Abstract
Desulfovibrio vulgaris was cultivated in a defined medium, and biomass was sampled for approximately 70 h to characterize the shifts in gene expression as cells transitioned from the exponential to the stationary phase during electron donor depletion. In addition to temporal transcriptomics, total protein, carbohydrate, lactate, acetate, and sulfate levels were measured. The microarray data were examined for statistically significant expression changes, hierarchical cluster analysis, and promoter element prediction and were validated by quantitative PCR. As the cells transitioned from the exponential phase to the stationary phase, a majority of the down-expressed genes were involved in translation and transcription, and this trend continued at the remaining times. There were general increases in relative expression for intracellular trafficking and secretion, ion transport, and coenzyme metabolism as the cells entered the stationary phase. As expected, the DNA replication machinery was down-expressed, and the expression of genes involved in DNA repair increased during the stationary phase. Genes involved in amino acid acquisition, carbohydrate metabolism, energy production, and cell envelope biogenesis did not exhibit uniform transcriptional responses. Interestingly, most phage-related genes were up-expressed at the onset of the stationary phase. This result suggested that nutrient depletion may affect community dynamics and DNA transfer mechanisms of sulfate-reducing bacteria via the phage cycle. The putative feoAB system (in addition to other presumptive iron metabolism genes) was significantly up-expressed, and this suggested the possible importance of Fe2+ acquisition under metal-reducing conditions. The expression of a large subset of carbohydrate-related genes was altered, and the total cellular carbohydrate levels declined during the growth phase transition. Interestingly, the D. vulgaris genome does not contain a putative rpoS gene, a common attribute of the δ-Proteobacteria genomes sequenced to date, and the transcription profiles of other putative rpo genes were not significantly altered. Our results indicated that in addition to expected changes (e.g., energy conversion, protein turnover, translation, transcription, and DNA replication and repair), genes related to phage, stress response, carbohydrate flux, the outer envelope, and iron homeostasis played important roles as D. vulgaris cells experienced electron donor depletion.
The underground corrosion of metal pipes used for gas or water and the generation of sulfide during digestion of domestic and agricultural wastes have been the economic and environmental processes that have historically driven the desire to understand the metabolism of sulfate-reducing bacteria (SRB) (16). The SRB have been a particular problem for the petroleum industry due to their roles in metal corrosion, petroleum souring, and the health hazards of hydrogen sulfide. In contrast, SRB can be advantageous for bioremediation processes. A variety of studies (6, 15, 17, 33, 39) have documented the ability of SRB, including Desulfovibrio spp., to reduce toxic metals, such as U(VI) and Cr(VI), enzymatically, a process that results in the production of species that are less water soluble. The modification of solubility properties caused by changing the redox state of the metal can be a potential avenue for bioremediation of contaminated groundwater and soils, and previous research specifically indicates that SRB are environmentally relevant experimental systems (1, 2, 32). Sulfate reducers have several advantages for heavy-metal reduction, including the presence of sulfate in a variety of environments and the protection of immobilized heavy metals from oxidation with iron sulfides (mackinawite) (3).
Desulfovibrio vulgaris Hildenborough has been studied extensively, and much is already known about the metabolic versatility of this bacterium (21, 53). D. vulgaris is capable of coupling the oxidation of a variety of electron donors (e.g., lactate, pyruvate, succinate, and ethanol) to the reduction of many different electron acceptors (e.g., sulfate, fumarate, uranium, chromium, and potentially O2) either directly or concomitantly. To effectively immobilize heavy metals and radionuclides, it is important to understand the cellular responses to adverse factors observed in contaminated subsurface environments, such as mixed contaminants and the changing ratios of electron donors and acceptors. As documented for other bacteria, the cellular responses to different stressors (e.g., osmolytes, heat, and pH) can overlap responses to the stationary phase (23). In addition, stimulation of microbial populations with carbon and/or energy sources to promote bioremediation can affect the physiology of indigenous microorganisms in relation to the available electron acceptors. The identification of stasis-induced genes and gene networks should provide fundamental information about the cellular processes needed for survival under pertinent field conditions (e.g., low nutrients, slow growth, and energy source variability) and a comparative framework for studies of additional stressors and environmental stimuli. Previous work has demonstrated that use of Desulfovibrio spp. for sulfate and metal reduction is well founded and that the genomic and proteomic tools developed for D. vulgaris provide the resources to obtain an in-depth understanding of cellular processes important for stress responses and bioremediation in relation to environmental conditions. In this study we focused on stasis-induced genes and gene networks as D. vulgaris transitioned from the exponential phase to the stationary phase. Our results demonstrated that the gene expression profiles of D. vulgaris cells changed in response to carbon and energy depletion and that gene expression during the stationary phase was not static.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
D. vulgaris ATCC 29579 was grown in a batch system that consisted of a glass column (diameter, 7 cm) with a diffuser, a septum, and a gas outlet. The column contained LS4D medium (1 liter) that was gassed with nitrogen at a rate of approximately 1 ml/min and was constantly stirred. LS4D medium is a defined medium containing approximately 50 mM Na2SO4, 60 mM NaC3H5O3, 8.0 mM MgCl2, 20 mM NH4Cl, 2.2 mM K2HPO4/KH2PO4 (pH 7.2), 0.6 mM CaCl2, Thauers vitamins (9), trace minerals (9), 30 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) buffer, 0.06 μM resazurin, and 10 mM NaOH (pH 7.2). Triplicate columns were inoculated with a 10% (vol/vol) inoculum from the same culture of D. vulgaris to obtain an optical density at 600 nm of approximately 0.06. Cultures were incubated in a 30°C water bath, and samples were collected aseptically at various times via a septum.
Sample collection.
During growth, samples were obtained from triplicate cultures at nine different times for RNA extraction and global expression analyses. The sampling times were selected based on monitored protein and lactate levels and represented the exponential phase (T1 to T3), the transition (T4 and T5), and the stationary phase (T6 to T9). Samples (approximately 5 mg protein) used for RNA extraction were collected at various times throughout the growth phases. Samples were collected in sterile, cold (−20°C) beakers and were then immediately pumped through a metal coil (inside diameter, 3 mm) kept at −20°C (CaCl2-H2O slurry) to chill them. After the samples were quickly cooled and collected, they were centrifuged for 6 min at 4,000 × g (4°C). Each supernatant was discarded, and the pellet was then submerged in liquid nitrogen to snap freeze it. The pellets were stored at −70°C until they were used for RNA extraction. Samples (1 to 5 ml) were also collected for determination of the protein and total carbohydrate associated with cell pellets, and supernatants were used for determination of lactate, sulfate, and acetate levels. Samples were stored at −20°C until processing.
Chemical analyses.
Protein was analyzed by the method of Lowry et al. (34), and bovine serum albumin (Pierce Biochemicals) was used as a standard. Carbohydrate contents were determined by a cysteine-sulfuric acid colorimetric assay as previously described (10). Lactate, acetate, and sulfate concentrations were determined by ion chromatography (Metrohm-Peak) with a Metrosep organic acid column and a Metrosep Anion Supp 5 column. All assays were done in duplicate, and the variation was less than 10%. The results for columns were averaged to obtain a complete protein and carbohydrate profile.
Oligonucleotide probe design and microarray construction.
DNA microarrays that included 3,482 of the 3,531 annotated protein-encoding sequences of the D. vulgaris genome were constructed with 70-mer oligonucleotide probes as previously described (18, 19, 20). Following examination of the entire probe set using the oligonucleotide probe design criteria (7), 3,471 (97.1%) specific oligonucleotide probes were obtained, and 103 probes(2.9%) were nonspecific. In addition, 10 oligonucleotides for 10 human genes and 10 oligonucleotides for 10 Arabidopsis genes were used with the D. vulgaris genome for positive (with mRNA spiked) or negative (without mRNA spiked) controls. All of the oligonucleotides that were designed were commercially synthesized without modification by MWG Biotech Inc., (High Point, NC). The concentrations of oligonucleotides were adjusted to 100 pmol/μl in 50% (vol/vol) dimethyl sulfoxide, and the oligonucleotides were spotted onto UltraGAPS glass slides (Corning Life Sciences, Corning, NY) using a BioRobotics Microgrid II microarrayer (Genomic Solutions, Ann Arbor, MI). A single slide contained duplicates of each oligonucleotide probe. In addition, six concentrations of genomic DNA were spotted (eight duplicates on a single slide) as positive controls. After printing, the oligonucleotide probes were fixed onto the slides by UV cross-linking (600 mJ of energy) according to the manufacturer's protocol (Corning Life Science, Corning, NY).
Total RNA extraction, purification, and labeling.
Total cellular RNA was isolated using the TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. RNA extracts were purified with an RNeasy mini kit (QIAGEN Valencia, CA) used according to the instructions, and on-column DNase digestion was performed with RNase-free DNase (QIAGEN, Valencia, CA) to remove genomic DNA.
To generate cDNA probes with reverse transcriptase (RTase), 10 μg of purified total RNA was used for each labeling reaction, as previously described (20, 50). Briefly, random hexamers (Invitrogen) were used for priming and were labeled with Cy3-dUTP or Cy5-dUTP (Amersham Biosciences, Piscataway, NJ). After labeling, RNA was removed by NaOH treatment, and cDNA was immediately purified with a QIAGEN PCR mini kit. The efficiency of labeling was routinely monitored by determining the absorbance at 260 nm (for DNA concentration), at 550 nm (for Cy3), or at 650 nm (for Cy5). Two samples of each total RNA preparation were labeled, one with Cy3-dUTP and the other with Cy5-dUTP, for microarray hybridization.
Microarray hybridization, washing, and scanning.
For determination of the overall hybridization signals, the sensitivity, and the number of genes detected, a genomic DNA or RNA sample labeled with a single dye was used as previously described (20). Hybridization was performed using a slide for each biological replicate, and there were duplicate probes for each gene. The microarrays were hybridized at 45°C overnight with 50% formamide. The labeled cDNAs were resuspended in 20 to 25 μl of hybridization solution that contained 50% (vol/vol) formamide, 1 mM dithiothreitol, 3× SSC, 0.3% (wt/vol) sodium dodecyl sulfate (SDS), and 0.8 μg/μl of herring sperm DNA (Invitrogen Life Technologies, CA) (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Each sample was incubated at 98°C for 5 min, centrifuged to collect condensation, and kept at 50 to 60°C. The sample was immediately applied to the microarray slide and covered with coverslip, and hybridization was carried out in a waterproof Corning hybridization chamber (Corning Life Science, Corning, NY) submerged in a 45°C water bath in the dark for 16 h. After hybridization, the coverslips were immediately removed and washed in a buffer containing 1× SSC and 0.1% (wt/vol) SDS for 5 min at 37°C. The microarrays were then washed in a new buffer containing 0.1× SSC and 0.1% (wt/vol) SDS for another 5 min at room temperature. Finally, the microarrays were washed with distilled water for 30 s at room temperature and dried with compressed air or by centrifugation. The microarrays were scanned using a ScanArray 5000 microarray analysis system (Packard BioChip Technologies, Massachusetts). Typically, 95 to 100% of laser power and 70 to 80% of photomultiplier tube were used for scanning.
Image quantification and data analysis.
To determine the signal fluorescence intensity for each spot, 16-bit TIFF scanned images were analyzed with ImaGene v6.0 (Biodiscovery, Marina Del Rey, CA) to quantify the spot signal, spot quality, and background fluorescent intensities. Empty spots, poor spots, and negative spots (e.g., spots without deposited probe) were flagged according to the instruction of the software and removed in the subsequent analysis (14).
The resulting data files were subjected to Lowess intensity-based normalization and were analyzed further using GeneSpring, version 5.1 (Silicon Genetics, Redwood City, CA). To assess the statistical significance of individual data points, the Student t test was used to calculate a P value in order to test the null hypothesis that the expression level was unchanged. A statistical model incorporating both per-gene variance (z values) and operon structure was used to compute the posterior probability that the expression level of each gene changed in the direction indicated by the mean value (40). Members of operons without a consistent signal for replicates were excluded, and genes with log2 ratios of | z| > 2.0 were considered significant. The sample from the first time (T1) was used as the control sample, and all subsequent samples were compared to the T1 sample in order to elucidate changes in gene expression during the transition to the stationary phase.
Hierarchical cluster analysis.
Microarray data sets for different times were analyzed by using average linkage hierarchical cluster analysis with Euclidean distance matrices and were visualized with TreeView as previously described (13, 47). If there were expression data for a gene for all sample times and the level of expression of log2(ratio) was ≥1.5 or ≤−1.5 at one or more times, the gene was used for cluster analysis, where the ratio is the ratio of the gene expression at one time (T2 to T9) to gene expression at T1. Gene expression with a log2(ratio) of ≥1.5 was considered up-expression, and gene expression with a log2(ratio) of ≤−1.5 was considered down-expression.
Quantitative PCR.
Quantitative PCR (qPCR) was performed with RNA extracts from samples obtained at 5, 20, 23, 29, 35, 43, and 51 h. Total RNA (5 μg) was used to prepare cDNA by reverse transcription. An RNA sample was mixed with 6 μg of random primers (Invitrogen). The mixture (total volume, 21 μl) was incubated at 70°C for 10 min and then placed on ice to cool. After cooling, 4 μl of 5× reverse transcription buffer, 2 μl of 0.1 M dithiothreitol, 1 μl of a solution containing each deoxynucleoside triphosphate at a concentration of 10 mM, 1 μl of RNase-Out inhibitor (Invitrogen), and 1 μl of reverse transcriptase (Invitrogen) were added to each tube. The tubes were incubated at 42°C for 2 h and then at 70°C for 15 min. The tubes were cooled on ice, and the absorbance at 260 nm was determined. The cDNA samples were diluted to obtain a concentration of 25 ng/μl, and a 2-μl sample of cDNA was used for qPCR.
The primers used for qPCR are described in Table 1. The primers were diluted to obtain 10 mM working stock solutions. Each qPCR mixture (final volume, 20 μl) contained 10 μl of SYBR Green Master Mix (Applied Biosystems), 0.7 μl of a 10 mM solution of the forward primer, 0.7 μl of a 10 mM solution of the reverse primer, 2 μl of cDNA template, and 6.6 μl of nuclease-free water. The real-time PCR was carried out with a Rotor-gene 6 (Corbett Research, Sydney, Australia) using the following conditions: one cycle of 95°C for 10 min and 40 cycles of 95°C for 15 s, 55°C for 30 s, and 60°C for 30 s. Standards were prepared from genomic DNA with a starting concentration of 108 copies/μl. Serial dilution was performed to obtain a standard curve for 108 to 101 copies/μl, and a negative control that did not contain RTase was included for all samples. When real-time PCR analysis was used to determine the levels of expression of nine genes, a high degree of correlation was observed for the results from microarrays and real-time PCR (R = 0.88), as reported in previous studies in which the workers used similar microarray procedures and techniques (11, 14, 18, 54).
TABLE 1.
Primers used for quantitative PCR for validation of whole-genome microarray results
| Gene | Sequence
|
Size of amplicon (bp) | |
|---|---|---|---|
| Forward primer (5′-3′) | Reverse primer (5′-3′) | ||
| DVU2776 | AAGTCACTTACAAGGGCAAG | GATTCCTTCACGTATTCCAC | 100 |
| DVU2571 | CCAGCTTGAAGACATGGT | GATGTAGCCGTAACGGTAGT | 100 |
| DVU1311 | TCACTGCCGAAGAGCTTA | ATGCTGGAGGTGTTCTCC | 103 |
| DVU3108 | GTACCTCGGAGACAAGATGT | GTCGTCCACATCATGGAA | 105 |
| DVU0014 | CAAGATGCGCAAGTTCTAC | TCATACGGTAGGTGATACGTC | 101 |
| DVU1858 | GATGACTTCGTGTGCAGAG | TGCACAACGTCATCTCAC | 101 |
| DVU2839 | CTTAGCCAAGCTACTAGACGAC | GCTGAGTTGATGCTCAGGTA | 100 |
| DVU2061 | AGAATCTGCTGGCTGACA | GTCGCAGTATGCCTTGAT | 101 |
Promoter element prediction.
Gibbs Recursive Sampler (31, 52) was used to predict transcription factor-binding motifs upstream of 136 (119 from the chromosome and 17 from the megaplasmid) differentially expressed D. vulgaris genes unique to the stationary phase (from T5 to T9) and not found at T1, as described previously (54). The Gibbs Recursive Sampler program was used with (i) 16-bp models allowed to fragment up to 24 bp; (ii) up to three sites per sequence, where P(no sites) = 0.2, P(one site) = 0.7, P(two sites) = 0.05, and P(three sites) = 0.05; (iii) a position-specific background model to account for variation in local base composition; and (iv) the Wilcoxon signed-rank test (http://bayesweb.wadsworth.org/gibbs/gibbs.html). The sequences used for alignment were intergenic regions that were ≥25 bp upstream of putative translation start codons, as defined by the D. vulgaris genome annotation (accession no. NC_002937 and NC_005863). Negative controls for the Wilcoxon test were randomly chosen from the set of D. vulgaris intergenic regions, provided that the sequence lengths matched those of the test sequences and excluded any sequence region that included a promoter for a gene whose expression changed at least twofold during the stationary phase of D. vulgaris compared with the expression at T1. The Wilcoxon test calculated a P value for the motif, given the null hypothesis that the sequences being studied (the test sequences) were not more likely to contain sites than the negative control sequences and also considered the score ranks of the predicted sites in the motif. The SCAN program was used to identify sites from a database that were described by the motif (35). SCAN calculated a P value for each of the identified sites that reflected the probability that a site with its score or better would occur in a random database of the same size.
RESULTS
Growth.
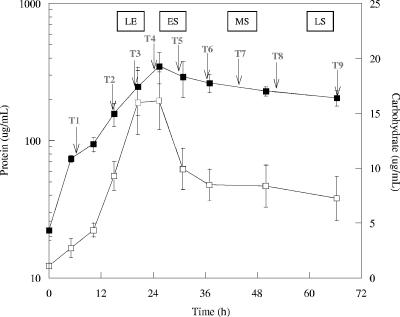
The three replicate cultures were grown in a defined medium with lactate and sulfate in batch mode, and the inoculum was in the exponential growth phase. Optical density at 600 nm was used to monitor growth for determination of sampling times. The biomass level was approximately 20 μg/ml protein initially and reached approximately 260 μg/ml protein in 25 h (Fig. 1).
FIG. 1.
Protein ▪ and carbohydrate (□) levels during growth of D. vulgaris cells with lactate and sulfate. The arrows indicate times at which biomass was removed for RNA extraction. The error bars indicate standard deviations. LE, late exponential phase; ES, early stationary phase; MS, mid-stationary phase; LS, late stationary phase.
The total carbohydrate levels increased steadily during the exponential phase and plateaued as the cells entered the stationary phase (Fig. 1). These results suggested that exhaustion of the electron donor also caused a shift in the carbon flux of the cells. The total carbohydrate levels declined during stationary-phase growth and were two- to threefold lower at the last sampling time. Preliminary results showed that the majority of measured carbohydrate in planktonic cells is internal (Fields and Clark, unpublished). These results suggested that the measured carbohydrate was glycogen, and previous results have documented that glycogen is present in Desulfovibrio spp. (44).
The exponential-phase growth rate was 0.11 h−1, the generation time was 6.41 h, and cessation of exponential growth coincided approximately with depletion of lactate (Fig. 2). Sulfate and lactate were depleted concomitantly; the lactate level declined at a higher rate, and the decline in the sulfate levels stopped when lactate was depleted (Fig. 2). The pH steadily increased during growth and reached approximately 9.0 in the early stationary growth phase.
FIG. 2.
Lactate levels (▪), sulfate levels (•), and pH (⧫) during growth of D. vulgaris with lactate and sulfate. The error bars indicate standard deviations. LE, late exponential phase; ES, early stationary phase; MS, mid-stationary phase; LS, late stationary phase.
Transcriptomics.
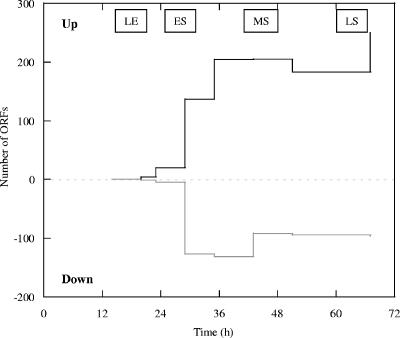
During growth, samples were obtained from triplicate cultures nine times for RNA extraction and global transcription analyses, four times in the exponential phase and five times in the stationary phase (Fig. 1). The sampling times are indicated in Fig. 1. For the samples from cultures in the exponential growth phase (T2 and T3) the expression of few genes was significantly altered compared to the expression at T1, and for a minimal number of open reading frames (ORFs) there were significant changes in transcription even at T4 (Fig. 3). At T5 through T9, approximately 130 to 250 presumptive ORFs were up-expressed as the cells experienced electron donor depletion and 90 to 130 ORFs were down-expressed (Fig. 3). In addition, the expression of the genes encoding many hypothetical proteins and conserved hypothetical proteins was altered at most of the times.
FIG. 3.
Total number of ORFs predicted to be up-expressed or down-expressed (|log2 ratio| ≥ 2 with | z| > 2.0) when the expression level at each time was compared to the expression level at T1. LE, late exponential phase; ES, early stationary phase; MS, mid-stationary phase; LS, late stationary phase.
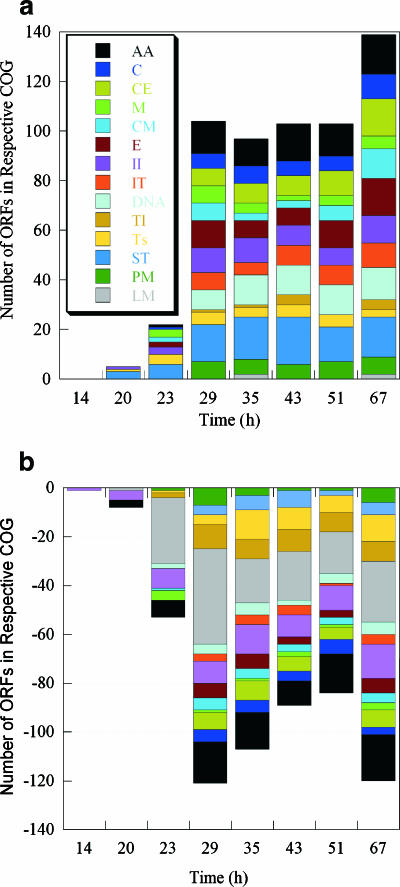
When the ORFs with significant changes were categorized into clusters of orthologous genes, several of the groups were represented predominantly by up- and down-expressed ORFs (Fig. 4). At 20 h and 23 h, genes predicted to be involved in amino acid transport and metabolism, energy production and conversion, and translation were the dominant genes that were down-expressed. These three groups of genes accounted for the largest fraction of down-expressed genes for all times, in addition to transcription and signal transduction (Fig. 4b).
FIG. 4.
Up-expressed (a) and down-expressed (b) genes (z ≥ 2.0) grouped as clusters of orthologous genes (COG) with respect to the sampling time. The following clusters of orthologous genes are indicated by different colors: amino acid transport and metabolism (AA), carbohydrate transport and metabolism (C), cell envelope biogenesis (CE), cell motility (M), coenzyme metabolism (CM), DNA replication and repair (DNA), energy production (E), inorganic ion transport (II), intracellular trafficking (IT), lipid metabolism (LM), posttranslational modification (PM), signal transduction (ST), transcription (Ts), and translation (Tl).
At 23 h (late exponential phase), approximately one-half of the down-expressed genes were involved in translation and transcription, and this trend continued at the remaining times. Genes involved in amino acid acquisition, carbohydrate metabolism, energy production, and cell envelope biogenesis showed both up- and down-expression (Fig. 4), and these results suggested that subsets of genes in the same cluster of orthologous genes may be specific to a particular growth phase.
Six small-subunit and 10 large-subunit rRNA protein genes were down-expressed more than twofold between 20 and 30 h postinoculation, but the levels of expression appeared to rebound once the stationary phase was established, although the levels were lower than those observed for exponential-phase growth (see Fig. S1 in the supplemental material). Other presumptive genes associated with translation were also down-expressed and included the nusG, EF-Ts, and IF-1 genes. Other proteins known to be involved in nutrient deprivation are SsrA and SmpB in Escherichia coli (26). The level of expression of the smpB homolog in D. vulgaris did not change more than twofold compared to the level at T1, but the levels did change over time. Finally, the Era protein of E. coli has recently been shown to specifically bind to small-subunit rRNA and the 30S ribosomal subunit and is thought to be a cell cycle check point for protein synthesis (45). DVU0052, annotated as a gene encoding a G-protein similar to Era, was drastically up-expressed in the 29-h sample.
Hierarchical cluster analysis of times.
The sampling times were sorted into clusters based upon similar patterns of expression across the entire genome. When the sampling times were compared, T2 (14 h), T3 (20 h), and T4 (23 h) were clustered, but sampling times later in the exponential phase of growth (20 and 23 h) were more closely grouped (Fig. 5). These results suggested that the gene expression profiles of cells were beginning to change 5 h before depletion of lactate, and this observation coincided with the down-expression of at least 14 ribosomal protein genes well before the lactate was completely depleted (see Fig. S1 in the supplemental material). Based upon cluster analysis, several genes were preferentially up-expressed at 20 and 23 h, and these genes included genes coding for a putative hemerythrin-cation binding protein (DVU0170), a putative TonB (DVU2390), a putative ABC transporter (DVU2380), and a conserved hypothetical protein (DVU0273). Interestingly, DVU0170 and DVU2390 were predicted to be involved in metal (i.e., Fe) transport, and DVU0273 and DVU2380 were predicted to have a FUR binding box upstream of the predicted translational start site (43).
FIG. 5.
Hierarchical cluster analysis of sampling times based upon expression profiles for all predicted ORFs with an expression level of log2(ratio) of ≥1.5 or ≤−1.5 (a) Analysis of 1,181 genes used to compare times. (b) Selected portion of the cluster analysis.
The cluster analysis indicated that the gene expression profile significantly shifted between the 23- and 29-h sampling times (Fig. 5), and this represented the transition between the exponential and stationary phases. The second major cluster contained the later times (29, 35, 43, 51, and 67 h), but the profile for 67 h (the last time) was dissimilar from the other profiles (Fig. 5). In addition, we observed a difference between the cells at 29 h and cells at 35 to 51 h. The results indicated that the transcriptional expression patterns were indicative of growth phase transitions and that there were distinct profiles for the relatively early stationary phase (approximately 5 h postexponential), the mid-stationary phase (approximately 10 to 30 h postexponential), and the late stationary phase (approximately 40 h postexponential).
A group of 33 genes were preferentially up-expressed at 29 h and could be placed into four major categories: lipoproteins, amino acid metabolism, metal binding, and phage related. Most phage-related genes were up-expressed at 29 h and remained up-expressed during the reminder of the experiment (see Fig. S2 in the supplemental material). A group of five putative lipoproteins (encoded by DVU0163, DVU1366, DVU2367, DVU2428, and DVU2496) were up-expressed, and at least two of these proteins were predicted to be in the outer membrane. The levels of expression of genes predicted to be involved in amino acid metabolism (DVU0601, DVU1012, DVU1331, DVU1413, DVU2297, and DVU3293) were more similar at 29 h than at other times. Two ORFs that may be involved in iron acquisition were preferentially up-expressed at 29 h, and they encoded a rubrerythrin with a ferritin-like domain (DVU0019) and a hemerythrin-cation binding protein (DVU0170).
The 35-, 43-, and 51-h times were grouped and most likely represented the mid-stationary phase (Fig. 5). Genes whose levels of expression were similar at these times could be categorized into the following groups: hypothetical proteins and conserved hypothetical proteins, phage-related genes, DNA-binding proteins, and control of translation. Three ORFs predicted to be involved in translation were DVU0084, encoding a putative alternative initiation factor that recycles GTP and GDP, DVU2218, encoding a putative EngC protein that is predicted to have a role in translation regulation, and DVU2486, encoding a putative acetyltransferase that acts upon ribosomal proteins.
Iron acquisition.
With respect to possible iron acquisition during carbon and energy source depletion, three ORFs predicted to be part of an Fe2+ transport system (DVU2571, DVU2572, and DVU2574) were up-expressed at 20 h, the expression peaked at 23 h, and then transcription declined after 29 h (see Fig. S3 in the supplemental material). The putative feo system is comprised of FeoAB, and a similar system is involved in Fe2+ transport in E. coli (25). Concurrently, nigerythrin (DVU0019) was up-expressed at 29 h, but then expression declined after 29 h. In the D. vulgaris genome there are nine ORFs that are annotated to have ferritin domains, and most were down-expressed or displayed little change under the growth conditions used. At the 67-h sampling time, the cells had experienced 42 h of electron donor depletion. Related to possible iron acquisition, two genes that were preferentially up-expressed at 67 h were DVU0103 and DVU3170. DVU0103 is a putative fepC gene that is predicted to encode an ABC-type cobalamin/Fe(III)-siderophore transport system, and DVU3170 is predicted to encode a precorrin-3B C17-methyltransferase involved in cobalamin biosynthesis.
Key genes for lactate and sulfate utilization.
D. vulgaris has six putative lactate permease genes (DVU2110, DVU2285, DVU2451, DVU2683, DVU3026, and DVU3284). Two presumptive permease genes were unchanged, three were down-expressed, and one was up-expressed (see Fig. S4 in the supplemental material). The levels of expression of the permease genes changed between 14 and 23 h, and this time represented the time from when the level of the lactate was one-half the initial level to the time when lactate was almost completely exhausted.
In the D. vulgaris genome there are three ORFs that are annotated as genes that encode sulfate permeases (DVU0053, DVU1999, and DVU0279), and the trend of expression of each of these genes was different as cells transitioned from the exponential phase to the stationary phase. The level of DVU0053 increased from 14 to 29 h and then decreased throughout the stationary phase. The trend for DVU0279 was the inverse of the trend for DVU0053, and the level decreased from 14 to 29 h and then remained relatively low. The level of DVU1999 was relatively low but gradually increased over the course of the experiment (see Fig. S5 in the supplemental material). It is important to note that the sulfate levels decreased initially up to 29 h and then remained constant around 15 mM throughout stasis (Fig. 2).
Three ORFs were annotated as putative lactate dehydrogenase genes (DVU0600, DVU2784, and DVU0253) in the D. vulgaris genome. The levels of two of these genes were elevated during exponential growth; however, the level of DVU0253 increased at a higher rate and decreased significantly after 23 h. The level of DVU0600 transcripts increased at a lower rate during the exponential growth phase and did not show down-regulation until after 29 h (data not shown). The level of ORF DVU2784 remained mostly unchanged. The expression of the putative dsrA and dvsB genes (encoding dissimilatory sulfite reductase subunits) remained largely unchanged, but there was slight down-expression over time starting at the transition to the stationary phase (data not shown).
Representative genes associated with stasis-induced stress.
D. vulgaris has 11 ORFs that are predicted to encode proteins annotated as universal stress proteins, based on relatedness to the Usp proteins of E. coli (29). Previous research with E. coli has shown that the levels of the UspA protein can become elevated in response to starvation for carbon, nitrogen, phosphate, sulfate, and amino acids, as well as exposure to heat, oxidants, metals, and antibiotics (29). Three ORFs (DVU0006, DVU0452, and DVU0893) were relatively stable throughout exponential growth, but there was down-expression between 23 and 29 h (see Fig. S6a in the supplemental material). In contrast, three ORFs (DVU1030, DVU3298, and DVU3336) were relatively stable throughout the exponential phase, but there was an increase in expression at 23 h (see Fig. S6b in the supplemental material). The results suggested that specific putative Usp proteins in D. vulgaris might have similar functions during growth transitions and nutrient depletion. Recently, a Usp protein in E. coli was shown to specifically complex with GroEL during the stationary phase (8), and the expression of both the putative GroEL and GroES proteins in D. vulgaris increased from 23 to 35 h (see Fig. S7 in the supplemental material).
Numerous genes predicted to be involved in stress responses were up-expressed as the cells transitioned into the stationary phase. For example, a putative C-terminal protease gene (DVU2336) predicted to be located in the periplasm was up-expressed between 23 and 29 h and remained up-expressed throughout the stationary phase (see Fig. S8 in the supplemental material). Two ORFs predicted to encode carbon starvation proteins (DVU0599 and DVU0598) were up-expressed between 20 and 29 h (see Fig. S8 in the supplemental material). These genes were similar to cstA in E. coli, and CstA is a predicted membrane protein that is involved in peptide utilization during carbon starvation (46).
Carbohydrate-related genes.
The increase in carbohydrate levels stopped as the cells transitioned to the stationary phase, and the levels actually began to decline (Fig. 2); the carbohydrate-to-protein ratio exhibited a similar trend. Genes predicted to be involved in carbohydrate metabolism were up-expressed during the phase transition (Fig. 4a). Representative genes that were significantly up-expressed included genes encoding a glycosyl transferase (DVUA0037), lysozyme (DVU1128), a polysaccharide deacetylase (DVUA0043), glycosyl transferase (DVU0351), a sugar dehydratase (DVU0448), an epimerase (DVU2455), glucokinase (DVU1035), and a sugar facilitator (DVUA0096). Interestingly, three of these genes were located on the megaplasmid of D. vulgaris, and preliminary results suggested that the carbohydrate profiles for a strain without the megaplasmid differed from wild-type profiles (Fields and Clark, unpublished results). The majority of these genes were up-expressed between 23 and 29 h, and the transcript levels remained elevated into the stationary phase (see Fig. S9 in the supplemental material).
The sugar dehydratase (DVU0448) is predicted to be a manno-dehydratase, the enzyme responsible for the first step in the conversion of GDP-mannose to GDP-fucose (28). In E. coli, GDP-l-fucose can act as a precursor for surface antigens (e.g., extracellular polysaccharide) (28). In addition, D. vulgaris has one putative glucokinase (DVU1035), and the gene was gradually up-expressed until 43 h. D. vulgaris has only four genes annotated as genes encoding probable sugar transporters, and DVUA0096 was the only one that exhibited up-expression during the growth transition to the stationary phase. Few studies have reported utilization of exogenous sugar by D. vulgaris, unlike Desulfovibrio fructosovorans, which can utilize some sugars (38). The total carbohydrate levels decreased more than twofold; however, significant amounts of extracellular carbohydrate associated with the cell or in the supernatant were not detected (data not shown). These results suggested that rerouting of internal carbohydrate could occur, but further work is needed to determine the possible role of carbon reserves in D. vulgaris.
In silico prediction of a stationary-phase promoter sequence.
In order to identify possible transcription factor-binding motifs for stationary-phase responsive genes, the upstream noncoding regions (200 bp) of 136 genes (up-expressed in the stationary phase) were searched for common motifs with the Gibbs Recursive Sampler (30, 52). The algorithms identified 5′-CxGCATGGG-3′ with statistical significance (P < 0.05). The base frequencies represented by the height of the stacked letter at each position are shown as a sequence logo in Fig. 6. The presumptive motif is quite different from a recently predicted σS consensus promoter element, 5′-TCTATACTTAA-3′, for E. coli (56). However, it should be noted that D. vulgaris does not have a predicted rpoS gene. The function of the possible nucleic acid motif needs to be characterized using experimental approaches.
FIG. 6.
Sequence logo of a predicted consensus sequence in the 200-bp regions upstream of up-expressed genes unique to stationary-phase growth in D. vulgaris.
DISCUSSION
In the cell cycle of bacteria there are periods of active growth and periods of slow growth that alternate depending on the nutrient levels and physicochemical conditions. In situ conditions rarely provide excess nutrients for the growth of bacteria, and short periods of growth alternate with prolonged periods of starvation. In the context of in situ bioremediation, nutrient scarcity and/or fluxes may be a common obstacle encountered by microorganisms due to the oligotrophic nature of most groundwater and sediment environments, and these conditions require the ability to transition between faster and slower growth. However, cells have mechanisms to survive cycles of nutrient depletion and starvation, as well as other stressful conditions commonly associated with prolonged starvation. The RpoS (σS) protein is a major transcriptional activator for cellular responses to the stationary phase in many bacteria, and σS induces approximately 100 genes in E. coli (27). In light of the predicted absence of rpoS in δ-Proteobacteria, our results demonstrated that D. vulgaris elicited a response to the stationary phase, and changes in gene transcription continued throughout growth.
Bacterial cultures grown in chemostats can provide a semisteady state for the cultivation of cells at a constant growth rate with one limiting nutrient. While this approach is powerful for physiological studies of an organism in a given state, it is not conducive for observations during transitions (e.g., changing levels of carbon and energy sources). Whole-genome transcriptomics have been used to compare regulatory gene mutants (5, 50, 54) and specific stressors (14, 18, 31), but only more recently have transcriptomics been used to compare different growth phases or the transitions (4, 7, 37, 47, 49, 55, 56). Most of the previous work has been with E. coli or pathogens, and few transcriptomic studies of growth phase transitions have been reported for “environmental” microorganisms. D. vulgaris is commonly used as a model SRB and is found in habitats that are exposed to continuously changing nutritional and environmental conditions. Because many stressors cause a decrease in the growth rate and/or altered nutrient utilization, it is important to elucidate responses during the transitions to better understand direct and indirect cellular adaptations.
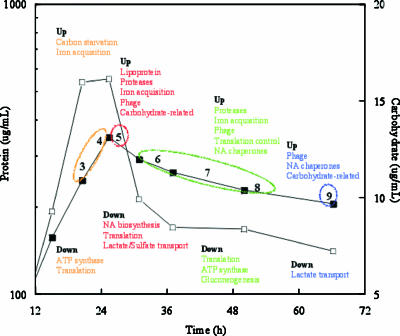
A summary of the major differentially expressed genes for the different times is shown in Fig. 7. T3 and T4 were combined and considered to be the late exponential phase (20 to 23 h) before the transition to the stationary phase. As expected, genes involved in ATP synthase, elongation factors, and ribosomal proteins were down-expressed. Presumptive genes that encode proteins related to carbon starvation and iron acquisition were significantly up-expressed as the cells transitioned. An annotated katA gene was also up-expressed, as was a putative pspA gene (encoding phage shock protein). PspA is considered part of a specialized extracytoplasmic stress response in gram-negative bacteria (12), and interestingly, most presumptive phage genes were significantly up-expressed at the next time examined (29 h) in the stationary phase. In E. coli, two important inducing conditions for pspA are dissipation of the proton motive force and mislocalization of secretin proteins that can be used for the extrusion of filamentous phage, and PspA may function to maintain the proton motive force (12). These results suggested that the depletion of an electron donor (a carbon and energy source) may have caused a decline in the proton motive force that affected protein secretion and/or the phage cycle.
FIG. 7.
Summary of significant up- and down-expressed genes at the late exponential (T3 and T4), early stationary (T5), mid-stationary (T6, T7, and T8), and late stationary phases (T9) of growth. NA, nucleic acid.
The cells transitioned into the stationary phase at 29 h (T5) and exhibited the greatest change in up- or down-expressed genes between successive times (T4 and T5). Between 35 and 51 h, 197 ± 7 and 105 ± 12 genes were up-expressed and down-expressed, respectively, and at the last sampling time (67 h) 250 and 96 genes were up- and down-expressed, respectively (Fig. 3). Temporal samples were not analyzed in recent studies of E. coli and Mycobacterium smegmatis, but similar levels of genes (252 and 137 genes, respectively) were described as up-expressed during the stationary phase (55, 56). Three times were analyzed during growth of Streptococcus pyogenes, and approximately 125 and 160 genes were up- and down-expressed, respectively (7).
In a recent report workers characterized cellular growth using genomic expression profiles for Clostridium acetobutylicum that transitioned from the exponential phase to the stationary phase (4). Similar to our results, ribosomal proteins were down-regulated in response to the transition, and cobalt and iron acquisition systems were up-expressed. In particular, the putative feoAB genes were up-expressed as C. acetobutylicum transitioned into the stationary phase, and the D. vulgaris feo genes exhibited the same trend (Table 2). In the case of D. vulgaris, Fe2+ levels may decline during growth due to the reduced conditions created by H2S production, and the feo system has been shown to be important for the acquisition of ferrous iron under anaerobic conditions in E. coli (25). Recent work with Legionella pneumophila revealed the importance of Fe2+ transport for intracellular replication as a human pathogen (42). The feo system may play a similar role in Fe2+ acquisition for environmental microorganisms under iron-limited conditions. These results suggested that Fe2+ may be an important nutrient when SRB are stimulated for bioremediation; however, further work is needed to determine the role of such systems under in situ conditions.
TABLE 2.
Representative genes that displayed significant up- or down-expression at the transition to and during the stationary phase of growth
| Gene | Putative designation or function | Change in expression ata:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| T2 | T3 | T4 | T5 | T6 | T7 | T8 | T9 | ||
| DVU2572 | feoA | 0.16 | 2.45 | 3.96 | 4.05 | 3.44 | 3.28 | 2.78 | 1.96 |
| DVU2571 | feoB | 0.18 | 2.47 | 4.83 | 2.47 | 1.71 | 1.83 | 1.54 | 0.83 |
| DVU1366 | Lipoprotein | 0.07 | 0.06 | 0.00 | 2.50 | 0.63 | 0.43 | −0.22 | −0.73 |
| DVU2861 | Phage | 0.01 | 0.36 | 0.21 | 2.48 | 3.36 | 2.68 | 2.54 | 2.66 |
| DVU2862 | Phage | −0.21 | 0.06 | 0.03 | 2.15 | 2.95 | 2.74 | 2.28 | 2.11 |
| DVU2869 | Phage | −0.05 | 0.29 | 0.30 | 2.67 | 3.83 | 3.68 | 3.35 | 2.99 |
| DVU1896 | rpsT | −0.62 | 0.00 | −0.72 | −4.26 | −3.86 | −3.75 | −4.15 | −3.22 |
| DVU1574 | rplY | −0.10 | −0.55 | −1.32 | −3.12 | −2.11 | −2.38 | −2.19 | −2.36 |
| DVU1575 | prsA | −0.29 | −0.84 | −1.72 | −3.42 | −1.54 | −1.44 | −1.70 | −1.86 |
| DVU1568 | ftn | 0.18 | 0.45 | 0.47 | −2.25 | −1.55 | −1.90 | −1.88 | −2.13 |
| DVU0019 | ngr | 0.39 | 0.66 | 0.74 | 1.90 | −0.06 | 0.37 | 0.44 | 0.74 |
| DVU2410 | sodB | 0.26 | 0.72 | 1.10 | 2.27 | 1.28 | NAb | 1.33 | 2.71 |
| DVU1858 | csp | 0.03 | 0.51 | 0.69 | 5.45 | 5.76 | 6.36 | 6.57 | 6.66 |
| DVU0938 | Isoamylase | 0.18 | 0.30 | 0.62 | 2.97 | 2.15 | 0.84 | 0.37 | 0.55 |
| DVUA0037 | Sugar transferase | −0.12 | −0.16 | −0.15 | 3.95 | NA | 4.68 | 5.09 | 3.84 |
| DVU1035 | glk | 0.35 | 0.48 | 0.19 | 4.59 | 3.17 | 4.57 | 5.49 | 7.14 |
| DVUA0043 | Polysaccharide deacetylase | −0.10 | NA | NA | 5.02 | 4.20 | 5.31 | 6.07 | NA |
| DVU2285 | Lactate permease | 0.06 | −0.46 | −1.06 | −2.34 | −2.94 | −1.27 | −1.43 | −2.51 |
| DVU0279 | SO4 permease | −0.64 | −0.53 | −1.02 | −2.28 | −2.29 | −2.49 | −1.16 | −1.56 |
| DVU0053 | SO4 permease | 0.32 | 0.38 | 0.70 | 1.49 | 0.92 | 0.26 | 0.15 | 0.51 |
Genes whose change in expression was at least 1.50 or −1.50 with a |z| value of ≥2.0 are indicated by boldface type.
NA, not applicable.
Two of the largest groups of genes for which there were significant changes in expression were ribosomal protein genes and phage-related genes. Some ribosomal protein genes started to exhibit down-expression at 20 h (one-half a generation time before cells entered the stationary phase), and these results suggested that actively growing cells were down-expressing some ribosomal proteins in response to the decrease in the level of the carbon and energy source. Interestingly, the levels of 16 ribosomal protein genes drastically decreased during the transition, but the levels rebounded to almost pre-stationary-phase levels by 35 h. This result suggested that even “resting” populations in the mid-stationary phase can have similar levels of ribosomal proteins. Other microorganisms (Helicobacter, Streptomyces, and Clostridium) have been shown to down-express ribosomal protein genes during the transition to later growth stages (4, 24, 51), but Chlamydia strains do not (36).
For many phage-related genes there was a dramatic increase in expression during the transition and in the stationary phase in D. vulgaris. In a recent study of S. pyogenes, phage-related genes were not significantly up-expressed as cells entered the stationary phase, and many genes were down-expressed (7). Recently, the lytic cycle of the Mu phage in E. coli was shown to be derepressed via the action of ClpXP and Lon proteases on the Rep repressor, and ClpXP and the Lon protease were up-expressed during the transition to the stationary phase in E. coli (41). The data indicated that phage-related genes were up-expressed in response to a decrease in the level of the carbon and energy source and/or in response to a decreased growth rate of the host cells. We inferred from these results that phage may play an important role in the ecophysiology of Desulfovibrio under nutrient-limited conditions. Interestingly, even though phage-related genes were up-expressed, 60% of the biomass remained after approximately 40 h in the stationary phase, and these results suggested that the phage did not cause complete lysis of the entire population.
Different trends of expression were observed for the different lactate and sulfate permeases as the cells transitioned between the growth phases. Based upon current annotation, we deduced that different permeases may be used in response to changing nutrient levels. For example, DVU3284 might be a low-Km lactate transporter that is up-expressed as the lactate levels decline, and DVU0053 and DVU0279 may be low-capacity, high-affinity and high-capacity, low-affinity sulfate transporters, respectively. An alternative explanation is that the sulfate permeases are regulated in a growth rate-dependent manner. A growth rate-dependent mechanism of regulation for the sulfate permeases could be explained by the fact that sulfate can be relatively abundant in most environments compared to electron donors and is the preferred electron acceptor for SRB. A major check point for growth control could be the presence and level of the electron donor and not necessarily the electron acceptor per se. Further work is needed to determine the individual roles of the different transporters and the environmental conditions under which each transporter is important.
The results indicated that a subset of approximately 110 genes were uniquely up-expressed as the cells transitioned to the stationary phase. The rest of the genes that were up-expressed during the stationary phase were also up-expressed with other stresses (i.e., NO2, NaCl, pH, and heat) (unpublished results). During the experiments described here, the majority of the pH change occurred during exponential growth; however, for a few genes there were significant changes in expression throughout exponential growth compared to the first time examined. In addition, the cluster analysis indicated that whole-genome expression profiles changed despite little change in the pH during the stationary phase. For particular hypothetical and conserved hypothetical proteins there was a trend of down-expression that was similar for the stationary-phase response and other stressors, but the exact role of the presumptive gene products is unknown.
The subset of up-expressed genes unique to the stationary phase was mainly involved with DNA repair, nucleic acid metabolism, amino acid metabolism, and carbohydrate metabolism. Fourteen of the unique stationary-phase genes that were up-expressed (13% of the total) were located on the megaplasmid. In a recent study of C. acetobutylicum, it was found that expression of the megaplasmid (pSOL1) genes increased at the onset of the stationary phase (4). Based on our preliminary results with D. vulgaris, a megaplasmid-free strain has different growth characteristics in the stationary phase and does not form significant biofilms compared to wild-type cells with the megaplasmid (Clark and Fields, unpublished results).
It should be noted that up to 17 genes on the megaplasmid (11% of megaplasmid ORFs) are predicted to play a role in carbohydrate metabolism, and the total carbohydrate levels decreased approximately twofold when the cells transitioned to the stationary phase. During the transition, the putative isoamylase and glucan phosphorylase genes were up-expressed, and the genes encoding a putative glucan synthase and glucan branching enzyme were down-expressed. In addition, the one putative fbp (fructose-1,6-bisphosphatase) gene of D. vulgaris was down-expressed, and this result suggested that there was a carbon flow in glycolysis from glucose toward pyruvate and not gluconeogenesis. Interestingly, the D. vulgaris genome has a putative fructose-2,6-biphosphatase (DVU3147), which is a unique characteristic for a bacterium. For DVU3147 there was an upward trend in expression during the transition, and this gene is located immediately upstream from a putative glucotransferase gene in both D. vulgaris and Desulfovibrio strain G20. The fructose-2,6-biphosphatase is thought to be important in the regulation of hepatic carbohydrate metabolism in metazoans (22), but it can have other functions in different organisms. The possible role or function in Desulfovibrio is not known.
In previously studied bacteria, the transcriptional regulation of gene expression in the stationary phase is primarily carried out by σS. Expression of σS is regulated at the level of transcription, translation, and protein degradation (27), and RpoS regulons have recently been delineated using global transcriptomics in E. coli, Pseudomonas fluorescens Pf-5, and M. smegmatis (48, 55, 56). It should be noted that D. vulgaris does not have an annotated rpoS gene, nor do other δ-Proteobacteria, including Desulfovibrio desulfuricans G20, Geobacter sulfurreducens PCA, Geobacter metallireducens, Bdellovibrio bacteriovorus HD100, Desulfotalea psychrophila, and Desulfuromonas acetoxidans. The D. vulgaris genome has four putative Rpo factor genes (rpoD, rpoH, rpoN, and fliA), but the expression of these genes did not change significantly during the growth experiment. These results indicated that D. vulgaris cells transitioned to the stationary phase after depletion of a carbon and energy source without a typical σS factor and suggested that other factors most likely control gene expression during growth transitions.
When 101 genera with sequenced genomes were checked for the presence of a putative rpoS gene, only 21 had a predicted rpoS factor, and these taxa included members of the following divisions: α-, β-, and γ-Proteobacteria, Bacteroidetes, Planctomycetes, and Spirochaetes. These data suggested that RpoS is not a universal transcriptional factor for bacteria, and there could be alternative mechanisms for the regulation of growth transitions in different microorganisms. It is feasible that D. vulgaris has a factor that functions as an RpoS but does not exhibit significant sequence similarity to previously reported sigma factors. Further work is needed to determine the control factor(s) in D. vulgaris involved in sensing environmental stimuli and what cellular systems are required for cells to survive the stationary phase. Elucidation of growth phase-dependent gene expression is essential for a general understanding of growth physiology, which is crucial for interpretation of data for stress-responsive genes. In addition, to effectively immobilize heavy metals and radionuclides via sulfate reduction, it is important to understand the cellular responses to adverse factors observed in contaminated subsurface environments, such as the changing ratios of electron donors and acceptors. Our results indicated that in addition to expected changes (e.g., energy conversion, protein turnover, translation, transcription, and DNA replication and repair) genes related to phage, carbohydrate flux, the outer envelope, and iron homeostasis played a major role in the cellular response to nutrient deprivation under the growth conditions tested.
Supplementary Material
Acknowledgments
This research was supported by the United States Department of Energy under the Environmental Remediation Sciences Program (DOE-ER63765) and by the Virtual Institute for Microbial Stress and Survival (http://VIMSS.lbl.gov) supported by the Office of Science, Genomics Program:GTL, through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U.S. Department of Energy. Oak Ridge National Laboratory is managed by University of Tennessee-Battelle LLC for the Department of Energy under contract DE-AC05-00OR22725.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abdelouas, A., Y. M. Lu, W. Lutze, and H. E. Nuttall. 1998. Reduction of U(VI) to U(IV) by indigenous bacteria in contaminated ground water. J. Contam. Hydrol. 35:217-233. [Google Scholar]
- 2.Abdelouas, A., W. Lutze, and H. E. Nuttall. 1999. Oxidative dissolution of uraninite precipitated on Navajo sandstone. J. Contam. Hydrol. 36:353-375. [Google Scholar]
- 3.Abdelouas, A., W. Lutze, W. Gong, E. H. Nuttall, B. A. Strietelmeier, and B. J. Travis. 2000. Biological reduction of uranium in groundwater and subsurface soil. Sci. Total Environ. 250:21-35. [DOI] [PubMed] [Google Scholar]
- 4.Alsaker, K. V., and E. T. Papoutsakis. 2005. Transcriptional program of early sporulation and stationary-phase event in Clostridium acetobutylicum. J. Bacteriol. 187:7103-7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beliaev, A. S., D. K. Thompson, M. W. Fields, L. Wu, D. P. Lies, K. H. Nealson, and J. Zhou. 2002. Microarray transcription profiling of a Shewanella oneidensis etrA mutant. J. Bacteriol. 184:4612-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beyenal, H., and Z. Lewandowski. 2004. Dynamics of lead immobilization in sulfate reducing biofilms. Water Res. 38:2726-2736. [DOI] [PubMed] [Google Scholar]
- 7.Beyer-Sehlmeyera, G., B. Kreikemeyera, A. Horsterb, and A. Podbielskia. 2005. Analysis of the growth phase-associated transcriptome of Streptococcus pyogenes. Int. J. Med. Microbiol. 295:161-177. [DOI] [PubMed] [Google Scholar]
- 8.Bockkareva, E. S., A. S. Girshovich, and E. Bibi. 2002. Identification and characterization of the Escherichia coli protein UP12, a putative in vivo substrate of GroEL. Eur. J. Biochem. 269:3032-3040. [DOI] [PubMed] [Google Scholar]
- 9.Brandis, A., and R. K. Thauer. 1981. Growth of Desulfovibrio species on hydrogen and sulfate as sole energy source. J. Gen. Microbiol. 126:249-252. [Google Scholar]
- 10.Chaplin, M. F. 1986. Monosaccharides, p. 1-2. In M. F. Chaplin and J. F. Kennedy (ed.), Carbohydrate analysis. IRL Press, Oxford, United Kingdom.
- 11.Chhabra, S. R., Q. He, K. H. Huang, S. P. Gaucher, E. J. Alm, Z. He, M. Z. Hadi, T. C. Hazen, J. D. Wall, J. Zhou, A. P. Arkin, and A. K. Singh. 2006. Global analysis of heat shock response in Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 188:1817-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Darwin, A. J. 2005. The phage-shock-protein response. Mol. Microbiol. 57:621-628. [DOI] [PubMed] [Google Scholar]
- 13.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao, H., Y. Wang, X. Liu, T. Yan, L. Wu, E. J. Alm, et al. 2004. Global transcriptome analysis of the heat shock response of Shewanella oneidensis. J. Bacteriol. 186:7796-7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorby, Y. A., and D. R. Lovley. 1992. Enzymatic uranium precipitation. Environ. Sci. Technol. 26:205-207. [Google Scholar]
- 16.Hamilton, W. A., and W. Lee. 1995. Biocorrosion, p. 243-264. In L. L. Barton (ed.), Sulfate-reducing bacteria. Plenum Press, New York NY.
- 17.Hazen, T. C., and H. H. Tabak. 2005. Developments in bioremediation of soils and sediments polluted with metals and radionuclides. 2. Field research on bioremediation of metals and radionuclides. Rev. Environ. Sci. Biotechnol. 4:157-183. [Google Scholar]
- 18.He, Q., K. H. Huang, Z. He, E. J. Alm, M. W. Fields, T. C. Hazen, A. P. Arkin, J. D. Wall, and J. Zhou. 2006. Energetic consequences of nitrite stress in Desulfovibrio vulgaris Hildenborough, inferred from global transcriptional analysis. Appl. Environ. Microbiol. 72:4370-4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He, Z., L. Wu, X. Li, M. W. Fields, and J. Zhou. 2005. Empirical establishment of oligonucleotide probe design criteria. Appl. Environ Microbiol. 71:3753-3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He, Z., L. Wu, M. W. Fields, and J. Zhou. 2005. Comparison of microarrays with different probe sizes for monitoring gene expression. Appl. Environ. Microbiol. 71:5154-5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heidelberg, J. F., R. Seshadri, S. A. Haveman, C. L. Hemme, I. T. Paulsen, J. F. Kolonay, et al. 2004. The genome sequence of the anaerobic, sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Nat. Biotechnol. 22:554-559. [DOI] [PubMed] [Google Scholar]
- 22.Heine-Suner, D., M. A. Diaz-Guillen, A. J. Lange, and S. Rodriguez de Cordoba. 1998. Sequence and structure of the human 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase heart isoform gene (PFKFB2). Eur. J. Biochem. 254:103-110. [DOI] [PubMed] [Google Scholar]
- 23.Hengge-Aronis, R. 2002. Signal transduction and regulatory mechanisms involved in control of the σS subunit of RNA polymerase in Escherichia coli. Microbiol. Mol. Biol. Rev. 66:373-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang, J., C. J. Lih, K. H. Pan, and S. N. Cohen. 2001. Global analysis of growth phase responsive gene expression and regulation of antibiotic biosynthetic pathways in Streptomyces coelicolor using DNA microarrays. Genes Dev. 15:3183-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kammler, M., C. Schon, and K. Hantke. 1993. Characterization of the ferrous iron uptake system of Escherichia coli. J. Bacteriol. 175:6212-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karzai, A. W., E. D. Roche, and R. T. Sauer. 2000. The SsrA-SmpB system for protein tagging, directed degradation, and ribosome rescue. Nat. Struct. Biol. 7:449-455. [DOI] [PubMed] [Google Scholar]
- 27.Khmel, I. A. 2005. Regulation of expression of bacterial genes in the absence of active cell growth. Russ. J. Genet. 41:968-984. [PubMed] [Google Scholar]
- 28.Kneidinger, B., M. Graninger, G. Adam, M. Puchberger, P. Kosma, S. Zayni, and P. Messner. 2001. Identification of two GDP-6-deoxy-d-lyxo-4-hexulose reductases synthesizing GDP-d-rhamnose in Aneurinibacillus thermoaerophilus L420-91T. J. Biol. Chem. 276:5577-5583. [DOI] [PubMed] [Google Scholar]
- 29.Kvint, K., L. Nachin, A. Diez, and T. Nystrom. 2003. The bacterial universal stress protein: function and regulation. Curr. Opin. Microbiol. 6:140-145. [DOI] [PubMed] [Google Scholar]
- 30.Lawrence, C. E., S. F. Altschul, M. S. Boguski, J. S. Liu, A. F. Neuwald, and J. C. Wootton. 1993. Detecting subtle sequence signals: a Gibbs sampling strategy for multiple alignment. Science 262:208-214. [DOI] [PubMed] [Google Scholar]
- 31.Liu, Y., W. Gao, L. Wu, X. Liu, T. Yan, E. J. Alm, A. P. Arkin, D. K. Thompson, M. W. Fields, and J. Zhou. 2005. Genomic expression profiling of Shewanella oneidensis MR-1 response to sodium salt stress. J. Bacteriol. 187:2501-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lloyd, J. R. 2003. Microbial reduction of metals and radionuclides. FEMS Microbiol. Rev. 27:411-425. [DOI] [PubMed] [Google Scholar]
- 33.Lovley, D. R. 1993. Dissimilatory metal reduction. Annu. Rev. Microbiol. 47:263-290. [DOI] [PubMed] [Google Scholar]
- 34.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 35.Neuwald, A. F., J. S. Liu, and C. E. Lawrence. 1995. Gibbs motif sampling: detection of bacterial outer membrane protein repeats. Protein Sci. 4:1618-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nicholson, T. L., L. Olinger, K. Chong, G. Schoolnik, and R. S. Stephens. 2003. Global stage-specific gene regulation during the developmental cycle of Chlamydia trachomatis. J. Bacteriol. 185:3179-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nielsen, K. K., and M. Boye. 2005. Real-time quantitative reverse transcription-PCR analysis of expression stability of Actinobacillus pleuropneumoniae housekeeping genes during in vitro growth under iron-depleted conditions. Appl. Environ. Microbiol. 71:2949-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ollivier, B., R. Cord-Ruwisch, E. C. Hatchikian, and J. L. Garcia. 1988. Characterization of Desulfovibrio fructosovorans sp. nov. Arch. Microbiol. 150:26-31. [Google Scholar]
- 39.Payne, R. B., L. Casalot, J. A. Ringbauer, Jr., B. Rapp-Giles, and J. D. Wall. 2002. Uranium reduction by cytochrome mutants of Desulfovibrio. Appl. Environ. Microbiol. 68:3129-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Price, M. N., K. H. Huang, E. J. Alm, and A. P. Arkin. 2005. A novel method for accurate operon predictions in all sequenced prokaryotes. Nucleic Acids Res. 33:880-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ranquet, C., A. Toussaint, H. de Jong, G. Maenhaut-Michel, and J. Geiselmann. 2005. Control of bacteriophage Mu lysogenic repression. J. Mol. Biol. 353:186-195. [DOI] [PubMed] [Google Scholar]
- 42.Robey, M., and N. P. Cianciotto. 2002. Legionella pneumophila feoAB promotes ferrous iron uptake and intracellular infection. Infect. Immun. 70:5659-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodionov, D. A., I. Dubchak, A. P. Arkin, E. J. Alm, and M. S. Gelfand. 2004. Reconstruction of regulatory and metabolic pathways in metal-reducing δ-Proteobacteria. Genome Biol. 5:R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Santos, H., P. Fareleira, A. V. Xavier, L. Chen, M. Y. Liu, and J. LeGall. 1993. Aerobic metabolism of carbon reserves by the obligate anaerobe Desulfovibrio gigas. Biochem. Biophys. Res. Commun. 195:551-557. [DOI] [PubMed] [Google Scholar]
- 45.Sayed, A., S. Matsuyama, and M. Inouye. 1999. Era, an essential Escherichia coli small G-protein, binds to the 30S ribosomal subunit. Biochem. Biophys. Res. Commun. 264:51-54. [DOI] [PubMed] [Google Scholar]
- 46.Schultz, J. E., and A. Matin. 1991. Molecular and functional characterization of a carbon starvation gene of Escherichia coli. J. Mol. Biol. 218:129-140. [DOI] [PubMed] [Google Scholar]
- 47.Seo, J., M. Bakay, Y.-W. Chen, S. Hilmer, B. Shneiderman, and E. P. Hoffman. 2004. Interactively optimizing signal-to-noise ratios in expression profiling: project-specific algorithm selection and detection p-value weighting in Affymetrix microarrays. Bioinformatics 20:2534-2544. [DOI] [PubMed] [Google Scholar]
- 48.Stockwell, V. O., and J. E. Loper. 2005. The sigma factor RpoS is required for stress tolerance and environmental fitness of Pseudomonas fluorescens Pf-5. Microbiol. 151:3001-3009. [DOI] [PubMed] [Google Scholar]
- 49.Tani, T. H., A. Khodursky, R. M. Blumenthal, P. O. Brown, and R. G. Matthew. 2002. Adaptation to famine: a family of stationary-phase genes revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 99:13471-13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thompson, D. K., A. S. Beliaev, C. S. Giometti, S. L. Tollaksen, T. Khare, D. P. Lies, et al. 2002. Transcriptional and proteomic analysis of a ferric uptake regulator (fur) mutant of Shewanella oneidensis: possible involvement of fur in energy metabolism, transcriptional regulation, and oxidative stress. Appl. Environ. Microbiol. 68:881-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thompson, L. J., D. S. Merrell, B. A. Neilan, H. Mitchell, A. Lee, and S. Falkow. 2003. Gene expression profiling of Helicobacter pylori reveals a growth-phase-dependent switch in virulence gene expression. Infect. Immun. 71:2643-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thompson, W., E. C. Rouchka, and C. E. Lawrence. 2003. Gibbs Recursive Sampler: finding transcription factor binding sites. Nucleic Acids Res. 31:3580-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Voordouw, G. 1995. The genus Desulfovibrio: the centennial. Appl. Environ. Microbiol. 61:2813-2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wan, X.-F., N. C. VerBerkmoes, L. A. McCue, D. Stanek, H. Connelly, L. J. Hauser, et al. 2004. Transcriptomic and proteomic characterization of the Fur modulon in the metal-reducing bacterium Shewanella oneidensis. J. Bacteriol. 186:8385-8400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang, R., J. T. Prince, and E. M. Marcotte. 2005. Mass spectrometry of the Mycobacterium smegmatis proteome: protein expression levels correlate with function, operons, and codon bias. Genome. Res. 15:1118-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weber, H., T. Polen, J. Heuveling, V. F. Wendisch, and R. Hengge. 2005. Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 187:1591-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.