Abstract
Waters from an extensive sulfide-rich aquifer emerge in the Frasassi cave system, where they mix with oxygen-rich percolating water and cave air over a large surface area. The actively forming cave complex hosts a microbial community, including conspicuous white biofilms coating surfaces in cave streams, that is isolated from surface sources of C and N. Two distinct biofilm morphologies were observed in the streams over a 4-year period. Bacterial 16S rDNA libraries were constructed from samples of each biofilm type collected from Grotta Sulfurea in 2002. β-, γ-, δ-, and ɛ-proteobacteria in sulfur-cycling clades accounted for ≥75% of clones in both biofilms. Sulfate-reducing and sulfur-disproportionating δ-proteobacterial sequences in the clone libraries were abundant and diverse (34% of phylotypes). Biofilm samples of both types were later collected at the same location and at an additional sample site in Ramo Sulfureo and examined, using fluorescence in situ hybridization (FISH). The biomass of all six stream biofilms was dominated by filamentous γ-proteobacteria with Beggiatoa-like and/or Thiothrix-like cells containing abundant sulfur inclusions. The biomass of ɛ-proteobacteria detected using FISH was consistently small, ranging from 0 to less than 15% of the total biomass. Our results suggest that S cycling within the stream biofilms is an important feature of the cave biogeochemistry. Such cycling represents positive biological feedback to sulfuric acid speleogenesis and related processes that create subsurface porosity in carbonate rocks.
Sulfidic caves form in carbonate rocks where sulfide-rich waters interact with oxygen at the water table or at subterranean springs. The caves form as a result of sulfuric acid production (equation 1) from microbial or abiotic sulfur oxidation. The sulfuric acid reacts with carbonate host rock to form gypsum and carbonic acid (equation 2).
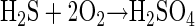 |
(1) |
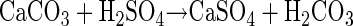 |
(2) |
Some of the longest caves known are thought to have formed by this process, including Lechugilla Cave in New Mexico, with 184 km of passages (17). Actively forming sulfidic caves are uncommon but intensely valuable as natural laboratories to understand factors influencing cave formation and resulting biological, geochemical, and isotopic signatures. Active sulfidic caves can host biogeochemically isolated ecosystems based entirely on microbial lithoautotrophic primary productivity (16, 38). These ecosystems are aphotic, terrestrial, subsurface environments comparable to sulfureta at hot springs and deep sea vents (10) and are of considerable interest as analogs for microbially dominated, early earth biotic communities such as those that might have developed after the initial rise of oxygen in the early Proterozoic era.
Available information from culturing, fluorescence in situ hybridization (FISH), and 16S rDNA libraries suggests that ɛ- and γ-proteobacteria are important biofilm-forming groups in the sulfidic cave waters studied to date. Microbial biofilms in Lower Kane Cave (Wyoming) springs and streams are dominated by filamentous ɛ-proteobacteria (6, 8). Microbial biofilms are also present in other active sulfidic caves, such as Movile Cave (Romania), Parker Cave (Kentucky), and Cesspool Cave (Virginia). Floating biofilms in Movile Cave contain aerobic and anaerobic sulfide-oxidizing lithoautotrophs (35, 39), methanotrophs (22), sulfate reducers, and organoheterotrophic bacteria and fungi (35). A 48-clone 16S rDNA library from a Sulfur River stream biofilm in Parker Cave included primarily ɛ-, γ-, and β-proteobacteria related to known sulfur-oxidizing bacteria, with over half of the reported sequences related to the ɛ-proteobacterium Thiomicrospira denitrificans (2). A 70-clone 16S rDNA library from a shallow pool below a Cesspool Cave spring was split roughly in half between ɛ-proteobacterial and Thiothrix-related γ-proteobacterial clones (7). The abundances of microbial populations in the Movile, Parker, and Cesspool Cave biofilms have not been determined.
The Grotta Grande del Vento-Grotta del Fiume (Frasassi) cave complex in central Italy hosts more than 23 km of passages and is a rare example of a large, actively forming sulfidic cave system. An important role for microorganisms in sulfide oxidation in the cave has been proposed (14). However, the microbial ecology of conspicuous microbial biofilms in sulfidic stream waters at Frasassi is unexplored. Here we report the molecular phylogeny and population structure of perennially abundant white microbial biofilms sampled from two cave streams over a 4-year period. Our data show that filamentous γ-proteobacteria dominate the biomass of the biofilms and that δ-proteobacteria are also abundant.
MATERIALS AND METHODS
Site description, sample collection, and geochemistry.
The Frasassi cave system is forming in the Jurassic Calcare Massiccio Formation (platform limestone) in the Appennine Mountains of the Marches Region, Central Italy (Fig. 1.). The active, sulfidic level of the cave is roughly at the elevation of the Sentino River, which flows 600 to 700 m below mountains on either side of the Frasassi gorge. A large area of outcropping sulfidic water in ramifying passages is accessible via technical caving routes inside the cave system. The water includes that of shallow cave streams, pools, and lakes and exhibits geochemical compositions that vary spatially and seasonally between well-defined end members (3, 37). Two biofilm samples with differing morphology (cottony versus feathery) were collected for clone library construction during a reconnaissance trip to Grotta Sulfurea in August 2002. Additional samples for FISH were collected in Grotta Sulfurea and Ramo Sulfureo on subsequent trips (samples RS03-46, RS03-10, GS04-15, RS05-6, RS05-21, and RS05-22). Samples for nucleic acid and elemental analyses were collected in sterile tubes, using sterile syringes or Pasteur pipettes, stored on ice, and processed or frozen within 4 to 6 h after collection. Water samples were collected in acid-washed polypropylene bottles. The conductivity, pH, and temperature of the stream waters were measured in situ using probes (WTW, Weilheim, Germany). Dissolved sulfide (methylene blue method), oxygen (indigo carmine method), and ammonium (salicylate method) concentrations were measured in the field, using a portable spectrophotometer according to the manufacturer's instructions (Hach Co., Loveland, Colo.). Oxygen measurements are reported as means of duplicate or triplicate tests and were reproducible within 10 to 15%. Anions were measured at the Osservatorio Geologico di Coldigioco Geomicrobiology lab, using a portable spectrophotometer within 12 h of collection (samples stored at 4°C) according to the manufacturer's instructions (Hach Co., Loveland Colo.). Total organic carbon (TOC) measurements were determined on triplicate samples, using a Shimadzu TOC-V series analyzer. Uncoated biofilm samples were examined with an FEI Quanta 200 ESEM with an Oxford INCA X-ray analytical system. Imaging and energy dispersive spectroscopy (EDS) analyses were performed in low vacuum mode at an accelerating voltage of 10 KeV.
FIG. 1.
Map of the Frasassi cave system showing major (named) caves in different shades of gray. Topographic lines and elevations (in meters) refer to the surface topography above the cave. Sampling locations in Grotta Sulfurea and at Ramo Sulfureo in Grotta del Fiume are shown as open circles.
Clone library construction.
Small subunit rDNA clone libraries were constructed from two white biofilm samples with differing morphologies (cottony versus feathery) collected within several meters of each other in the Grotta Sulfurea stream in August 2002 (Fig. 1). DNA was extracted using the MoBio Soil DNA extraction kit (MoBio) according to the manufacturer's instructions. Bacterial 16S rRNA genes were amplified by PCR from purified environmental DNA. Each 50-μl reaction mixture contained DNA template, 1.25 U ExTaq polymerase (TaKaRa Bio Inc., Shiga, Japan), 0.2 mM (each) deoxynucleoside triphosphate, 1× ExTaq PCR buffer, 0.2 μM 1492r universal primer (5′-GGTTACCTTGTTACGACTT-3′), and 0.2 μM 27f bacterial domain primer (5′-AGAGTTTGATCCTGGCTCAG-3′). Reactions were incubated in a thermal cycler as follows: 5 min at 94°C for initial denaturation, followed by 25 cycles of PCR consisting of 1 min at 94°C, annealing for 45 s at 45°C, and elongation for 1 min at 72°C, with a final elongation for 20 min at 72°C. Nonspecific amplification of DNA from the feathery biofilm DNA extract necessitated the use of touchdown PCR. Reactions were incubated in a thermal cycler as follows: 5 min at 94°C for initial denaturation, followed by 32 cycles of touchdown PCR consisting of 45 s at 94°C, annealing for 1 min at temperatures ranging from 64 to 48°C, and elongation for 90 s at 72°C, with a final elongation for 20 min at 72°C. PCR products were checked for size and concentration by gel electrophoresis, cloned into the pCR4-TOPO plasmid, and used to transform chemically competent OneShotTOP10 Escherichia coli cells as specified by the manufacturer (TOPO TA cloning kit; Invitrogen). Colonies containing plasmids with inserts were isolated by streak plating onto LB agar containing 50 μg/ml of kanamycin and subsequently inoculated into LB plus kanamycin liquid broth. Plasmids were isolated from overnight liquid cultures, using the Wizard Plus SV Minipreps DNA purification kit (Promega Corp., Wis.). An aliquot of each culture was preserved in 15% (wt/vol) sterile glycerol and stored at −80°C.
Sequencing and phylogenetic analysis.
Plasmid inserts were sequenced at the Penn State University Biotechnology Center or the University of Wisconsin Biotechnology Center, using T3 and T7 plasmid-specific primers and a 536f internal SSU rDNA primer (5′-CAGCMGCCGCGGTAATWC-3′). Partial sequences were assembled using CodonCode Aligner, version 1.2.4 (CodonCode Corp.), with Phred base calling and were manually checked. Sequences were aligned using ARB (31) and manually checked. Possible chimeras were identified by a partial treeing approach as described in reference 20. Briefly, aligned sequences were added to a database of 341 representative bacterial species and minimized with the Lane mask (27). The minimized alignment was split into 5′ and 3′ halves, and neighbor-joining trees were constructed from each half, using ARB. These partial trees were checked for branching incongruities. Partial treeing results were compared with the online analyses Bellerophon (18) and CHIMERA_CHECK version 2.7 at the Ribosomal Database Project II website (4). Putative chimeras were excluded from subsequent analyses.
Nearly full-length 16S rDNA sequences were grouped into phylotypes with >98% nucleotide similarity, and one representative of a phylotype was included in each phylogenetic analysis. Analyses included closely related BLAST matches (1), full-length sequences of clones from sulfidic caves and springs, and cultured representatives of major clades within bacterial subdivisions (aligned in ARB). Alignments were minimized using Lane's bacterial mask as described above (1,286 nucleotide positions) (27). Phylogenetic trees were constructed using the Bayesian analysis implemented in MrBayes, version 3.0b4 (19), using the Kimura two-parameter substitution model (25) and a gamma distribution of site-to-site rate variation with six discrete rate categories and run for 500,000 generations. The first 20% of sampled trees were discarded, and consensus trees were computed by 50% majority rule, using version 4.0b10 of PAUP* (42). Bayesian trees were compared against parsimony bootstrap consensus trees (heuristic search; 500 bootstrap replicates; PAUP*, version 4.0b10) and maximum likelihood analyses (fastDNAml algorithm implemented in ARB). Rarefaction analyses were computed using EstimateS version 7.5.0 (5).
Probe design and FISH.
Samples for FISH were collected in sterile microcentrifuge tubes, stored on ice or at 4°C, and fixed within 24 h after collection. Samples and isolates grown in the lab for use as control cells were fixed in three volumes of freshly prepared 4% (wt/vol) paraformaldehyde in 1× phosphate-buffered saline (PBS) for 3 to 4 h and stored in a 1:1 PBS/ethanol solution at 4°C or −20°C. Fixed samples and control cells were applied to multiwell, Teflon-coated glass slides, air dried, and dehydrated by successive immersion in 50%, 80%, and 90% ethanol washes (3 min each). Hybridizations were carried out in 8 μl/well of buffer containing 0.9 M NaCl, 20 mM Tris/HCl (pH 7.4), 0.01% sodium dodecyl sulfate (SDS), 25 to 50 ng of each oligonucleotide probe, and the formamide concentrations given in Table 1. Oligonucleotide probes were synthesized and labeled at the 5′ end with fluorescent dyes (Cy3, Cy5, and FLC) at Sigma-Genosys. The slides were incubated for 2 h at 46°C in sealed chambers equilibrated with the hybridization buffer. The slides were immersed for 15 min at 48°C in wash buffer (20 mM Tris/HCl [pH 7.4], 0.01% SDS, 5 mM EDTA, and NaCl concentrations determined by Lathe's formula [29]). The slides were then rinsed with distilled water, air dried, and counterstained with 4′,6′-diamidino-2-phenylindole (DAPI). The slides were mounted with Vectashield (Vectashield Laboratories) and viewed on a Nikon E800 epifluorescence microscope.
TABLE 1.
Oligonucleotide probes used in this study
| Probe | Target group | Sequence (5′ to 3′)c | % Formamide | Target site | Reference |
|---|---|---|---|---|---|
| EUB338a | Most bacteria | GCT GCC TCC CGT AGG AGT | 0-50 | 16S (338-355) | 2 |
| EUB338-IIa | Planctomycetales | GCA GCC ACC CGT AGG TGT | 0-50 | 16S (338-355) | 7 |
| EUB338-IIIa | Verrucomicrobiales | GCT GCC ACC CGT AGG TGT | 0-50 | 16S (338-355) | 7 |
| ARCH915 | Archaea | GTG CTC CCC CGC CAA TTC CT | 20 | 16S (915-934) | 44 |
| GAM42a | γ-Proteobacteria | GCC TTC CCA CAT CGT TT | 35 | 23S (1027-1043) | 34 |
| cGAM42a | Competitor | GCC TTC CCA CTT CGT TT | 35 | 23S (1027-1043) | 34 |
| DELTA495a | Most δ-proteobacteria, some Gemmatimonas | AGT TAG CCG GTG CTT CCT | 45b | 16S (495-512) | 31 |
| cDELTA495a | Competitor | AGT TAG CCG GTG CTT CTT | 45 | 16S (495-512) | This study |
| SRB385 | Some δ-proteobacteria, some Actinobacteria, and Gemmatimonas | CGG CGT CGC TGC GTC AGG | 35 | 16S (385-402) | 2 |
| EP404 | ɛ-Proteobacteria | AAA KGY GTC ATC CTC CA | 30b | 16S (404-420) | Adapted from ref. 49 |
| EP404mis | Negative control for EP404 | AAA KGY GTC TTC CTC CA | 30 | 16S (404-420) | This study |
| BEG811 | Frasassi Beggiatoa clade | CCT AAA CGA TGG GAA CTA | 35b | 16S (811-828) | This study |
Combined in equimolar amounts to make EUBMIX.
Stringency optimized in this study.
Letters in bold indicate mismatches between probes and competitors or controls.
The probes used in this study are described in Table 1. Probes EUBMIX, ARCH 915, GAM42a (including competitor probe cGAM42a), and SRB385 have been used extensively to characterize environmental samples and were used as described in the references in Table 1. One new probe (BEG811) and two modified probes (DELTA495a and EP404) were designed using the Probe_Design function of the ARB software package. Probes were designed and tested exactly as described in reference 21, including searches against publicly available sequences, using BLAST, the greengenes workbench (http://greengenes.lbl.gov/), and the online database probeBase (http://www.microbial-ecology.de/probeBase). Hybridization stringencies were determined using positive and negative controls in experiments with formamide concentrations from 0 to 50%. Optimal formamide concentrations for BEG811 were determined using Beggiatoa alba (negative control; 1-bp mismatch). The EP404 probe was adapted from the EP402-423 probe targeting ɛ-proteobacteria described by Takai et al. (44) but was shortened for use with the lower hybridization temperature in our FISH protocol. Optimal formamide concentrations were determined using Thiomicrospira denitrificans (positive control), an environmental sample containing a Thiovulum species with a 1-bp mismatch to the probe (negative control), and a fluorescently labeled oligonucleotide (EP404mis) with a 1-bp mismatch (negative control). Probe DELTA495a was designed to target most δ-proteobacteria in a microarray reverse hybridization protocol (30). The probe was modified for use with our FISH protocol, including design of a competitor oligonucleotide (cDELTA495a) with a 1-bp mismatch. The optimal formamide concentration for the DELTA495a probe was determined in the presence of equimolar cDELTA495a, using Desulfovibrio gigas and Desulfosarcina variabilis as positive controls and E. coli as a negative control (1-bp mismatch).
Microscopy.
Samples fixed with 4% paraformaldehyde as described above were viewed using phase contrast, differential interference contrast, and epifluorescence microscopy using a Nikon E800 epifluorescence microscope. The presence of sulfur inclusions was assayed on samples which had not been exposed to ethanol during preservation or dehydration. The samples were examined with phase contrast optics before and after incubation in 90% ethanol (3 or 10 min, 25°C) to check for sulfur dissolution (34). For each FISH experiment (sample-probe combination), between four and six microscope slide wells were examined. Because the biofilms were composed primarily of long filaments, the biomass of cells hybridizing with each probe relative to DAPI-stained cells was estimated visually and recorded as one of five categories: not detected, <5%, 5 to 15%, 15 to 60%, or 60 to 100%.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences determined in this study have been submitted to GenBank with accession numbers DQ133908 to DQ133940 and DQ415745 to DQ415869.
RESULTS
Biofilm morphology and geochemistry.
The cave stream biofilms that we observed over a 4-year period in Grotta Sulfurea and Ramo Sulfureo had two distinct morphologies. Cottony biofilms were found on the surface of fine, dark gray sediment lining slow-flowing reaches or margins of streambeds (Fig. 2A and B). Feathery biofilms were found attached to coarse limestone particles (coarse sand to boulder sized) in rapidly flowing reaches of the same streams (Fig. 2C and D). Cottony biofilms commonly had a tufted appearance, as in Fig. 2A, and were observed to reorganize themselves within minutes after a sampling disturbance, concentrating at the sediment/water interface in collection tubes. This behavior was not observed for feathery biofilms. The two biofilm morphologies formed a patchwork in the stream channels, with patches ranging in size from 10 to 100 cm in diameter. Geochemical data for bulk stream water surrounding the biofilms are shown in Table 2.
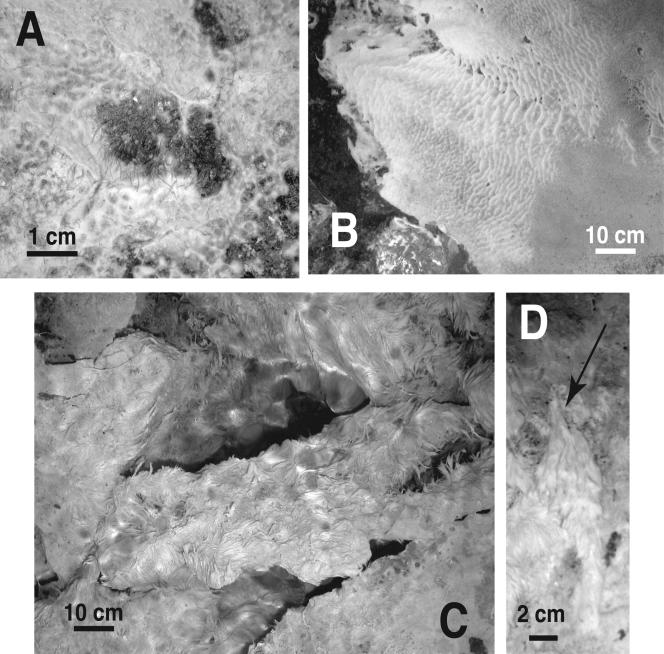
FIG. 2.
Examples of cave stream biofilm morphologies in situ at sampling sites. Cottony biofilms are shown in panels A (Ramo Sulfureo; RS05-21) and B (Grotta Sulfurea; GS04-15). Dark gray, fine sediment is visible underneath the biofilms. Note the tufted appearance of the biofilm in panel A. Numerous red worms visible in panel A extend approximately 0.5 cm above the dark gray sediment surface. Feathery biofilms are shown in panels C (Ramo Sulfureo; RS05-22) and D (Grotta Sulfurea; cottony biofilm used for clone library construction). The mottled appearance of the images in panels C and D is due to the turbulent water surface interacting with the camera flash. The large objects in panel C are biofilm-coated limestone boulders. In panel D, an 8-cm-long biofilm is attached to a limestone pebble in the streambed at the arrow.
TABLE 2.
Morphology, FISH results, and geochemical parameters for stream biofilms used in FISH experimentsa
| Sample | Date taken | Biofilm morphology | Biomass of cells for indicated probe:
|
Temp (°C) | Cond (μS/cm) | pH | S2− (μM) | O2 (μM) | NH4+ (μM) | SO42− (μM) | NO2− (μM) | NO3− (μM) | TOC (mg/liter) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ARCH915 | EUBMIX | GAM42a | BEG811 | EP404 | SRB385 | DELTA495a | |||||||||||||
| RS03-46 | 10/28/03 | Cottony | + | ++++ | + | ++++ | ++ | + | +++ | 13.6 | 1,964 | 7.3 | NM | NM | NM | NM | NM | NM | NM |
| RS03-10 | 10/28/03 | Feathery | ND | ++++ | ++++ | + | + | + | + | 13.6 | 1,964 | 7.3 | NM | NM | NM | NM | NM | NM | NM |
| GS04-15 | 7/07/04 | Cottony | ND | ++++ | ++ | ++++ | ++ | + | +++ | 13.6 | 1,693 | 7.4 | NM | 6.0 | 74 | 1333 | <2.0 | <0.7 | NM |
| RS05-6 | 5/27/05 | Feathery | + | ++++ | ++++ | ++ | ND | + | ND | 13.4 | 1,370 | 7.4 | 0.2 | 12.6 | 35 | 1427 | <2.0 | NM | 2.8 |
| RS05-21 | 8/07/05 | Feathery | + | ++++ | ++++ | ++ | + | + | ND | 13.5 | 1,823 | 7.3 | 212 | 2.6 | 67 | 1302 | <2.0 | <0.7 | 2.5 |
| RS05-22 | 8/07/05 | Cottony | ND | ++++ | ++ | ++++ | ++ | + | + | 13.5 | 1,823 | 7.3 | 212 | 2.6 | 67 | 1302 | <2.0 | <0.7 | 2.5 |
The biomass of cells hybridizing to each probe relative to DAPI-stained cells in FISH experiments is indicated by ND, not detected; +, <5%; ++, 5 to 15%; +++, 15 to 60%; or ++++, 60 to 100%. Cond, specific conductivity; TOC, total organic carbon; NM, not measured.
Biofilms in the cave streams were white, presumably due to the presence of small particles of elemental sulfur. Large bright inclusions were visible under phase contrast microscopy inside microbial filaments in both biofilm types, exemplified by the images shown in Fig. 3. Bright inclusions less than 1.5 μm in diameter disappeared after the microscope slide was soaked in 90% ethanol for 3 min. Soaking for 10 min removed even the largest (∼5-μm diameter) inclusions. EDS analyses confirmed the presence of sulfur accumulations within microbial filaments. The intensity of sulfur peaks in EDS spectra collected at intervals along the length of filaments ranged from background levels (measured outside the filaments) to more than an order of magnitude higher, indicating localized accumulations of sulfur within the cells (data not shown).
FIG. 3.
Phase contrast micrographs showing abundant bright sulfur inclusions in filamentous cells in cottony (A) and feathery (B) biofilm samples. The scale bars are 10 microns.
Clone libraries.
A total of 87 (feathery biofilm) and 70 (cottony biofilm) nonchimeric 16S rDNA sequences were retrieved from the Grotta Sulfurea stream biofilms collected in August 2002. Rarefaction analyses and abundances of clones in major bacterial lineages are compared in Fig. 4A and B, respectively. The large majority of clones from both biofilms were related to sulfur-cycling Proteobacteria, with fewer clones from Verrucomicrobia, Cytophaga-Flexibacter-Bacteroides, and other lineages (Fig. 4B). β-Proteobacterial sequences were related to Thiomonas species (99% identity) and Thiobacillus denitrificans (97% identity). γ-Proteobacterial clones included close relatives of Thiothrix species, Beggiatoa species, and a sulfidic cave and spring clone group, as well as a distant relative of Coxiella burnetii (Fig. 5). Beggiatoa-related clones were present in both biofilms and formed a monophyletic clade within the Thiotrichaceae. This clade accounted for almost half of the total sequences retrieved from the cottony biofilm. δ-Proteobacterial clones related to sulfur-reducing and disproportionating isolates were abundant and diverse in both biofilms, comprising 19 phylotypes in the feathery biofilm and 14 phylotypes in the cottony biofilm (Fig. 6). Roughly half of δ-proteobacterial clones from each library were close relatives of Desulfocapsa species. ɛ-Proteobacterial sequences (Fig. 7) were retrieved only from the feathery biofilm. They comprised eight phylotypes and were phylogenetically related to Arcobacter species or to a symbiont/sulfidic cave clone group that includes the isolate Sulfurovum lithotrophicum.
FIG. 4.
Rarefaction curves (A) and percentage of clones in major bacterial lineages (B) for stream biofilm 16S rDNA clone libraries. WM and zEL are clones from this study. OTU, operational taxonomic unit.
FIG. 5.
Bayesian phylogenetic analysis of Grotta Sulfurea white biofilm 16S rDNA clones grouping within the γ-proteobacteria. Clones from this study are indicated in large bold type (WM and zEL clones). Filled black circles indicate nodes supported by Bayesian posterior probabilities of ≥90% and maximum parsimony bootstrap values of ≥90%. Filled gray circles indicate nodes supported by Bayesian posterior probabilities of ≥75% and maximum parsimony bootstrap values of ≥75%. Open circles indicate nodes supported by Bayesian posterior probabilities of ≥75% or maximum parsimony bootstrap values of ≥75%. CFB, Cytophaga-Flexibacter-Bacteroides.
FIG. 6.
Bayesian phylogenetic analysis of Grotta Sulfurea white biofilm 16S rDNA clones grouping within sulfur-reducing clades of the δ-proteobacteria. Clones from this study are indicated in large bold type (WM and zEL clones). Filled black circles indicate nodes supported by Bayesian posterior probabilities of ≥90% and maximum parsimony bootstrap values of ≥90%. Filled gray circles indicate nodes supported by Bayesian posterior probabilities of ≥75% and maximum parsimony bootstrap values of ≥75%. Open circles indicate nodes supported by Bayesian posterior probabilities of ≥75% or maximum parsimony bootstrap values of ≥75%.
FIG. 7.
Bayesian phylogenetic analysis of Grotta Sulfurea white biofilm 16S rDNA clones grouping within the ɛ-proteobacteria. Clones from this study are indicated in large bold type (WM clones). No ɛ-proteobacterial clones were retrieved from the cottony biofilm sample (zEL clones). Filled black circles indicate nodes supported by Bayesian posterior probabilities of ≥90% and maximum parsimony bootstrap values of ≥90%. Filled gray circles indicate nodes supported by Bayesian posterior probabilities of ≥75% and maximum parsimony bootstrap values of ≥75%. Open circles indicate nodes supported by Bayesian posterior probabilities of ≥75% or maximum parsimony bootstrap values of ≥75%.
FISH probe design and optimization.
Probe BEG811 was designed to target a narrow clade containing Beggiatoa-related sequences retrieved in the two stream biofilm 16S rDNA clone libraries (35 sequences total) (Fig. 5). Other Thiotrichaceae, including Beggiatoa and Thioploca species, have one or more mismatches to the probe. Based on comparisons with all available sequences in public databases, the probe is specific for the Frasassi Beggiatoa clade, with the exception that it matches one Leptospirillum-related environmental phylotype (Nitrospira lineage) retrieved from an extremely acidic environment. At 35% formamide stringency, the probe hybridized with Beggiatoa-like cells in both feathery and cottony biofilms but not with Beggiatoa alba control cells (1-bp mismatch).
Probe EP404 targets diverse environmental and sulfur-cycling ɛ-proteobacteria and is complementary to all ɛ-proteobacteria retrieved in Frasassi stream biofilm clone libraries (Fig. 7), as well as to >90% of ɛ-proteobacteria in sulfur-oxidizing clades that are available from public databases, including the Arcobacter, Sulfurospirillum, Thiomicrospira, and symbiont clades and unaffiliated sequences from sulfur-rich environments. At 30% formamide stringency, EP404 produced bright fluorescence from positive-control Thiomicrospira denitrificans cells. Negative-control experiments using fluorescently labeled probe EP404mis (1-bp mismatch to the control cells) (Table 1) produced no signal at 30% formamide. EP404 was also tested with an environmental sample containing a Thiovulum population with a 1-bp mismatch to the probe. No fluorescence was detected from the negative-control Thiovulum cells at 30% formamide.
Probe DELTA495a is identical to the microarray oligonucleotide with the same name described in reference 30 but has not previously been adapted for FISH. The probe targets all major groups of sulfate-reducing δ-proteobacteria as well as environmental clones within the newly proposed bacterial lineage Gemmatimonadetes. BLAST searches indicated that numerous nontarget Proteobacteria and Actinobacteria have a 1-bp mismatch to the probe near the 3′ end. A competitor oligonucleotide (cDELTA495a) was designed in order to eliminate hybridization of the probe with these groups (Table 1). In the presence of an equimolar competitor probe at 45% formamide, DELTA495a produced bright fluorescence from positive-control cells (Desulfovibrio gigas and Desulfosarcina variabilis) and no fluorescence from negative-control E. coli cells (1-bp mismatch).
Probe SRB385 targets a broad range of δ-proteobacteria (50% of matches in public databases) but also matches some Actinobacteria (20% of matches), some Gemmatimonadetes (7% of matches), and isolated sequences scattered throughout the bacterial tree (23% of matches).
Microscopy and FISH.
Both cottony and feathery biofilms were composed primarily of long filamentous bacteria. The biomass of cells that hybridized with specific oligonucleotide probes in FISH experiments is shown in Table 2. Archaeal cells detected using the ARCH915 probe were rare (typically <3% of the biomass) or not detected. In contrast, almost all cells in the biofilms fluoresced brightly in experiments with the EUBMIX probe set. DAPI staining confirmed that all cells in the biofilms hybridized with either EUBMIX or ARCH915 probes.
Further FISH experiments revealed two distinct microbial communities correlated with the macroscopic morphologies of the biofilms. Cottony biofilms (GS04-15, RS03-46, and RS05-22) contained abundant large filaments (5 to 7 μm in diameter; 20 to 500 μm long) that hybridized with the newly designed probe BEG811, targeting Beggiatoa-related Frasassi clones (Fig. 8). The large filaments lacked vacuoles and had rounded termini and abundant sulfur inclusions (Fig. 3A and 8). Interestingly, the Beggiatoa-like filaments did not hybridize with the GAM42a probe targeting 23S rRNA of γ-proteobacteria. Two of the cottony biofilm samples (GS04-15 and RS03-46) had an additional filamentous bacterial population closely intertwined with the Beggiatoa-like filaments. The smaller filaments (1 to 4 μm in diameter; 50 to 500 μm long) hybridized with probes EUBMIX and DELTA495a (Fig. 9A and B) and lacked sulfur inclusions. In addition to the long filaments, DELTA495a also hybridized with dense populations of large vibrios (3 um long) and smaller populations of rods and cocci (0.5 to 1 um in diameter). EP404-positive cells were relatively few, comprising 5 to 15% of the biomass (Table 2), and included short rods (<1 μm in diameter) in dense colonies and rare, thin, short filaments (1 μm in diameter; 20 to 50 μm long). Probe SRB385 hybridized with scattered rods and cocci cells in all cottony biofilm samples.
FIG. 8.
FISH micrographs of cottony biofilm samples GS04-15 (A), RS03-46 (B), and RS05-22 (C) hybridized with EUBMIX (green) and BEG811 (red) probes. Cells which hybridized with both probes appear yellow/orange. The scale bars are 10 μm.
FIG. 9.

FISH micrographs of cottony biofilm samples GS04-15 (A) and RS03-46 (B) hybridized with EUBMIX (green) and DELTA495a (red) probes. Cells which hybridized with both probes appear yellow/orange. The scale bars are 10 μm.
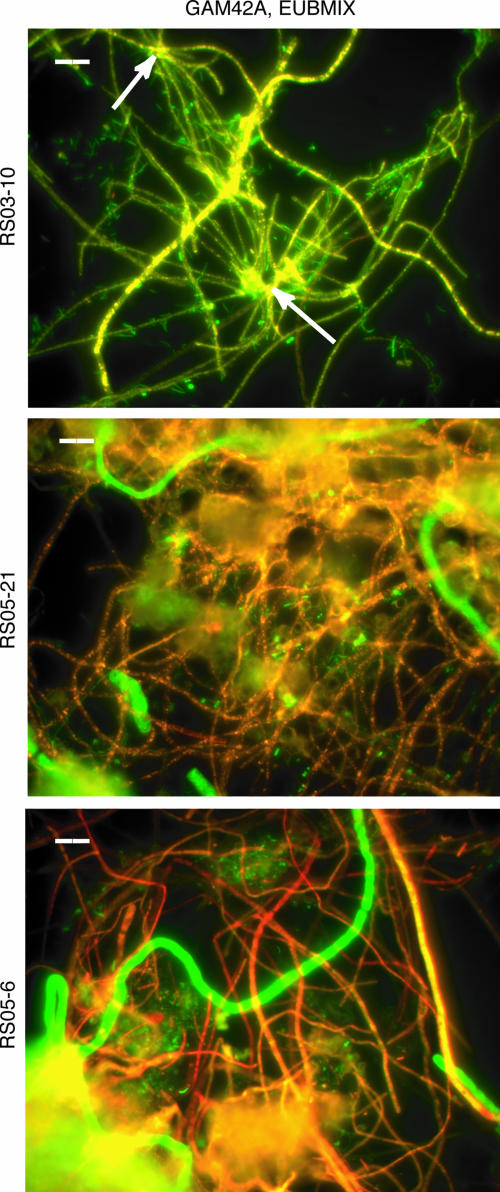
Feathery biofilm samples (RS03-10, RS05-6, and RS05-21) were dominated by long, thin filaments with sulfur inclusions that hybridized with the GAM42a probe (1 to 2.5 μm in diameter; 50 to 500 μm long) (Fig. 3B and 9). These filaments had no observable vacuoles under phase, transmitted-light, or fluorescence microscopy. The filaments were often observed in rosettes (Fig. 10) or attached to mineral holdfasts at one end. Thick filaments identical to Beggiatoa-like cells in the cottony biofilms were also present and made up ∼4 to 15% of the biomass. Probe EP404, targeting ɛ-proteobacteria, detected isolated rods and cocci (∼1 um in diameter) contributing 0 to 5% of the biomass. Probe SRB385 hybridized with scattered rods and cocci cells in all samples. Probe DELTA495a detected isolated rods in sample RS03-10 but no cells in the other two feathery biofilm samples.
FIG. 10.
FISH micrographs of feathery biofilm samples hybridized with EUBMIX (green) and GAM42a (red) probes. Cells which hybridized with both probes appear yellow/orange. The scale bars are 10 μm.
DISCUSSION
Sulfur oxidizers.
Neutral-pH cave streams at Frasassi are sites of intense limestone corrosion, equivalent to roughly 15 mg CaCO3/cm2/year, or 5 cm/1,000 years (15). In the present study, we found perennially abundant white biofilms attached to corroding limestone surfaces and to thin organic-rich sediments in the streams. These biofilms are dominated by filamentous γ-proteobacteria with Beggiatoa-like (cottony biofilms) or Thiothrix-like (feathery biofilms) cell morphologies and abundant sulfur inclusions. β- and ɛ-proteobacteria related to Thiobacillus, Arcobacter, and other sulfur-oxidizing groups were retrieved in clone libraries but made up a small fraction of the biomass in both biofilm types, based on FISH experiments (Table 2).
Freshwater and small marine Beggiatoa species are typically described as gradient microorganisms which colonize steep oxygen and sulfide gradients near the sediment/water interface (46). The newly designed probe BEG811 hybridized with 5- to 7-μm-diameter filaments present in cottony biofilms from both the Ramo Sulfureo and Grotta Sulfurea sample sites at Frasassi. The morphology and behavior of the filaments were consistent with descriptions of freshwater Beggiatoa species (40, 46), including evidence of gliding motility. The Beggiatoa-like filaments were present in all of the stream biofilm samples we analyzed but dominated in cottony biofilms collected from the surfaces of fine, dark gray sediment near the margins or in slowly flowing reaches of streams.
Feathery biofilms were dominated by thin, GAM42a-positive filaments, often in rosettes or attached to mineral particles by holdfasts at one end. Compared to recently described rosettes of vacuolate sulfur oxidizers growing at shallow marine hydrothermal vents (4 to 10 μm) (24), the feathery biofilm filaments were smaller in diameter (1.0 to 2.5 μm) and nonvacuolate. Feathery biofilms were found attached to limestone rocks in rapidly flowing water, typical of Thiothrix habitats where sulfide and oxygen are turbulently mixed in a strong current (28).
Cottony and feathery biofilm types were commonly observed adjacent to each other in the same streams, separated by distances of 10 to 100 cm. This pattern is exemplified by the two Grotta Sulfurea biofilms used for clone library construction and FISH sample pairs RS03-46 (cottony)/RS03-10 (feathery) and RS05-21 (cottony)/RS05-22 (feathery). This phenomenon suggests that bulk stream water geochemical measurements are not sensitive enough to detect ecologically meaningful variations in sulfide and oxygen concentrations which may control biofilm spatial and temporal distributions. Measured sulfide concentrations at Ramo Sulfureo ranged from 0.2 μM (May) to 212 μM (August), consistent with the seasonal range of sulfide concentrations observed in previous long-term hydrologic studies at the Ramo Sulfureo stream sample site (3).
Oxygen concentrations in the bulk stream waters varied over a much smaller range (2.6 to 12.6 μM). Data from Lower Kane Cave (8) show that Thiothrix-related 16S rDNA clones are not common in biofilm communities until dissolved oxygen concentrations reach at least 10 to 20 μM. In contrast, the dissolved oxygen concentration in the growth zone of Beggiatoa species is reported to be lower, in the range of 1 to 2.5 μM (33). Notably, feathery Thiothrix-like biofilms were observed only in turbulent water, where oxygen diffusion from the cave atmosphere would be much faster than in slowly flowing water hosting fine sediment and cottony biofilms. We also observed thick accumulations (∼2 cm) of Thiothrix-like filaments at a sulfidic spring outflow draining the cave system (J. L. Macalady, unpublished results). Oxygen concentrations in the spring water were high (84 μM) compared to those for streams inside the cave. We conclude that oxygen availability is an important factor controlling the spatial distributions of Frasassi Beggiatoa and Thiothrix populations and that Beggiatoa populations thrive in microenvironments with slightly lower oxygen availability than Thiothrix.
The ɛ-proteobacterial clones retrieved from the feathery stream biofilm sample are closely related to sulfur-oxidizing strains or clones from sulfide- and sulfur-rich environments, suggesting that ɛ-proteobacteria play a role in S oxidation at Frasassi. Although ɛ-proteobacteria were not retrieved in the cottony biofilm clone library, ɛ-proteobacteria were consistently detected in cottony biofilms in FISH experiments using probe EP404. These results are not contradictory, since our clone libraries did not catalog the bacterial diversity in the samples exhaustively (Fig. 4A), and since PCR bias may have influenced the composition of the clone libraries. Regardless, ɛ-proteobacteria contributed only a minor fraction (0 to 15%) of the biomass in the six biofilms we examined using FISH. The EP404 probe used in this study matches more than 90% of ɛ-proteobacteria sequences in sulfur-oxidizing clades that are available from public databases, including sequences from sulfur-rich environments such as sulfidic caves. Although it is possible that the EP404 probe failed to detect one or more ɛ-proteobacteria populations in the stream biofilms, other FISH results dictate that these populations would represent a small fraction of the biomass compared to Beggiatoa-like or Thiothrix-like cells (Table 2). In addition, the EP404 probe hybridized with abundant ɛ-proteobacteria filaments in an isolated veil-like biofilm sampled from a stagnant Frasassi cave pool with very low dissolved oxygen content (0.6 μM) (Macalady, unpublished), confirming that it detects populations of ɛ-proteobacteria in the cave system.
Previous carbon isotope studies conducted at the Ramo Sulfureo stream site further support the conclusion that ɛ-proteobacteria are minor contributors to the biomass of the stream biofilms colonizing the site. Sarbu et al. (37) found that isotopic fractionation (Δδ13C) between dissolved bicarbonate and stream biofilm biomass carbon ranged from −23 to −30.5‰ (37), consistent with autotrophs detected using the pentose phosphate cycle. The observed fractionation is too large for autotrophs detected using the reductive tricarboxylic acid cycle, for which Δδ13C is ∼−3 to −13 ‰ (49). Recent studies using enzyme assays and genome sequencing methods strongly suggest that ɛ-proteobacterial autotrophs use the reverse tricarboxylic acid cycle and lack enzymes for carbon fixation via the pentose phosphate cycle (23, 41, 43).
A likely explanation for the dominance of γ-proteobacteria in the stream biofilms is that oxygen concentrations in the streams are maintained above the range preferred by ɛ-proteobacteria. This hypothesis is supported by hydrologic studies showing that the cave streams are mixtures containing upwelling sulfidic water and oxygenated percolating water, with the oxygen-saturated water contribution ranging from 35 to 65% (3). Our results are consistent with data from Lower Kane Cave, which hosts biofilms dominated by filamentous ɛ-proteobacteria in spring water with <0.2 μM dissolved oxygen (8).
Engel et al. (9) showed that microorganisms directly influence the rate of limestone dissolution in Lower Kane Cave springs because of their close physical proximity to limestone surfaces and because they are microaerophiles which can consume sulfide before it comes into contact with high concentrations of dissolved oxygen. Sulfur-oxidizers using nitrate as an electron acceptor (equation 3) could extend the zone of sulfide oxidation into anoxic niches but would produce one-fifth as much acid per mole of sulfide than sulfide oxidation with oxygen (equation 4):
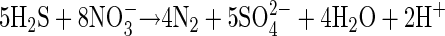 |
(3) |
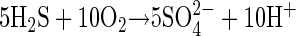 |
(4) |
Nitrate and nitrite concentrations in Frasassi cave streams are below detection limits (<2 and <0.7 μM, respectively) despite the presence of abundant dissolved ammonium (35 to 74 μM) and detectable oxygen (2.6 to 12.6 μM), suggesting that nitrate is effectively scavenged by anaerobic microorganisms. Many sulfur oxidizers are facultative anaerobes, including Beggiatoa and Thiothrix species (34) and some β- and ɛ-proteobacteria (45). Denitrifying sulfide-oxidizers are numerically abundant in floating white biofilms in Movile Cave, based on culturing experiments (35). Future work will investigate whether nitrate plays a major role in S-oxidizer metabolism in Frasassi stream biofilm communities.
Sulfur reducers.
Sulfidic cave biofilms examined to date using molecular methods either did not contain detectable δ-proteobacteria (Cesspool Cave [7] and Parker Cave [2]) or contained a small percentage of clones related to Desulfocapsa thiozymogenes (Lower Kane Cave [8]). In contrast to previous sulfidic cave studies, δ-proteobacteria in Frasassi cave stream biofilms were abundant and diverse and included numerous phylotypes related to Desulfocapsa (14 phylotypes) and Desulfonema (7 phylotypes) species, in addition to Syntrophobacter species, Syntrophus species, Desulfoarculum baarsii, Desulfobacter postgatei, Desulfomonile species, and the Geobacteraceae. Probes SRB385 and DELTA495a detected bacterial populations with diverse morphologies in both biofilm types. Although it is possible that some of the populations identified by these probes are not sulfur reducers, the FISH results are consistent with the abundance and diversity of clones retrieved in 16S rDNA libraries.
Desulfonema-related clones were the second-most-abundant δ-proteobacterial group retrieved in clone libraries, and their close relationships to sequences from cultivated Desulfonema spp. strains is supported by phylogenetic analyses (Fig. 6). In two of the cottony biofilm samples, Beggiatoa filaments were associated with densely intertwined, filamentous bacteria that hybridized with probe DELTA495a (Fig. 10). Although probe DELTA495a hybridizes with members of the poorly known lineage Gemmatimonadetes in addition to sulfate-reducing δ-proteobacteria, it is highly likely, based on morphology, that the filaments are related to members of the genus Desulfonema. Desulfonema species strains oxidize short-chain aliphatic acids and/or alcohols to CO2. Close physical associations between filamentous sulfur-oxidizing bacteria and filamentous δ-proteobacteria have been observed in organic-rich marine and lacustrine surface sediments, where Desulfonema filaments are epibionts on large sulfur-oxidizing filaments such as Thioploca spp. (12). Desulfonema spp. are the dominant sulfate-reducing bacteria in permanently oxic regions of hypersaline cyanobacterial biofilms (32) and are widespread in freshwater and marine environments, where their gliding motility allows them to exploit oxic-anoxic interfaces (36, 47). Both cottony and feathery biofilm clone libraries contained sequences related to Desulfonema species. However, the greater abundance of putative Desulfonema filaments we observed in cottony biofilms using FISH probe DELTA495a (Table 2) is consistent with their preference for steep oxic-anoxic interfaces, gliding motility, and previously noted associations with Beggiatoa.
Pure cultures of Desulfocapsa spp. grow autotrophically by disproportionation of sulfite, thiosulfate, or elemental sulfur and require low sulfide concentrations for efficient growth (11). Sulfide scavenging in nature can be provided by sulfide-complexing metals or by sulfide-oxidizing bacteria living in close proximity. Desulfocapsa-like bacteria have been identified in meromictic Lake Cadagno, where they likely derive an energy benefit by living in aggregates with sulfide-oxidizing phototrophs (48). Desulfocapsa isolates can also grow as either sulfur or sulfate reducers (11). Based on strong bootstrap and Bayesian posterior probability support for the placement of Frasassi clones within the Desulfocapsa clade (Fig. 6) and the fact that many are distant from cultivated representatives of the genus (95% identity), they likely represent novel ecological types warranting further study.
The abundance and phylogenetic diversity of sulfate- and sulfur-reducing bacteria in Frasassi stream biofilms indicates that they are the locus of intensive S cycling within the cave system. Such cycling has been suggested by previous S isotopic studies (13). The association of sulfur oxidizers and sulfur reducers has special implications for the microbial ecology of Frasassi biofilms and for the geochemistry of sulfidic caves. Based on geochemical data, sulfide is at least transiently scarce (as low as 0.2 uM) in the bulk stream water at Ramo Sulfureo due to seasonal changes in hydrology, whereas dissolved sulfate is always abundant (1 to 2 mM). The presence of sulfur-disproportionating and sulfate-reducing bacteria in the stream biofilms thus provides a buffer for sulfur oxidizers against temporal swings in the availability of sulfide. Sulfate reduction also represents a pathway by which primary productivity from electron donors other than sulfide (e.g., ammonium, hydrocarbons, etc.) can be recycled to fuel sulfuric acid production and dissolution at limestone surfaces. An analogous process driven by photosynthetic primary productivity causes globally significant sulfuric acid carbonate dissolution in productive marine shelf sediments (26). The formation of biofilms containing both S oxidizers and S reducers therefore represents biological feedback to sulfuric acid cave formation and similar processes which create subsurface porosity in limestone rocks.
Acknowledgments
We thank Alessandro Montanari for logistical support and the use of facilities and laboratory space at the Osservatorio Geologico di Coldigioco. We also thank Laura Cleaveland, Annaliese Eipert, Paola D'Eugenio, Dan Jones, and Ani Kameenui for field assistance, Joel Moore for TOC analyses, and Yohey Suzuki for insightful discussions. E.H.L. (Carleton College), L.K.A. (Brown University), B.K. (Carleton College), and K.M. (Carleton College) contributed to this work while undergraduate students.
B.K. and K.M. thank Carleton College for financial support. L.K.A. was supported in part by undergraduate research funds from the Penn State Astrobiology Research Center (PSARC; NASA award NNA04CC06A). This study was funded by the Biogeosciences Program of the National Science Foundation (EAR 0311854).
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tools. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Angert, E. R., D. E. Northup, A.-L. Reysenbach, A. S. Peek, B. M. Goebel, and N. R. Pace. 1998. Molecular phylogenetic analysis of a bacterial community in Sulphur River, Parker Cave, Kentucky. Am. Mineral. 83:11-12. [Google Scholar]
- 3.Cocchioni, M., S. Galdenzi, V. Amici, L. Morichetti, and L. Nacciarriti. 2003. Studio idrochimico delle acque nelle Grotte di Frasassi. Grotte Ital. 4:49-61. [Google Scholar]
- 4.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The ribosomal database project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colwell, R. K. 2005. EstimateS: statistical estimation of species richness and shared species from samples, version 7.5. [Online.] http://purl.oclc.org/estimates.
- 6.Engel, A. S., N. Lee, M. L. Porter, L. A. Stern, P. C. Bennett, and M. Wagner. 2003. Filamentous “Epsilonproteobacteria” dominate microbial mats from sulfidic cave springs. Appl. Environ. Microbiol. 69:5503-5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engel, A. S., M. L. Porter, B. K. Kinkle, and T. C. Kane. 2001. Ecological assessment and geological significance of microbial communities from Cesspool Cave, Virginia. Geomicrobiol. J. 18:259-274. [Google Scholar]
- 8.Engel, A. S., M. L. Porter, L. A. Stern, S. Quinlan, and P. C. Bennett. 2004. Bacterial diversity and ecosystem function of filamentous microbial mats from aphotic (cave) sulfidic springs dominated by chemolithoautotrophic “Epsilonproteobacteria” FEMS Microbiol. Ecol. 51:31-53. [DOI] [PubMed] [Google Scholar]
- 9.Engel, A. S., L. A. Stern, and P. C. Bennett. 2004. Microbial contributions to cave formation: new insights into sulfuric acid speleogenesis. Geology 32:269-372. [Google Scholar]
- 10.Forti, P., S. Galdenzi, and S. M. Sarbu. 2002. Hypogenic caves: a powerful tool for the study of seeps and their environmental effects. Contin. Shelf Res. 22:2373-2386. [Google Scholar]
- 11.Frederiksen, T.-M., and K. Finster. 2004. The transformation of inorganic sulfur compounds and the assimilation of organic and inorganic carbon by the sulfur disproportionating bacterium Desulfocapsa sulfoexigens. Antonie Leeuwenhoek 85:141-149. [DOI] [PubMed] [Google Scholar]
- 12.Fukui, M., A. Teske, B. Assmus, G. Muyzer, and F. Widdel. 1999. Physiology, phylogenetic relationships, and ecology of filamentous sulfate-reducing bacteria (genus Desulfonema). Arch. Microbiol. 172:193-203. [DOI] [PubMed] [Google Scholar]
- 13.Galdenzi, S., and T. Maruoka. 2003. Gypsum deposits in the Frasassi Caves, central Italy. J. Cave Karst Studies 65:111-125. [Google Scholar]
- 14.Galdenzi, S., M. Menicheti, S. M. Sarbu, and A. Rossi. 1999. Frasassi caves: a biogenic hypogean karst system, p. 101-106. In Proceedings of the European Conference, Karst 99, Grands Causses, Vercors, France vol. 20. Universite de Provence, Aix-en-Provence, France. [Google Scholar]
- 15.Galdenzi, S., M. Menichetti, and P. Forti. 1997. La corrosione di placchette calcaree ad opera di acque sulfuree: dati sperimentali in ambiente ipogeo, p. 187-190. In Proceedings of the Twelfth International Congress of Speleology, vol 1. Swiss Institute of Speleology and Karst Studies, La Chaux-de-Fonds, Switzerland.
- 16.Galdenzi, S., and S. M. Sarbu. 2000. Chemiosintesi e speleogenesi in un ecosistema ipogeo: i rami sulfurei delle grotte di frasassi. Grotte Italia 1:3-18. [Google Scholar]
- 17.Hill, C. A. 1995. Sulfur redox reactions: hydrocarbons, native sulfur, Mississippi Valley-type deposits, and sulfuric acid karst in the Delaware Basin, New Mexico and Texas. Environ. Geol. 25:16-23. [Google Scholar]
- 18.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon; a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 19.Huelsenbeck, J. P. 2000. MrBayes: Bayesian inferences of phylogeny. University of Rochester, Rochester, N.Y.
- 20.Hugenholtz, P., and T. Huber. 2003. Chimeric 16S rDNA sequences of diverse origin are accumulating in the public databases. Int. J. Syst. Evol. Microbiol. 53:289-293. [DOI] [PubMed] [Google Scholar]
- 21.Hugenholtz, P., G. W. Tyson, and L. L. Blackall. 2001. Design and evaluation of 16S rRNA-targeted oligonucleotide probes for fluorescent in situ hybridization, p. 29-42. In M. Aquino de Muro and R. Rapley (ed.), Gene probes: principles and protocols. Humana Press, London, United Kingdom.
- 22.Hutchens, E., S. Radajewski, M. G. Dumont, I. R. McDonald, and J. C. Murrell. 2004. Analysis of methanotrophic bacteria in Movile Cave by stable isotope probing. Environ. Microbiol. 6:111-120. [DOI] [PubMed] [Google Scholar]
- 23.Hügler, M., C. O. Wirsen, G. Fuchs, C. D. Taylor, and S. M. Sievert. 2005. Evidence for autotrophic CO2 fixation via the reductive tricarboxylic acid cycle by members of the ɛ subdivision of proteobacteria. J. Bacteriol. 187:3020-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalanetra, K. M., S. L. Huston, and D. C. Nelson. 2004. Novel, attached, sulfur-oxidizing bacteria at shallow hydrothermal vents possess vacuoles not involved in respiratory nitrate accumulation. Appl. Environ. Microbiol. 70:7487-7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 26.Ku, T. C. W., L. M. Walter, M. L. Coleman, R. E. Blake, and A. M. Martini. 1999. Coupling between sulfur recycling and syndepositional carbonate dissolution: evidence from oxygen and sulfur isotope composition of pore water sulfate, South Florida Platform, USA. Geochim. Cosmochim. Acta 63:2529-2546. [Google Scholar]
- 27.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Wiley, New York, N.Y.
- 28.Larkin, J. M., and W. R. Strohl. 1983. Beggiatoa, Thiothrix, and Thioploca. Ann. Rev. Microbiol. 37:341-367. [DOI] [PubMed] [Google Scholar]
- 29.Lathe, R. 1985. Synthetic oligonucleotide probes deduced from amino acid sequence data: theoretical and practical considerations. J. Mol. Biol. 183:1-12. [DOI] [PubMed] [Google Scholar]
- 30.Loy, A., A. Lehner, N. Lee, J. Adamczyk, H. Meier, J. Ernst, K.-H. Schleifer, and M. Wagner. 2002. Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl. Environ. Microbiol. 68:5064-5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüβmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Minz, D., S. Fishbain, S. J. Green, G. Muyzer, Y. Cohen, B. E. Rittmann, and D. A. Stahl. 1999. Unexpected population distribution in a microbial mat community: sulfate-reducing bacteria localized to the highly oxic chemocline in contrast to a eukaryotic preference for anoxia. Appl. Environ. Microbiol. 65:4659-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nelson, D. C., N. P. Revsbech, and B. B. Jørgensen. 1986. Microoxic-anoxic niche of Beggiatoa spp.: microelectrode survey of marine and freshwater strains. Appl. Environ. Microbiol. 52:161-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nielsen, P. H., M. A. de Muro, and J. L. Nielsen. 2000. Studies on the in situ physiology of Thiothrix spp. present in activated sludge. Environ. Microbiol. 2:389-398. [DOI] [PubMed] [Google Scholar]
- 35.Rohwerder, T., W. Sand, and C. Lascu. 2003. Preliminary evidence for a sulphur cycling in Movile Cave, Romania. Acta Biotechnol. 23:101-107. [Google Scholar]
- 36.Rooney-Varga, J. N., R. Devereux, R. S. Evans, and M. E. Hines. 1997. Seasonal changes in the relative abundance of uncultivated sulfate-reducing bacteria in a salt marsh sediment and in the rhizosphere of Spartina alterniflora. Appl. Environ. Microbiol. 63:3895-3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sarbu, S., S. Galdenzi, M. Menichetti, and G. Gentile. 2000. Geology and biology of the Frasassi caves in central Italy: an ecological multi-disciplinary study of a hypogenic underground karst system, p. 359-378. In H. Wilkens (ed.), Ecosystems of the world, vol. 30. Elsevier, New York, N.Y. [Google Scholar]
- 38.Sarbu, S. M., T. C. Kane, and B. K. Kinkle. 1996. A chemoautotrophically based cave ecosystem. Science 272:1953-1955. [DOI] [PubMed] [Google Scholar]
- 39.Sarbu, S. M., B. K. Kinkle, L. Vlasceanu, T. C. Kane, and R. Popa. 1994. Microbiological characterization of a sulfide-rich groundwater ecosystem. Geomicrobiol. J. 12:175-182. [Google Scholar]
- 40.Schulz, H. N., and B. B. Jorgensen. 2001. Big bacteria. Annu. Rev. Microbiol. 55:105-137. [DOI] [PubMed] [Google Scholar]
- 41.Suzuki, Y., T. Sasaki, M. Suzuki, Y. Nogi, T. Miwa, K. Takai, K. H. Nealson, and K. Horikoshi. 2005. Novel chemoautotrophic endosymbiosis between a member of the Epsilonproteobacteria and the hydrothermal-vent gastropod Alviniconcha aff. hessleri (Gastropoda: Provannidae) from the Indian Ocean. Appl. Environ. Microbiol. 71:5440-5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swofford, D. L. 2000. PAUP*: phylogenetic analysis using parsimony and other methods. Sinauer Associates, Sunderland, Mass.
- 43.Takai, K., B. J. Campbell, S. C. Cary, M. Suzuki, H. Oida, T. Nunoura, H. Hirayama, S. Nakagawa, Y. Suzuki, F. Inagaki, and K. Horikoshi. 2005. Enzymatic and genetic characterization of carbon and energy metabolisms by deep-sea hydrothermal chemolithoautotrophic isolates of Epsilonproteobacteria. Appl. Environ. Microbiol. 71:7310-7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takai, K., H. Hirayama, T. Nakagawa, Y. Suzuki, K. H. Nealson, and K. Horikoshi. 2004. Thiomicrospira thermophila sp nov., a novel microaerobic, thermotolerant, sulfur-oxidizing chemolithomixotroph isolated from a deep-sea hydrothermal fumarole in the TOTO caldera, Mariana Arc, Western Pacific. Int. J. Syst. Evol. Microbiol. 54:2325-2333. [DOI] [PubMed] [Google Scholar]
- 45.Takai, K., F. Inagaki, S. Nakagawa, H. Hirayama, T. Nunoura, Y. Sako, K. H. Nealson, and K. Horikoshi. 2003. Isolation and phylogenetic diversity of members of previously uncultivated ɛ-proteobacteria in deep-sea hydrothermal fields. FEMS Microbiol. Lett. 218:167-174. [DOI] [PubMed] [Google Scholar]
- 46.Teske, A., and D. C. Nelson. 2004. The genera Beggiatoa and Thioploca. In M. Dworkin (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed., vol. 3.17. Springer-Verlag, New York, N.Y.
- 47.Teske, A., N. B. Ramsing, K. Habicht, M. Fukui, J. Kuver, B. B. Jorgensen, and Y. Cohen. 1998. Sulfate-reducing bacteria and their activities in cyanobacterial mats of Solar Lake (Sinai, Egypt). Appl. Environ. Microbiol. 64:2943-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tonolla, M., A. Demarta, S. Peduzzi, D. Hahn, and R. Peduzzi. 2000. In situ analysis of sulfate-reducing bacteria related to Desulfocapsa thiozymogenes in the chemocline of meromictic Lake Cadagno (Switzerland). Appl. Environ. Microbiol. 66:820-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zerkle, A. L., C. H. House, and S. L. Brantley. 2005. Biogeochemical signatures through time as inferred from whole microbial genomes. Am. J. Sci. 305:467-502. [Google Scholar]