Abstract
The PCR-based methodology applied to multiple-locus variable numbers of tandem repeat (VNTR) analysis was recently shown to be a useful technique for the molecular typing of clinical isolates of several bacterial species. We have adopted this method for the molecular typing of methicillin-resistant Staphylococcus aureus. Five staphylococcal VNTR loci (sdr, clfA, clfB, ssp, and spa) were subjected to analysis, and it was shown that the method allows typing of S. aureus strains with the discriminatory power and reproducibility of pulsed-field gel electrophoresis while at the same time being rapid and applicable to analysis of large numbers of isolates.
During the past few years, the most remarkable advances in molecular typing were achieved by analysis of variable numbers of tandem repeat (VNTR) loci identified in the genomes of eukaryotic and prokaryotic species during genome sequencing projects. The number of repeat units at the same locus often varies from strain to strain and can be detected by PCR with flanking primers, a technique also commonly used for DNA fingerprinting of eukaryotic and prokaryotic species (1, 5, 7, 9, 11). However, to the best of our knowledge, this method has never been used to type Staphylococcus aureus strains, although several genes with repetitive sequences were analyzed for the purpose of S. aureus typing (16).
The sequencing of the S. aureus genome indicated the presence of several VNTR loci, including sdr, clfA, clfB, ssp, coa, and spa. The sdr locus (10) comprises two or three closely linked and tandemly arrayed open reading frames containing sdrC, sdrD, and sdrE, which encode Sdr proteins. The Sdr proteins, together with clumping factor A (ClfA) (12) and clumping factor B (ClfB) (13), are members of a family of surface proteins which are characterized by the presence of an R region containing various numbers of the repeated Ser-Asp dipeptides encoded by an 18-nucleotide DNA repeat at the 3′ region of the sdr genes. The ssp locus contains a gene (sspA) encoding a serine protease (V8 protease) (14), the C-terminal fragment of which is built of multiple, variable numbers of tripeptide repeats encoded by 9-nucleotide repeating units (2). Finally, coa and spa genes coding for collagen binding protein and protein A, respectively, have various numbers of degenerated repeats of 81 and 24 bp, respectively (15). This polymorphism has been commonly used for differentiating S. aureus isolates (6, 8). In this paper, we describe a multiple-locus VNTR analysis (MLVA) system to discriminate among different S. aureus clinical isolates based on the analysis of five (sdr, clfA, clfB, ssp, and spa) tandem repeat loci composed of seven individual genes. This method of exploring the VNTR polymorphism enables the typing of clinical methicillin-resistant Staphylococcus aureus (MRSA) isolates, determination of their diversity and evolutionary relationships with discriminatory power, and a reproducibility matching the commonly used pulsed-field gel electrophoresis (PFGE) technique.
The strain collection encompassed 34 human MRSA isolates (Table 1). It included (i) five isolates with indistinguishable PFGE patterns (isolates 1 to 5); (ii) two groups of epidemiologically related strains composed of four isolates from hospitals in four different locations in Poland displaying closely related PFGE patterns (isolates 6 to 9, differing by ≤3 DNA fragments) and eight isolates from a single hospital exhibiting similar PFGE patterns which differ by ≤7 DNA fragments (isolates 10 to 17); (iii) seven genetically unrelated isolates from Polish hospitals showing PFGE patterns which differ by ≥8 DNA fragments (isolates 18 to 24); and (iv) ten isolates representing different geographical origins (isolates 25 to 34). The last group includes the reference S. aureus strain NCTC 8325/0.
TABLE 1.
Results of S. aureus clinical isolate typing
| Isolate no. | Origin (country, yr) or control strain | PFGE type | MLVA pattern |
|---|---|---|---|
| 1 | Poland, 1997 | A | 1 |
| 2 | Poland, 1997 | A | 1 |
| 3 | Poland, 1997 | A | 1 |
| 4 | Poland, 1997 | A | 1 |
| 5 | Poland, 1997 | A | 1 |
| 6 | Poland, 1992 | B1 | 2 |
| 7 | Poland, 1994 | B2 | 3 |
| 8 | Poland, 1992 | B3 | 4 |
| 9 | Poland, 1992 | B4 | 5 |
| 10 | Poland, 1992 | C1 | 6 |
| 11 | Poland, 1992 | C2 | 7 |
| 12 | Poland, 1992 | C3 | 7 |
| 13 | Poland, 1992 | C4 | 8 |
| 14 | Poland, 1992 | C5 | 7 |
| 15 | Poland, 1992 | C6 | 9 |
| 16 | Poland, 1992 | C7 | 7 |
| 17 | Poland, 1992 | C8 | 7 |
| 18 | Poland, 1992 | D | 10 |
| 19 | Poland, 1994 | E | 11 |
| 20 | Poland, 1994 | F | 12 |
| 21 | Poland, 1992 | G | 13 |
| 22 | Poland, 1996 | H | 14 |
| 23 | Poland, 1994 | I | 15 |
| 24 | Poland, 1996 | J | 16 |
| 25 | Turkey, 1996 | K | 17 |
| 26 | Slovenia, 1996 | L | 18 |
| 27 | Russia, 1998 | M | 19 |
| 28 | Lithuania, 1998 | N | 20 |
| 29 | Slovenia, 1998 | O | 21 |
| 30 | Bulgaria, 1998 | P | 22 |
| 31 | Bulgaria, 1998 | R | 23 |
| 32 | Czech Republic, 1996 | S | 24 |
| 33 | England, 1992 | T | 25 |
| 34 | 8325/0 | U | 26 |
Preparation of genomic DNA and PFGE were performed as previously described (3). The PFGE patterns with the Dice coefficient of ≥0.77 (corresponding to ≤7 DNA fragment differences between patterns) were assigned to the same type. Total DNA for MLVA was prepared with five to ten colonies lifted from tryptic soy agar plates incubated for 20 h at 37°C and suspended in 50 μl of lysis buffer (0.006 M Tris-HCl [pH 7.6], 1 M NaCl, 0.1 M Na2EDTA, 0.5% Polyoxyethylene 20 cetyl ether [Brij 58], 1% N-Lauroylsarcosine sodium salt). Lysostaphin and RNase A were added to concentrations of 50 and 60 μg/ml, respectively, and the mixture was incubated for 30 min at 37°C. Isolation of total DNA was then performed by using the Genomic DNA Prep Plus kit (A&A Biotechnology, Gdynia, Poland). Briefly, proteinase K was added to a concentration of 1 mg/ml, and the mixture incubated for 0.5 h at 37°C. After vigorous vortexing, the total solution was transferred to a spin minicolumn, and DNA was adsorbed onto a silica membrane during this centrifugation step. The minicolumn was washed twice to remove any residual contaminants, and the bound DNA was eluted with 500 μl of preheated (75°C) elution buffer (10 mM Tris-HCl [pH 8.5]). The purified DNA was diluted 1:10 with sterile water and then kept at 20°C until PCR.
A set of PCR primers (Table 2) to simultaneously amplify the hypervariable VNTR regions of the spa, sspA, clfA, clfB, sdrC, sdrD, and sdrE genes was designed based on the genome sequence of six strains available through the S. aureus genome sequence projects (Table 2). A similarity in the DNA sequence flanking the SD repeats (region R) of the sdr genes allowed selection of single pairs of primers for amplification of all three individual genes in the sdr locus. The ClustalW software was used to align appropriate gene fragments necessary for the generic primer design. The PCR mix contained 1× PCR buffer, 1.5 mM MgCl2, deoxynucleoside triphosphate mix (dNTPs; 0.2 mM each), 0.05 unit of Taq recombinant polymerase (MBI Fermentas, Vilnius, Lithuania) per μl, 0.5 μM each ClfB-F and ClfB-R, 1 μM each ClfA-F, ClfA-R, SdrCDE-F, SdrCDE-R, Spa-F, Spa-R, SspA-F, and SspA-R, and 1 μl of template DNA, all in a final volume of 20 μl. Amplification of the DNA fragments was performed with predenaturation at 94°C for 5 min followed by 20 cycles of 30 s at 94°C, 30 s at 55°C, 30 s at 72°C, with a final extension at 72°C for 5 min. Five microliters of PCR products was electrophoresed in 2% agarose Micropor GAMMA (Prona) gels in the presence of ethidium bromide in 0.5× TBE (Tris-borate-EDTA) (ICN, Aurora, Ohio, or Harlan Scientific, Indianapolis, Ind.) buffer for 3.5 h at 2.5 V/cm. As a DNA size standard, a 50-bp DNA ladder (Amersham Pharmacia Biotech, Little Chalfont, Buckinghamshire, England) was included in each run. The DNA bands were visualized on an UV transilluminator, photographed, and scanned, and the RFLP patterns then were both visually evaluated and analyzed as TIFF files by using Quantity One software (version 4.2 for Windows and Macintosh; Bio-Rad, Munich, Germany). Any two MLVA patterns differing by one or more bands were considered distinct types.
TABLE 2.
Primers used in this studya
| Marker locus | Primer designation | Primer sequence |
|---|---|---|
| clfA | ClfA-F | 5′-GATTCTGACCCAGGTTCAGA |
| ClfA-R | 5′-CTGTATCTGGTAATGGTTCTTT | |
| clfB | ClfB-F | 5′-ATGGTGATTCAGCAGTAAATCC |
| ClfB-R | 5′-CATTATTTGGTGGTGTAACTCTT | |
| sdr | SdrCDE-F | 5′-GTAACAATTACGGATCATGATG |
| SdrCDE-R | 5′-TACCTGTTTCTGGTAATGCTTT | |
| spa | Spa-F | 5′-AGCACCAAAAGAGGAAGACAA |
| Spa-R | 5′-GTTTAACGACATGTACTCCGT | |
| ssp | SspA-F | 5′-ATCMATTTYGCMAAYGATGACCA |
| SspA-R | 5′-TTGTCTGAATTATTGTTATCGCC |
The primers were designed based on the completed and unfinished S. aureus genomes available from The Institute for Genomic Research (http://www.tigr.org) (COL strain), the University of Oklahoma (http://www.genome.ou.edu) (strain 8325), Sanger Institute (http://www.sanger.ac.uk) (strains MRSA252 and MSSA476), and Juntendo University (http://www.juntendo.ac.jp) (strains Mu50 and N315).
All of the strains in the collection were typeable with extraordinary reproducibility regardless of the genomic DNA preparations.
Analysis of reliability of the MLVA fingerprinting was further performed on isolate 9 (Table 1), as it appeared that all amplicons generated for the individual locus were present in the MLVA profile (data not shown). Apparently, however, due to some deviation in electrophoretic migration of a single DNA fragment in comparison to multiple fragments, the amplicons slightly mismatch.
The in vivo stability of the MLVA patterns was determined with nine S. aureus isolates obtained during a 6-week period from specimens collected at different sites from a single MRSA carrier. Despite the fact that the PFGE pattern of these isolates showed occasional differences of up to 2 fragments, the MLVA analysis yielded an identical fingerprint in all nine isolates (data not shown). The in vivo stability of the MLVA analysis was further confirmed with a pair of isolates with indistinguishable PFGE patterns, collected 3 months apart from the throat of a single MRSA carrier. In parallel with the identical PFGE patterns, the MLVA fingerprinting also yielded indistinguishable results (data not shown). To test the in vitro stability of the MLVA patterns, four epidemiologically unrelated S. aureus strains were subjected to 20 rounds of passage on sheep blood agar plates. Total DNA extracted from the first, tenth, and last cultures and after analysis using the MLVA system yielded precisely identical band patterns. Taken together, these data clearly indicate the high stability of the MLVA patterns for S. aureus strains maintained both in vitro and in vivo.
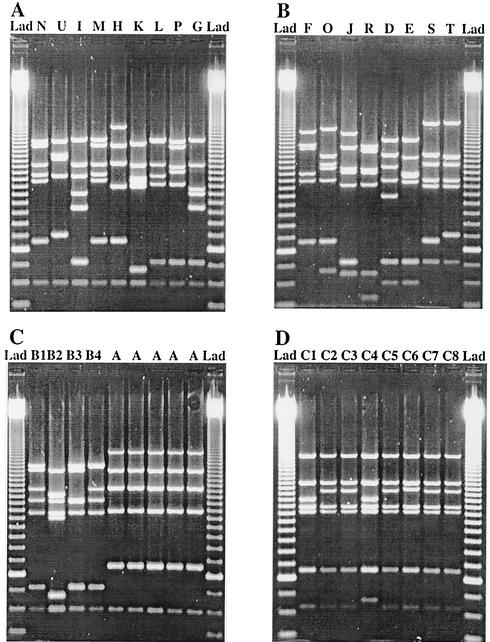
A comparison of the MLVA patterns for every isolate with the PFGE results is summarized in Table 1. Isolates 1 to 5, with indistinguishable PFGE patterns, also showed identical MLVA patterns (Fig. 1C). Isolates 6 to 9 (Fig. 1C) and 10 to 17 (Fig. 1D), representing genetically related strains according to PFGE typing, displayed MLVA patterns that differed in 2 to 8 bands and in no more than 4 bands, respectively. Unrelated isolates 18 to 34 (Fig. 1A and B) gave MLVA patterns that differed by between 2 and 14 bands.
FIG. 1.
MLVA patterns of isolates 18 to 34 (A and B), isolates 6 to 9 and 1 to 5 (C), and isolates 10 to 17 (D), indicated by PFGE types above the lanes (see Table 1). The order of isolates is concordant with the genetic relationship of isolates obtained by PFGE analysis (data not shown). Lanes Lad, 50-bp ladder; bottom band, 100 bp.
Initially, the analysis of all six loci (sdr, clfA, clfB, ssp, coa, and spa) was employed, but it was noticed that the molecular clock of the coagulase locus runs much more slowly than that of other loci reported here, and this locus shows little variation among unrelated strains (data not shown). Therefore, to simplify the analyzed band pattern, the coa locus analysis was withdrawn from MLVA. This, however, did not compromise the differentiation power of MLVA.
The differentiation power is one of the most important parameters of any bacteria typing method. It is defined as the ability to clearly differentiate among unrelated isolates but at the same time to demonstrate the relationship of all organisms isolated from individuals infected through the same source. This criterion is fulfilled by MLVA as a typing system, since all 17 unique, geographically distinct strains were clearly discriminated one from another. On the other hand, epidemiologically linked, and therefore genetically related, S. aureus isolates showed little variation of repeat units. Moreover, the MLVA method allows the performance of an analysis of strains based on a simple, easy-to-interpret pattern of bands on the gel. In the case of other DNA-based typing methods of comparably high differentiation power, these patterns are far more complex.
Any method applied to classification and/or subtyping of bacterial strains must enable the typing of all organisms within a species. In the case of S. aureus, the major drawback of phage typing is the fact that some strains lack a bacteriophage receptor and remain untypeable, because they fail to become infected with the typing phage. Also, the DNA-based typing methods relying on DNA fragment amplification fail to type some strains due to differences in the DNA sequence to which primers anneal. In the case of MLVA, however, this is unlikely, since the probability that all seven primer-annealing regions of DNA are mutated is extremely low. Accordingly, we have applied the MLVA method to 176 strains, and all were shown to be typeable, since the primers designed in this study allowed amplification from the total DNA of every locus (sdr, clfA, clfB, ssp, and spa) from all tested isolates of S. aureus (data not shown).
As with any other typing method, MLVA requires a specific setup. The hardware includes a thermocycler, gel box, low-voltage power supplies, and a photographic system, which constitute the standard equipment of each molecular biology laboratory. Other supplies required are PCR amplification reagents and restriction enzymes. Together, these minimal requirements ensure low setup and operation costs for the MLVA-based S. aureus typing method. The procedure, however, requires appropriate standardization of the PCR, which must be performed individually with each pair of primers on five to six nonrelated S. aureus isolates, as well as the commonly available laboratory reference strain 8325/0, and then compared with the results of multiplex DNA amplification. Once the multiplex PCR yields identification of all individual amplicons, the method is set.
The simultaneous analysis of the variation in number of repeat units in seven individual genes endows MLVA with discriminatory power and reproducibility comparable to those of both PFGE and randomly amplified polymorphic DNA. At the same time, the method described here is simple, inexpensive, easy to interpret, and sensitive, and it should be reliable for the comparison of data obtained in different laboratories. Moreover, based on the multiplex PCR primers tagged with different fluorescent dyes, an automated genotyping can be easily developed. This adaptation may annihilate inherent difficulty in the comparison of results from different laboratories obtained by methods relying on a gel separation of DNA fragments (4).
In summary, the MLVA system, as described here, seems to be at the very least comparable to any other high-discriminatory-power DNA-based or classical method applied currently to S. aureus typing. In this paper, only 34 isolates were analyzed, but these isolates were carefully selected to compare directly the discriminatory powers of MLVA and PFGE. Clearly, the data obtained with the MLVA system fulfills all the criteria of a broadly useful typing method, and it can be adapted to large-scale epidemiological studies of staphylococcal infections. Moreover, the remarkable sensitivity of the method may be implemented to develop S. aureus typing directly in clinical specimens. However, the method needs further validation on a larger number of S. aureus strain collections.
Acknowledgments
These studies were supported by grant 6P04A 08320 from Committee of Scientific Research (KBN, Warsaw, Poland) (to A.S. and K.K.).
REFERENCES
- 1.Adair, D. M., P. L. Worsham, K. K. Hill, A. M. Klevytska, P. J. Jackson, A. M. Friedlander, and P. Keim. 2000. Diversity in a variable-number tandem repeat from Yersinia pestis. J. Clin. Microbiol. 38:1516-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carmona, C., and G. L. Gray. 1987. Nucleotide sequence of the serine protease gene of Staphylococcus aureus, strain V8. Nucleic Acids Res. 15:6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Lencastre, H., I. Couto, I. Santos, J. Melo-Cristino, A. Torres-Pereira, and A. Tomasz. 1994. Methicillin resistance Staphylococcus aureus disease in a Portuguese hospital: characterization of clonal types by a combination of DNA typing methods. Eur. J. Clin. Microbiol. Infect. Dis. 13:64-73. [DOI] [PubMed] [Google Scholar]
- 4.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farlow, J., K. L. Smith, J. Wong, M. Abrams, M. Lytle, and P. Keim. 2001. Francisella tularensis strain typing using multiple-locus, variable-number tandem repeat analysis. J. Clin. Microbiol. 39:3186-3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frenay, H. M., J. P. Theelen, L. M. Schouls, C. M. Vandenbroucke-Grauls, J. Verhoef, W. J. van Leeuwen, and F. R. Mooi. 1994. Discrimination of epidemic and nonepidemic methicillin-resistant Staphylococcus aureus strains on the basis of protein A gene polymorphism. J. Clin. Microbiol. 32:846-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frothingham, R., and W. A. Meeker-O'Connell. 1998. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology 144:1189-1196. [DOI] [PubMed] [Google Scholar]
- 8.Goh, S. H., S. K. Byrne, J. L. Zhang, and A. W. Chow. 1992. Molecular typing of Staphylococcus aureus on the basis of coagulase gene polymorphisms. J. Clin. Microbiol. 30:1642-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeffreys, A. J., V. Wilson, and S. L. Thein. 1992. Hypervariable ‘minisatellite’ regions in human DNA. Bio/Technology 24:467-472. [PubMed] [Google Scholar]
- 10.Josefsson, E., K. W. McCrea, D. Ni Eidhin, D. O'Connell, J. Cox, M. Hook, and T. J. Foster. 1998. Three new members of the serine-aspartate repeat protein multigene family of Staphylococcus aureus. Microbiology 144:3387-3395. [DOI] [PubMed] [Google Scholar]
- 11.Keim, P., L. B. Price, A. M. Klevytska, K. L. Smith, J. M. Schupp, R. Okinaka, P. J. Jackson, and M. E. Hugh-Jones. 2000. Multiple-locus variable-number tandem repeat analysis reveals genetic relationships within Bacillus anthracis. J. Bacteriol. 182:2928-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McDevitt, D., P. Francois, P. Vaudaux, and T. J. Foster. 1994. Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol. Microbiol. 11:237-248. [DOI] [PubMed] [Google Scholar]
- 13.Ni Eidhin, D., S. Perkins, P. Francois, P. Vaudaux, M. Hook, and T. J. Foster. 1998. Clumping factor B (ClfB), a new surface-located fibrinogen-binding adhesin of Staphylococcus aureus. Mol. Microbiol. 30:245-257. [DOI] [PubMed] [Google Scholar]
- 14.Rice, K., R. Peralta, D. Bast, J. de Azavedo, and M. J. McGavin. 2001. Description of staphylococcus serine protease (ssp) operon in Staphylococcus aureus and nonpolar inactivation of sspA-encoded serine protease. Infect. Immun. 69:159-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Belkum, A., S. Scherer, L. van Alphen, and H. Verbrugh. 1998. Short-sequence DNA repeats in prokaryotic genomes. Microbiol. Mol. Biol. Rev. 62:275-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wichelhaus, T. A., K. P. Hunfeld, B. Boddinghaus, P. Kraiczy, V. Schafer, and V. Brade. 2001. Rapid molecular typing of methicillin-resistant Staphylococcus aureus by PCR-RFLP. Infect. Control. Hosp. Epidemiol. 22:294-298. [DOI] [PubMed] [Google Scholar]