
In contrast to the active bacteria, fungal populations in the soil appeared to be much more dynamic in biomass turnover. Here, the addition of the E. coli cells caused a dramatic increase of fungal rRNA detectable in relation to bacterial templates within 1 h (Fig. 1). However, intriguingly, most fungal rRNA was intrinsically distributed within “heavier” gradient fractions than bacterial templates in both 13C-labeled and unlabeled E. coli incubation series. Distinctions between the two treatments were not significant, with most fungal templates after 1 h banding at ∼1.81 g/ml in the 13C gradient and slightly below in the 12C gradient. The amounts of fungal rRNA measured were largest after 1 week of incubation but decreased subsequently, indicating a decline of the stimulatory effect by labile biomass addition.
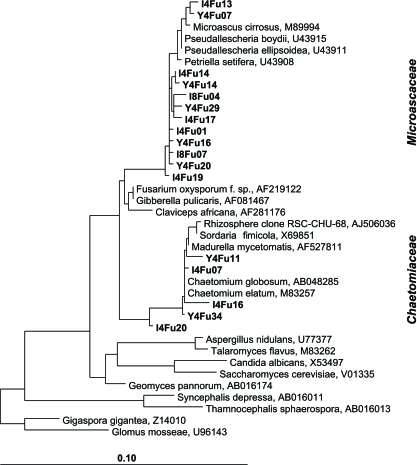
In order to preliminarily identify the detected fungi, three small clone libraries were generated from different fractions after 1 h and after 2 weeks (Table 2). All libraries were dominated by Microascaceae closely related to Petriella and Pseudallescheria spp. (Fig. 4). A further frequent group of clones was related to Chaetomium spp., but significant differences between density-resolved fungal communities were not evident from these libraries.
TABLE 2.
Composition of fungal clone librariesa
| Phylogenetic lineage | No. of sequences in libraryb:
|
||
|---|---|---|---|
| Y4Fu at 1 h (1.817) | I4Fu at 2 wk (1.815) | I8Fu at 2 wk (1.788) | |
| Sordariomycetes | |||
| Microascaceae | 22 | 8 | 9 |
| Hypocreales | 1 | ||
| Chaetomiaceae | 4 | 5 | |
| Inserta sedis | 1 | 2 | 3 |
| Eyrotiomycetes | 1 | ||
| Dothideomycetes | 3 | 1 | |
| Basidiomycota | 1 | ||
| Zygomycota | 2 | 2 | |
Phylogenetic affiliations and numbers of fungal 18S rRNA sequences retrieved in clone libraries generated from density-resolved nucleic acids at various sampling time points after 13C-biomass amendment.
BD values (g ml−1) are in parentheses.
FIG. 4.
Phylogenetic affiliation of fungal 18S rRNA clones from gradient fractions. Representative clones within the Sordariomycetes retrieved from “heavy” (Y04Fu and I04Fu clones) and “light” (I08Fu clones) fractions are shown in boldface. The scale bar represents 10% sequence divergence. GenBank accession numbers of reference sequences are given.
DISCUSSION
Here, the incorporation of isotopically labeled biomass carbon from the gram-negative bacterium E. coli into discrete members of a soil detrivore food web was traced by means of RNA-SIP. This application of SIP to the analysis of carbon sequestration by a soil microbial trophic network provides strong evidence for the utility of this technique in unraveling the turnover and flow of complex carbon pools in natural systems. To date, this has been successfully demonstrated for root exudate-based trophic networks (10, 20) and now is extended to microbial biomass itself.
The experiment was conducted using only one soil type and one bacterial biomass substrate. Also, due to the laborious nature of rRNA-SIP, replicate centrifugation gradients or data from replicate incubations are not presented. Therefore, and without testing other soils and settings, quantitative estimates of the role of the identified microbes in terrestrial C fluxes cannot be deduced at present. The amount of 13C label amended to the soil (36 μg biomass 13C per g soil) corresponded to only 2.8 μmol of 13C per g soil and thus represents a small amount of label in comparison to other rRNA-SIP studies (32). However, the amount of biomass added certainly will cause alterations in the activity and composition of intrinsic microbiota (e.g., priming effects [8]), as shown here in both unlabeled and 13C-labeled time series, for bacteria and fungi. Nevertheless, the amount of labeled biomass added represented only 0.2% of the indigenous soil organic carbon and therefore can still be considered “close to in situ”, especially regarding the rather patchy applications of labile bacterial biomass introduced into agricultural soil environments by, e.g., manure or feces.
Despite this small substrate pulse, diverse members of the Myxococcales, the Xanthomonadaceae, and the Bacteroidetes were identified to have incorporated the added carbon with high specificity after 1 week of incubation. All of these indigenous soil bacteria are more or less closely related to organisms capable of gliding motility. Many members of these lineages have been described as micropredators, capable of actively lysing their living bacterial prey (22-24). It has been suggested that they play a key role in the turnover of biomass carbon and other labile organic material in soil ecosystems (21), but direct and cultivation-independent evidence has not been available yet. Evidently, it is not possible to infer a functional trait as complex as gliding motility by mere 16S rRNA sequence similarity. However, this functional trait would provide the detected bacteria with preferential access to the added cellular substrate and thus a unifying competitive advantage, despite being phylogenetically diverse. The data suggest that these microbes were selectively incorporating 13C from the added cellular organic material, regardless of whether actively lysing the added 13C-E. coli cells or degrading dead biomass.
In contrast to bacteria, the 13C label could not be clearly tracked into fungal components of the soil microbial food web. Intriguing questions are why the detected fungal rRNA was mostly distributed in fractions “heavier” than those for bacterial rRNA also in the 12C control incubations and why the addition of 13C-E. coli did not cause a more pronounced shift in BD for fungal templates. Earlier studies report the location of unlabeled Fusarium sp. rRNA at densities similar to unlabeled bacterial rRNA (13), and shifts in BD for indigenous soil fungal communities upon 13C labeling have been observed (15). The contrasting observations of our present report illustrate the urgent need for a more exhaustive investigation of the distribution of unlabeled and 13C-labeled eukaryotic rRNA templates than can be expected for CsTFA centrifugation gradients.
Fraction-specific differences in the qualitative assemblage of fungal rRNA templates were not evident. It is unlikely that rRNA-based fungal fingerprinting techniques (for a recent summary, see reference 1) would have revealed clear distinctions. Rather, we hypothesize that the total indigenous fungal community was stimulated from the decomposing E. coli biomass, and the Microascaceae and Chaetomiaceae were most abundant. However, they did not incorporate the added label into rRNA to an extent clearly differentiable from the controls. This clearly prevents any predatory interpretation of these fungi, albeit this has been described for others (27) and cannot be totally excluded. Information on the ecological role of the Microascaceae in natural habitats is scarce (2), and such fungi are mainly known as opportunistic pathogens (26). But generally, fungi are as likely as bacteria to utilize decomposing biomass. Here, the most surprising finding was the apparent small time span required by the Petriella relatives to form increased amounts of rRNA after addition of E. coli biomass, thus highlighting a very dynamic role of these fungi in the degradation of available microbial biomass in the soil.
A lack of specific quantification and fingerprinting assays has prevented us from including a further group of microeukaryotes in the analyses: the protozoa. In addition to bacteria and fungi, protozoa would certainly have contributed to the degradation of the added E. coli biomass, but unfortunately detailed information cannot be provided for this experiment. Current efforts focus on the development of suitable assays that make distinct protozoan lineages accessible for (RNA-) SIP studies, and future work will show whether 13C label stemming from added bacterial biomass can be traced also to these eukaryotic lineages. Already in an earlier study, terrestrial cercomonads were shown to have incorporated carbon stemming from 13C-labeled methylotrophs (15); however, their detection was not targeted.
In conclusion, with the help of rRNA-SIP and a complex 13C substrate, we provide insight into the “black box” of a soil microbial food web and direct evidence for the identity of specific key players therein. Putatively motile bacterial micropredators were shown to play a pivotal role in the sequestration and turnover of the added labile biomass. Bacterial gliding motility may be a key trait controlling ecological niche positioning of these microbes, as it grants preferential access to attractive nondiffusible substrates. In consequence, the detected “heavy” bacterial lineages could be considered an ecologically unified functional clade. Further investigations regarding other soil types and water availabilities may show whether this holds true as a general concept for soil bacterial biomass turnover. Also, experiments with more-recalcitrant carbon sources may show whether RNA-SIP is applicable to the turnover of more-stable carbon pools, such as humified soil organic matter.
Acknowledgments
This work was supported by grants from the Deutsche Forschungsgemeinschaft within the priority program SPP 1090 (Soils as Source and Sink for CO2—Mechanisms and Regulation of Organic Matter Stabilization in Soils) to M.K. and within the priority program SFB 395 (Interactions, Adaptations, and Catalytic Capabilities of Soil Microorganisms) to M.W.F., as well as by the Helmholtz and Max Planck Societies.
We thank Bianca Pommerenke (MPI Marburg) for dedicated technical assistance and an anonymous reviewer for valuable suggestions.
REFERENCES
- 1.Anderson, I. C., and J. W. G. Cairney. 2004. Diversity and ecology of soil fungal communities: increased understanding through the application of molecular techniques. Environ. Microbiol. 6:769-779. [DOI] [PubMed] [Google Scholar]
- 2.April, T. M., S. P. Abbott, J. M. Foght, and R. S. Currah. 1998. Degradation of hydrocarbons in crude oil by the ascomycete Pseudallescheria boydii (Microascaceae). Can. J. Microbiol. 44:270-278. [DOI] [PubMed] [Google Scholar]
- 3.Bonkowski, M. 2004. Protozoa and plant growth: the microbial loop in soil revisited. New Phytol. 162:617-631. [DOI] [PubMed] [Google Scholar]
- 4.Dumont, M. G., and J. C. Murrell. 2005. Stable isotope probing-linking microbial identity to function. Nat. Rev. Microbiol. 3:499-504. [DOI] [PubMed] [Google Scholar]
- 5.Hairston, N. G., F. E. Smith, and L. B. Slobodkin. 1960. Community structure, population control, and competition. Am. Nat. 94:421-425. [Google Scholar]
- 6.Jeon, C. O., W. Park, P. Padmanabhan, C. DeRito, J. R. Snape, and E. L. Madsen. 2003. Discovery of a bacterium, with distinctive dioxygenase, that is responsible for in situ biodegradation in contaminated sediment. Proc. Natl. Acad. Sci. USA 100:13591-13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kindler, R., A. Miltner, H. H. Richnow, and M. Kästner. 2 June 2006. Fate of gram-negative bacterial biomass in soil—mineralization and contribution to SOM. Soil Biol. Biochem. [Online.] doi: 10.1016/j.soilbio.2006.04.049. [DOI]
- 8.Kuzyakov, Y., J. K. Friedel, and K. Stahr. 2000. Review of mechanisms and quantification of priming effects. Soil Biol. Biochem. 32:1485-1498. [Google Scholar]
- 9.Loreau, M., S. Naeem, P. Inchausti, J. Bengtsson, J. P. Grime, A. Hector, D. U. Hooper, M. A. Huston, D. Raffaelli, B. Schmid, D. Tilman, and D. A. Wardle. 2001. Biodiversity and ecosystem functioning: current knowledge and future challenges. Science 294:804-808. [DOI] [PubMed] [Google Scholar]
- 10.Lu, Y., and R. Conrad. 2005. In situ stable isotope probing of methanogenic Archaea in the rice rhizosphere. Science 309:1088-1090. [DOI] [PubMed] [Google Scholar]
- 11.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lueders, T., K. J. Chin, R. Conrad, and M. Friedrich. 2001. Molecular analyses of methyl-coenzyme M reductase alpha-subunit (mcrA) genes in rice field soil and enrichment cultures reveal the methanogenic phenotype of a novel archaeal lineage. Environ. Microbiol. 3:194-204. [DOI] [PubMed] [Google Scholar]
- 13.Lueders, T., M. Manefield, and M. W. Friedrich. 2004. Enhanced sensitivity of DNA- and rRNA-based stable isotope probing by fractionation and quantitative analysis of isopycnic centrifugation gradients. Environ. Microbiol. 6:73-78. [DOI] [PubMed] [Google Scholar]
- 14.Lueders, T., B. Pommerenke, and M. W. Friedrich. 2004. Stable-isotope probing of microorganisms thriving at thermodynamic limits: syntrophic propionate oxidation in flooded soil. Appl. Environ. Microbiol. 70:5778-5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lueders, T., B. Wagner, P. Claus, and M. W. Friedrich. 2004. Stable isotope probing of rRNA and DNA reveals a dynamic methylotroph community and trophic interactions with fungi and protozoa in oxic rice field soil. Environ. Microbiol. 6:60-72. [DOI] [PubMed] [Google Scholar]
- 16.Manefield, M., A. S. Whiteley, R. I. Griffiths, and M. J. Bailey. 2002. RNA stable isotope probing, a novel means of linking microbial community function to phylogeny. Appl. Environ. Microbiol. 68:5367-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller, L. G., K. L. Warner, S. M. Baesman, R. S. Oremland, I. R. McDonald, S. Radajewski, and J. C. Murrell. 2004. Degradation of methyl bromide and methyl chloride in soil microcosms: use of stable C isotope fractionation and stable isotope probing to identify reactions and the responsible microorganisms. Geochim. Cosmochim. Acta 68:3271-3283. [Google Scholar]
- 18.Miltner, A., H.-H. Richnow, F.-D. Kopinke, and M. Kästner. 2004. Assimilation of CO2 by soil microorganisms and transformation into soil organic matter. Org. Geochem. 35:1015-1024. [Google Scholar]
- 19.Radajewski, S., P. Ineson, N. R. Parekh, and J. C. Murrell. 2000. Stable-isotope probing as a tool in microbial ecology. Nature 403:646-649. [DOI] [PubMed] [Google Scholar]
- 20.Rangel-Castro, J. I., K. Killham, N. Ostle, G. W. Nicol, I. C. Anderson, C. M. Scrimgeour, P. Ineson, A. Meharg, and J. I. Prosser. 2005. Stable isotope probing analysis of the influence of liming on root exudate utilization by soil microorganisms. Environ. Microbiol. 7:828-838. [DOI] [PubMed] [Google Scholar]
- 21.Reichenbach, H. 1999. The ecology of the myxobacteria. Environ. Microbiol. 1:15-21. [DOI] [PubMed] [Google Scholar]
- 22.Reichenbach, H. 1992. The genus Lysobacter, p. 3256-3275. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes, 2nd ed. Springer, New York, N.Y.
- 23.Reichenbach, H. 1992. The order Cytophagales, p. 3631-3675. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, 2nd ed. Springer, New York, N.Y.
- 24.Reichenbach, H., and M. Dworkin. 1992. The Myxobacteria, p. 3416-3487. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, 2nd ed. Springer, New York, N.Y.
- 25.Rozen, Y., A. Nejidat, K. H. Gartemann, and H. Belkin. 1999. Specific detection of p-chlorobenzoic acid by Escherichia coli bearing a plasmid-borne fcbA′::lux fusion. Chemosphere 38:633-641. [DOI] [PubMed] [Google Scholar]
- 26.Summerbell, R. C., S. Krajden, and J. Kane. 1989. Potted plants in hospitals as reservoirs of pathogenic fungi. Mycopathologia 106:13-22. [DOI] [PubMed] [Google Scholar]
- 27.Thorn, G. 1997. The fungi in soil, p. 63-108. In J. D. van Elsas, J. T. Trevors, and E. M. H. Wellington (ed.), Modern soil microbiology, 1st ed. Marcel Dekker Inc., New York, N.Y.
- 28.Tiedje, J. M., S. Asuming-Brempong, K. Nusslein, T. L. Marsh, and S. J. Flynn. 1999. Opening the black box of soil microbial diversity. Appl. Soil Ecol. 13:109-122. [Google Scholar]
- 29.Wardle, D. A., R. D. Bardgett, J. N. Klironomos, H. Setala, W. H. van der Putten, and D. H. Wall. 2004. Ecological linkages between aboveground and belowground biota. Science 304:1629-1633. [DOI] [PubMed] [Google Scholar]
- 30.Wardle, D. A., and G. W. Yeates. 1993. The dual importance of competition and predation as regulatory forces in terrestrial ecosystems: evidence from decomposer food-webs. Oecologia 93:303-306. [DOI] [PubMed] [Google Scholar]
- 31.Weiss, M., R. Geyer, R. Russow, H. H. Richnow, and M. Kästner. 2004. Fate and metabolism of [15N]2,4,6-trinitrotoluene in soil. Environ. Toxicol. Chem. 23:1852-1860. [DOI] [PubMed] [Google Scholar]
- 32.Whiteley, A. S., M. Manefield, and T. Lueders. 2006. Unlocking the ‘microbial black box’ using RNA-based stable isotope probing technologies. Curr. Opin. Biotechnol. 17:67-71. [DOI] [PubMed] [Google Scholar]