Abstract
We report molecular evidence that ammonia-oxidizing archaea (AOA) occur in activated sludge bioreactors used to remove ammonia from wastewater. Using PCR primers targeting archaeal ammonia monooxygenase subunit A (amoA) genes, we retrieved and compared 75 sequences from five wastewater treatment plants operating with low dissolved oxygen levels and long retention times. All of these sequences showed similarity to sequences previously found in soil and sediments, and they were distributed primarily in four major phylogenetic clusters. One of these clusters contained virtually identical amoA sequences obtained from all five activated sludge samples (from Oregon, Wisconsin, Pennsylvania, and New Jersey) and accounted for 67% of all the sequences, suggesting that this AOA phylotype may be widespread in nitrifying bioreactors.
Nitrogen discharges into the environment pose multiple threats to ecosystem health, including toxicity (NH3), oxygen depletion (NH4+, NO2−, and organic N), and stimulation of algal blooms (NH4+, NO2−, NO3−, and organic N) (23). To prevent these adverse impacts, wastewater treatment plants (WWTPs) use bioreactors to oxidize ammonium to nitrate, and, where nitrate removal is also required, design features for denitrification are also included (12). Nitrification, the key and often rate-limiting step in N removal (23), entails the two-step microbial oxidation of ammonia to nitrate via nitrite. The two steps are catalyzed by chemolithotrophic ammonia-oxidizing bacteria (AOB) and nitrite-oxidizing bacteria. While some heterotrophic bacteria (18) and anaerobic ammonia-oxidizing (anammox) bacteria (19) oxidize ammonia, AOB are thought to be largely responsible for the oxidation of ammonia in WWTPs (and natural environments). In particular, members of the betaproteobacterial genera Nitrosomonas and Nitrosospira are thought to be the most important AOB in activated sludge (11).
Recently, however, it was discovered that autotrophic oxidation of ammonia is not restricted to the domain Bacteria. Könneke et al. (8) isolated an ammonia-oxidizing archaeon (AOA) named Nitrosopumilus maritimus from the rocky substratum of a tropical marine aquarium tank. N. maritimus is the first cultivated representative of the ubiquitous marine “group 1” Crenarchaeota and, like AOB, grows chemolithoautotrophically by oxidizing ammonia to nitrite under mesophilic conditions. In addition, N. maritimus contains putative genes for all three subunits (amoA, amoB, and amoC) of ammonia monooxygenase, the key enzyme used for bacterial ammonia oxidation. Thus, a critical link connecting archaeal ammonia monooxygenase genes originally identified through metagenomic studies and the process of aerobic ammonia oxidation has now been established (15, 22, 24). Treusch et al. (22) recently demonstrated the presence and expression of archaeal amoA genes in soil, while Francis et al. (5) demonstrated the ubiquity of AOA in marine and estuarine sediments, as well as in oxic and suboxic water columns. Considering that domestic wastewater contains 1 to 2 mM ammonium (∼25 mg N/liter) along with ∼1 mM N as organic nitrogen (∼15 mg N/liter) (20), it seemed possible that activated sludge bioreactors could also harbor AOA.
Accordingly, in this paper, we summarize the results of a search for archaeal amoA gene sequences in activated sludge bioreactor samples using a PCR primer set specific for the archaeal amoA gene (5). Below we describe the discovery of sequences that are most closely related to AOA sequences previously recovered from samples from sediments and soils. This is the first evidence that AOA occur in activated sludge bioreactors and opens avenues for study of AOA abundance and activity in activated sludge bioreactors, the largest application of environmental biotechnology.
WWTP description and sample collection.
Activated sludge samples were collected from nine WWTPs (Table 1) where nitrification was active. The Palo Alto WWTP employs a trickling filter followed by an aerated bioreactor for organic compound removal and nitrification (http://www.city.palo-alto.ca.us/depts/pubworks/waterquality/index.html). The San Jose WWTP is equipped with a four-stage process (anoxic-aerobic-anoxic-aerobic) that simultaneously removes organic compounds and nitrogen. The Dane-Iowa, McMinnville, Hammonton, Chalfont, American Bath, and Evesham WWTPs operate with the aerated-anoxic Orbal process (4), while the Chambersburg WWTP operates with a variation of the Orbal process called the VLR process (17). In these processes, oxygen is supplied to the anoxic tank at a rate that is less than the oxygen uptake rate, resulting in extremely low dissolved oxygen (DO) concentrations (<0.2 mg/liter) and enabling simultaneous nitrification and denitrification (4, 10). Grab samples were collected from the aeration basin of the Palo Alto WWTP, from the second aerobic stage of the San Jose WWTP, and from the end of the third channel in all aerated-anoxic plants. All samples were immediately frozen before transport to the laboratory. In addition, for comparison, duplicate sediment samples were collected from two sites in South San Francisco Bay; site C1 was located near the outfall from the Palo Alto WWTP, while site CR was located nearby but was relatively unaffected by the treatment plant outfall (data not shown). All samples were stored at −80°C until DNA extraction.
TABLE 1.
Wastewater treatment plants analyzed in this study and operational data for them
| WWTP | Location | Flow rate (m3/day) | Solids retention time (days) | Hydraulic retention time (h) | Concn of influent organic compounds (mg/liter) | Concn of influent NH3 + NH4+ (mg N/liter) | Concn of effluent NH3 + NH4+ (mg N/liter) | DO concn (mg/liter) | AOA amoA PCR amplification | AOB amoA PCR amplification |
|---|---|---|---|---|---|---|---|---|---|---|
| Chambersburg | Chambersburg, PA | 19,000 | 15 | 36 | NAa | 20.6 | 0.31 | NA | + | − |
| Dane-Iowa | Mazomanie, WI | 1,000 | 20 | 61 | 237b | 35.3 | <0.05 | 0.1, 0.3, 7.0c | + | + |
| McMinnville | McMinnville, OR | 11,400 | 15 | 25 | 540d | 17.0 | <0.15 | 0.0, 0.4, 2.0c | + | + |
| Hammonton | Hammonton, NJ | 3,500 | 22 | 52 | NA | 44.8 | 0.12 | 0.1, 0.3, 2.5c | + | + |
| Chalfont | New Britain, PA | 8,500 | 15 | 24 | 306b | 25.0 | 0.17 | <0.5, 0.5-2.0, 2.0c | + | + |
| American Bath | Lima, OH | 4,200 | 10 | 26 | 252b | NA | 0.065 | 0, 2-3, 6-7c | − | + |
| Evesham | Evesham, NJ | 5,500 | 18 | 48 | 150b | 30.2 | 0.47 | 0.1, 0.3, 1.7c | − | + |
| Palo Alto | Palo Alto, CA | 114,000 | 7 | 5.7 | 177d | 19.0 | 0.59 | 2.5e | − | + |
| San Jose | San Jose, CA | 450,000 | 9 | 11 | 360b | 24.2 | 0.40 | 2.8, 4.5f | − | + |
NA, not available.
Five-day biochemical oxygen demand.
DO concentrations of first, second, and third channels.
Chemical oxygen demand.
DO concentration of aeration tank.
DO concentrations of first and second aeration tanks.
DNA extraction, PCR, cloning, and sequencing.
Genomic DNA was extracted from 1.5 ml of activated sludge or 0.25 g of sediment using an UltraClean soil extraction kit (Mobio Laboratories, Solana Beach, CA). Approximately 20 to 50 ng of genomic DNA was used for each PCR. Archaeal amoA gene fragments (635 bp) were PCR amplified using primers Arch-amoAF (5′-STAATGGTCTGGCTTAGACG-3′) and Arch-amoAR (5′-GCGGCCATCCATCTGTATGT-3′), as previously described (5). Duplicate PCR products were pooled and purified by gel electrophoresis using a QIAEXII gel extraction kit (QIAGEN Inc., Valencia, CA). The purified PCR products were cloned using the pGEM-T Easy vector system (Promega, Madison, WI). For each library, 10 to 20 clones were randomly selected for sequencing with the SP-6 primer using ABI 3100 and 3730 capillary sequencers (PE Applied Biosystems). To screen samples for the presence of AOB, bacterial amoA genes were amplified as previously described (13), but the products were not cloned or sequenced.
Phylogenetic and statistical analyses.
To generate the phylogenetic tree in Fig. 1, archaeal amoA sequences were aligned using ClustalX, version 1.83 (21); in this analysis we included 30 representative activated sludge sequences, 9 sequences from South San Francisco Bay sediments, and 46 sequences from the GenBank database. A neighbor-joining tree (14) was generated, and a bootstrap analysis was performed using the same software with 1,000 resampling trials. For comparison, parsimony and maximum likelihood trees were generated using PAUP (version 4.0b10). Archaeal amoA-based richness was estimated (using the Chao1 richness estimator) and operational taxonomic units (OTUs) were defined at multiple cutoffs using the DOTUR program (16). To facilitate comparison with archaeal amoA sequences obtained from natural environments (5), 2% and 5% nucleotide cutoffs were used to define OTUs.
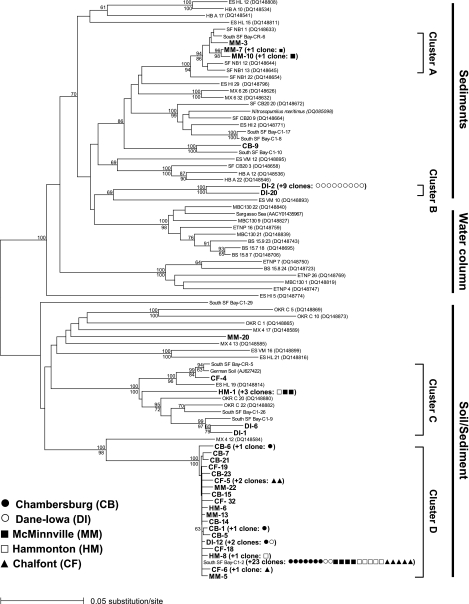
FIG. 1.
Unrooted neighbor-joining tree showing phylogenetic relationships among archaeal amoA sequences originating from activated sludge samples (CB, DI, MM, HM, and CF [this study]), water columns, soil, or sediments, including South San Francisco Bay (South SF Bay) C1 and CR sediments (this study); OKR soil; SF NB, SF CB, MX, HB, ES HI, ES VM, and ES HL sediments (5); water columns (Sargasso Sea [24]; BS, MB, and ETNP [5]); German soil (22); and the pure-culture ammonia-oxidizing archaeon N. maritimus (8). Activated sludge archaeal amoA sequences are indicated by boldface type, and sequences exhibiting >99% identity are indicated by symbols in parentheses. Distance bootstrap values (above the lines) and parsimony bootstrap values (below the lines) providing >60% support are indicated at the nodes. The major clusters indicated in this distance tree were also supported using maximum likelihood analysis.
Distribution and diversity of AOA in activated sludge.
Activated sludge samples from nine geographically distributed WWTPs in which nitrification was active were investigated to determine the presence of AOA (Table 1). We retrieved a total of 75 archaeal amoA sequences from five of these plants (Chambersburg, Dane-Iowa, McMinnville, Hammonton, and Chalfont), all of which utilize the aerated-anoxic process (see below); archaeal amoA sequences were not detected in the other four bioreactors analyzed. The activated sludge amoA sequences exhibited 69 to 87% and 79 to 96% identity with the N. maritimus amoA sequence (GenBank accession number DQ085098) (8) at the nucleotide and amino acid levels, respectively. For the 75 archaeal amoA sequences recovered from the activated sludge bioreactors, 11 and 9 unique OTUs were observed at the 2% and 5% nucleotide cutoffs, respectively. The Chao1 richness estimates (3) for the various WWTPs ranged from 2 to 8 OTUs per sample at the 2% cutoff (and from 2 to 4 OTUs at the 5% cutoff), which is on the low end of the range previously observed for water column samples (5 to 37 OTUs) and below the values estimated for soil and sediment samples (11 to 48 OTUs) (5). Phylogenetic analysis (Fig. 1) revealed that all of the archaeal amoA sequences retrieved from activated sludge samples were most closely related to sequences previously classified as sequences obtained from “sediments” and “soil/sediment” by Francis et al. (5). Most of the sequences fell in four major clusters (clusters A, B, C, and D); the exceptions were two clones designated MM-20 and CB-9. Cluster A is a well-supported clade (bootstrap value, 100%) comprised primarily of sequences retrieved from sediments of a low-salinity North San Francisco Bay site (SF NB) (5). Cluster C includes sequences originating from German soil (22), suboxic Tennessee soil (OKR clones [5]), and South San Francisco Bay sediment (C1 and CR clones [this study]). Clusters B and D are most distinct from previously reported environmental sequences and may be specific for activated sludge bioreactors. Cluster B sequences are more than 16% divergent from the next closest sequences in the database. The sequence nearest cluster D is MX 4 12 (5), a clone obtained from surface sediments in a heavily nitrogen-impacted coastal bay in Mexico (1). Although this clone differs by 13% from cluster D, there is strong bootstrap support (100%) for the phylogenetic placement of this sequence. Interestingly, cluster D includes virtually identical (98 to 100% nucleotide identity) clones from all five activated sludge samples, despite the fact that these samples were collected from WWTPs that are widely separated geographically (Table 1). In addition, cluster D contains by far the largest number of sequences of the four clusters (comprising 67% of all amoA clones retrieved in this study), suggesting that AOA belonging to cluster D may be widespread in activated sludge bioreactors at wastewater treatment plants.
Despite the presence of the same dominant sequence type (cluster D) at five WWTPs, at least one unique sequence type was recovered from each plant. However, this study was not designed to rigorously identify factors controlling AOA diversity and community composition. The Chambersburg WWTP library exhibited the least diversity, and 18 of 19 clones fell into cluster D. In the Hammonton and Chalfont WWTPs, clones were distributed solely in clusters C and D; the Dane-Iowa WWTP clones were distributed in clusters B, C, and D; and in the McMinnville WWTP, the clones were distributed in clusters A, C, and D. Not all of the activated sludge bioreactor samples examined in this study (Table 1) showed PCR amplification of archaeal amoA. Thus, AOA are either absent or present at levels below the detection levels in some WWTPs, or these plants may harbor additional archaeal amoA sequence types that were not amplified by our primers. Interestingly, although archaeal amoA could not be amplified from the Palo Alto WWTP (despite our screening of AOB-containing samples from 35 weeks of operation), sequences closely related or identical to other sludge AOA sequences (including sequences in cluster D) were recovered from sediments located at the plant's outfall in South San Francisco Bay (site C1 clones). All of the PCR-positive samples were collected from WWTPs operating with aerated-anoxic processes (i.e., the Orbal and VLR processes [17]), in which extremely low DO concentrations are maintained, enabling simultaneous nitrification and denitrification (4, 10). Additionally, as shown in Table 1, AOA-positive samples were collected from WWTPs operating with long retention times (>15 days of solids retention time, >24 h of hydraulic retention time). Thus, it is possible that either or both of these features (low DO levels and long retention times) facilitate the growth of AOA; however, archaeal amoA could not be amplified from two Orbal plants (American Bath and Evesham). Although some AOB have been reported to have anaerobic metabolism (2), efficient nitrification in environments with minimal aeration and low DO concentrations (i.e., aerated-anoxic conditions) has long been a mystery (9). The answer to whether AOA play a role in low-DO nitrification or “nitrifier denitrification” awaits quantitative information regarding the abundance and activity of AOA in these plants. However, it is worth noting that AOA sequences have frequently been recovered from suboxic sediments and water columns, including the Black Sea (5). It is also notable that bacterial amoA could not be amplified using bacterial amoA primers (13) from the Chambersburg WWTP (data not shown), while archaeal amoA was amplified.
AOA may well play a key role in both nitrogen and carbon cycling in marine water columns, sediments, and soils (5-8, 22). Although we have not yet examined the relative abundance or activity of AOA in activated sludge environments, this study clearly demonstrates the presence of molecular markers for AOA, including an archaeal amoA cluster (cluster D) that may be widespread in activated sludge bioreactors. Most importantly, this study provides a basis for future research on AOA in wastewater treatment bioreactors, including assessment of the relative contribution of AOB and AOA to ammonia oxidation, operational conditions that facilitate AOA growth, and the stability of AOA activity.
Nucleotide sequence accession numbers.
The archaeal amoA sequences reported in this study have been deposited in the GenBank database under accession numbers DQ278494 to DQ278568 (activated sludge) and DQ278569 to DQ278592 (sediments).
Acknowledgments
We thank Daniel R. Noguera of the University of Wisconsin for sending Orbal activated sludge samples, Charles S. Applegate of USFilter-Envirex (Waukesha, WI) and plant operators for providing operational data for the Orbal WWTPs, and Tom Curtis (University of Newcastle upon Tyne) and members of the Francis lab for providing useful comments on the manuscript.
This study was funded by a grant from the Stanford Institute for the Environment (to C.A.F. and C.S.C.) and by the Palo Alto Water Quality Control Plant.
REFERENCES
- 1.Beman, J. M., K. R. Arrigo, and P. A. Matson. 2005. Agricultural runoff fuels large phytoplankton blooms in vulnerable areas of the ocean. Nature 434:211-214. [DOI] [PubMed] [Google Scholar]
- 2.Bock, E., I. Schmidt, R. Stuven, and D. Zart. 1995. Nitrogen loss caused by denitrifying Nitrosomonas cells using ammonium or hydrogen as electron-donors and nitrite as electron-acceptor. Arch. Microbiol. 163:16-20. [Google Scholar]
- 3.Chao, A. 1984. Nonparametric-estimation of the number of classes in a population. Scand. J. Stat. 11:265-270. [Google Scholar]
- 4.Daigger, G. T., and H. X. Littleton. 2000. Characterization of simultaneous nutrient removal in staged, closed-loop bioreactors. Water Environ. Res. 72:330-339. [Google Scholar]
- 5.Francis, C. A., K. J. Roberts, J. M. Beman, A. E. Santoro, and B. B. Oakley. 2005. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. USA 102:14683-14688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hallam, S. J., T. J. Mincer, C. Schleper, C. M. Preston, K. Roberts, P. M. Richardson, and E. F. DeLong. 2006. Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol. 4:520-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ingalls, A. E., S. R. Shah, R. L. Hansman, L. I. Aluwihare, G. M. Santos, E. R. M. Druffel, and A. Pearson. 2006. Quantifying archaeal community autotrophy in the mesopelagic ocean using natural radiocarbon. Proc. Natl. Acad. Sci. USA 103:6442-6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Könneke, M., A. E. Bernhard, J. R. de la Torre, C. B. Walker, J. B. Waterbury, and D. A. Stahl. 2005. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543-546. [DOI] [PubMed] [Google Scholar]
- 9.Park, H.-D., and D. R. Noguera. 2004. Evaluating the effect of dissolved oxygen on ammonia-oxidizing bacterial communities in activated sludge. Water Res. 38:3275-3286. [DOI] [PubMed] [Google Scholar]
- 10.Park, H.-D., J. M. Regan, and D. R. Noguera. 2002. Molecular analysis of ammonia-oxidizing bacterial populations in aerated-anoxic Orbal processes. Water Sci. Technol. 46:273-280. [PubMed] [Google Scholar]
- 11.Purkhold, U., A. Pommerening-Röser, S. Juretschko, M. C. Schmid, H.-P. Koops, and M. Wagner. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl. Environ. Microbiol. 66:5368-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rittmann, B. E., and P. L. McCarty. 2000. Environmental biotechnology: principles and applications. McGraw-Hill Higher Education, New York, N.Y.
- 13.Rotthauwe, J. H., K. P. Witzel, and W. Liesack. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63:4704-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 15.Schleper, C., G. Jurgens, and M. Jonuscheit. 2005. Genomic studies of uncultivated archaea. Nat. Rev. Microbiol. 3:479-488. [DOI] [PubMed] [Google Scholar]
- 16.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith, G. 1996. Presented at the WEFTEC 69th Annual Conference and Exposition, Dallas, Tex.
- 18.Stouthamer, A. H. 1992. Metabolic pathways in. Paracoccus denitrificans and closely related bacteria in relation to the phylogeny of prokaryotes. Antonie Leeuwenhoek Int. J. Gen. Mol. Microbiol. 61:1-33. [DOI] [PubMed] [Google Scholar]
- 19.Strous, M., J. A. Fuerst, E. H. M. Kramer, S. Logemann, G. Muyzer, K. T. van de Pas-Schoonen, R. Webb, J. G. Kuenen, and M. S. M. Jetten. 1999. Missing lithotroph identified as new planctomycete. Nature 400:446-449. [DOI] [PubMed] [Google Scholar]
- 20.Tchobanoglous, G., F. L. Burton, and H. D. Stensel. 2003. Wastewater engineering: treatment and reuse, 4th ed. Metcalf and Eddy Inc. McGraw-Hill, New York, N.Y.
- 21.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Treusch, A. H., S. Leininger, A. Kletzin, S. C. Schuster, H.-P. Klenk, and C. Schleper. 2005. Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ. Microbiol. 7:1985-1995. [DOI] [PubMed] [Google Scholar]
- 23.U.S. Environmental Protection Agency. 1993. Manual: nitrogen control. U.S. Environmental Protection Agency, Cincinnati, Ohio.
- 24.Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Y. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y. H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]