Abstract
Many microorganisms produce surface-active substances that enhance the availability of water-insoluble substrates. Although many of these biosurfactants have interesting potential applications, very little is known about their biosynthesis. The basidiomycetous fungus Ustilago maydis secretes large amounts of mannosylerythritol lipids (MELs) under conditions of nitrogen starvation. We recently described a putative glycosyltransferase, Emt1, which is essential for MEL biosynthesis and whose expression is strongly induced by nitrogen limitation. We used DNA microarray analysis to identify additional genes involved in MEL biosynthesis. Here we show that emt1 is part of a gene cluster which comprises five open reading frames. Three of the newly identified proteins, Mac1, Mac2, and Mat1, contain short sequence motifs characteristic for acyl- and acetyltransferases. Mutational analysis revealed that Mac1 and Mac2 are essential for MEL production, which suggests that they are involved in the acylation of mannosylerythritol. Deletion of mat1 resulted in the secretion of completely deacetylated MELs, as determined by mass spectrometry. We overexpressed Mat1 in Escherichia coli and demonstrated that this enzyme acts as an acetyl coenzyme A-dependent acetyltransferase. Remarkably, Mat1 displays relaxed regioselectivity and is able to acetylate mannosylerythritol at both the C-4 and C-6 hydroxyl groups. Based on these results, we propose a biosynthesis pathway for the generation of mannosylerythritol lipids in U. maydis.
Many microorganisms produce extracellular amphipathic compounds that act as biosurfactants. This structurally diverse group of surface-active molecules has diverse functions in microbial physiology. In general, they increase the availability of hydrophobic nutrients and enhance attachment to nonpolar surfaces. In addition to these more general functions, some biosurfactants also bind heavy metals, display antimicrobial activity, or play an important role in pathogenic development or biofilm formation (28). The chemical properties and the biological activity of these natural surfactants have prompted widespread application in the food and chemical industries but also in environmental protection and as biocontrol agents (2).
The basidiomycetous fungus Ustilago maydis is known to produce large amounts of extracellular glycolipids (1, 6, 23, 32). U. maydis is unique among fungal producers of biosurfactants because it produces two structurally different classes of glycolipids. Ustilagic acid is a cellobiose lipid in which the disaccharide is O-glycosidically linked to the ω-hydroxyl group of 2,15,16-trihydroxy- or 15,16-dihydroxyhexadecanoic acid (22). In addition to ustilagic acid, which displays antibiotic activity (9), U. maydis secretes an extracellular oil which is heavier than water and consists of mannosylerythritol lipids (MELs) (1, 6, 21). MELs consist of a mannosylerythritol disaccharide which is acylated with short-chain (C2 to C8) and medium-chain (C10 to C18) fatty acids at the mannosyl moiety (Fig. 1A). Depending on the number of acetyl groups, mannosylerythritol lipids can be differentiated into MEL A (fully acetylated), MEL B and MEL C (monoacetylated at R-6 and R-4, respectively), and the fully deacetylated MEL D (18). Mannosylerythritol-containing lipids have been identified in a variety of fungal species, e.g., Schizonella melanogramma, Candida antarctica, Kurtzmanomyces sp., and Pseudozyma aphidis (5, 15, 18, 27). Besides their high surface activity, mannosylerythritol lipids display interesting biological activities. MELs induce neuronal differentiation in mammalian PC12 cells (35) and have been identified in a screen for inhibitors of dopamine receptors (21). In addition, mannosylerythritol lipids exhibit high affinity to immunoglobulins (12) and increase significantly the transfection efficiency of liposomes (14). Thus, MELs belong to the most interesting candidates of glycolipid biosurfactants for biotechnological applications (19).
FIG. 1.
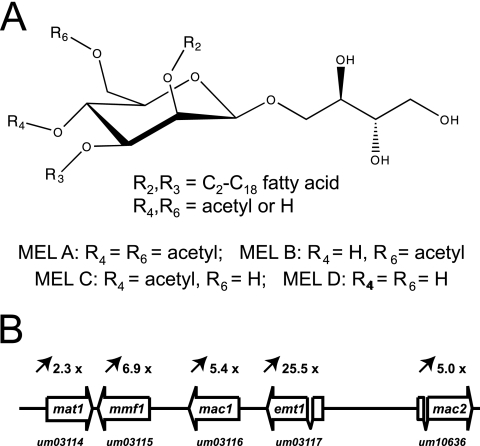
Molecular structure of MELs and organization of the MEL biosynthesis gene cluster. (A) MELs consist of the disaccharide mannosylerythritol, which is esterified with short-chain (C2 to C8) and medium-chain (C10 to C18) fatty acids at positions R-2 and R-3. Depending on the degree of acetylation at positions R-4 and R-6, MELs are differentiated into MEL A, MEL B, MEL C, and MEL D. (B) The complete MEL biosynthesis gene cluster comprises the mat1 acetyltransferase gene, the mmf1 gene, which specifies a member of the major facilitator family, mac1 and mac2, encoding putative acyltransferases, and the previously identified glycosyltransferase gene emt1. Induction of transcription under conditions of nitrogen limitation is indicated above the open reading frames. emt1 and mac2 each contain a single intron. The MUMDB entry numbers are shown.
Here we describe the identification of a gene cluster for mannosylerythritol lipid biosynthesis which consists of five open reading frames. Mutational analysis and in vitro enzyme assays allow prediction of the biosynthesis pathway leading to the production of mannosylerythritol lipids.
MATERIALS AND METHODS
Strains, plasmids, and culture conditions.
U. maydis strains FB1, MB215 and MB215Δcyp1 have been described previously (10). U. maydis strains were grown at 28°C in liquid yeast extract-peptone-sucrose (1% yeast extract, 2% peptone, 2% sucrose) or on solid potato dextrose agar. Solid medium contained 1.5% (wt/vol) Bacto agar. For selection of transformants, potato dextrose plates containing 200 μg/ml hygromycin were used. To induce glycolipid production, strains were grown at 28°C in liquid yeast extract-peptone-sucrose medium to exponential phase and then shifted to nitrogen starvation medium containing 1.7 g/liter yeast nitrogen base without ammonium sulfate and 5% glucose as the carbon source. Glycolipids were isolated after cells were cultivated for 4 days at 28°C on the rotary shaker as described previously (10). Transformation of U. maydis was performed as described previously (30).
Escherichia coli strain DH5α was used for all DNA manipulations. Strain BL21(DE3) was used for overexpression of the acetyltransferase Um03114 (Mat1).
Generation and characterization of mutants defective for glycolipid production.
Mutants were generated by a PCR-based method (16). One-kilobase flanking regions of the candidate genes were amplified by PCR (left flank, primers L1 and L2; right flank, primers R1 and R2). The primers L2 and R1 each contain a characteristic SfiI site as indicated by italic letters in the list of primers below. After SfiI digestion, PCR fragments were ligated to a hygromycin resistance cassette with compatible SfiI ends and directly transformed into U. maydis.
Primers.
The following primers were used: Um03114-L1 (GCGCATTTGCTCACATGTATCGC), Um03114-L2 (CACGGCCTGAGTGGCCGATCAGCGACAGCTCGATGTGC), Um03114-R1 (GTGGGCCATCTAGGCCCAACCTAAGCAGTCACGTTTCCAG), Um03114-R2 (GTGCAAACCTTTGTGCACGGCG), Um03115-L1 (GAGCTGGCTGCCTGGGCTGCGTC), Um03115-L2 (CACGGCCTGAGTGGCCGTATACGATCAGTGTACGATCCTG), Um03115-R1 (GTGGGCCATCTAGGCCCCAGCCCACTGCGTGCGCAGCG), Um03115-R2 (CTATCTACCACAAGCTAAGGTGG), Um03116-L1 (CACGACGAGTCAAGTTGTGCCG), Um03116-L2 (CACGGCCTGAGTGGCCGCCAAAGAGGTAGATTTGAACC), Um03116-R1 (GTGGGCCATCTAGGCCCGCTGTTGGCACTTTACGTTTG), Um03116-R2 (GCTGTGGCCTGGTTCAACGTCC), Um10636-L1 (CACGAATGTTAGCGCGATCGC), Um10636-L2 (CACGGCCTGAGTGGCCGTTAACCGTGATTTGTGCTACAAC), Um10636-R1 (GTGGGCCATCTAGGCCCACTCTGCGACTGTTTTCTG), and Um10636-R2 (GGAAGTTGGGGACTTTAAGCC).
Total RNA preparation.
Wild-type strain MB215 was grown overnight at 28°C to an optical density at 600 nm of 1.0 in yeast nitrogen base medium containing 5% glucose and 0.2% ammonium sulfate. After centrifugation, the cells were resuspended in the same volume of fresh medium lacking a nitrogen source. After 24 h, RNA was prepared as described previously (29).
Microarray analysis.
cDNA was synthesized from 5 μg of total RNA (MB215) with the Superscript Choice system (Invitrogen). For first-strand synthesis, a GeneChip T7-Oligo(dT) promoter primer kit (Affymetrix) was used. After second-strand synthesis, the cDNA was purified by the GeneChip sample cleanup module (QIAGEN). During the in vitro transcription reaction, cRNA was biotin labeled using a BioArray HighYields RNA transcript labeling kit (ENZO). For this reaction, 5 μl of the purified cDNA was used. The synthesized cRNA was purified by the GeneChip sample cleanup module (QIAGEN). Twenty micrograms of cRNA was fragmented into 35- to 200-bp fragments (GeneChip sample cleanup module; QIAGEN). Fifteen micrograms of the fragmented cRNA was hybridized to the MPI-UstilagoA array for 16 h. The arrays were stained with streptavidin-phycoerythrin (EukGe-WS2 manual, GeneChip Fluidics station 400; Affymetrix).
Data analysis.
Gene chips were scanned, and the resulting image files were used to calculate, normalize, and compare hybridization intensity data by using MICROARRAY SUITE 4.0 software (Affymetrix). The program dChip1.2 was used for global normalization (24). During comparison of experimental and control arrays, all genes which showed a less-than-twofold change in expression level were eliminated (24).
Isolation of glycolipids.
Extracellular glycolipids were extracted from suspension cultures with ethyl acetate and separated by thin-layer chromatography (TLC) as described previously (10).
Overexpression of Um03114 (Mat1) in E. coli and cell-free acetyltransferase assays.
The Um03114 (mat1) open reading frame was amplified by PCR using the primers 5′-ACATATGAAGAGCAACGTGGATACTG-3′ (the NdeI site is in italics) and 5′-ACTCGAGCTATTCGACAAAGATGTACCTTCC-3′ (the XhoI site is in italics). The amplified DNA was digested with NdeI and XhoI and cloned into the expression vector pET-15b (Novagen). The plasmid pET15b-Mat1 was transformed into the E. coli strain BL21(DE3) and used for overexpression as described previously (17). Two hours after induction with 1 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG), cells were harvested and suspended in 0.01 volume of 10 mM Tris-HCl (pH 7.5). Cells were lysed by lysozyme treatment, and cell debris was removed by centrifugation at 12,000 × g for 20 min. The supernatant was used for enzyme assays (crude enzyme fraction). The reaction mixture contained 5 μl of the crude enzyme fraction, 10 μl of deacetylated mannosylerythritol lipid in 50% ethanol (extracted MELs of a 0.1-ml culture), 2.5 μl of acetyl coenzyme A (CoA) in water (100 mM), and 182.5 μl of 10 mM Tris-HCl (pH 7.5). The reaction mixtures were incubated for 1 h at 37°C and extracted with 200 μl ethyl acetate. The MEL products were analyzed by TLC and liquid chromatography coupled to tandem mass spectrometry (LC/MSn).
Mass spectrometry.
High-performance liquid chromatography (HPLC) separation of the extracted MELs (100 μl) was performed with a 1100-HPLC system (Agilent, Germany) equipped with a 3-μm Nucleosil 250/3 C8 column (Macherey-Nagel, Germany). The gradient applied at a flow rate of 0.4 ml/min and a column temperature of 45°C was as follows (buffer A is water with 0.05% formic acid; buffer B is methanol with 0.045% formic acid): linear gradient from 60% buffer B to 95% buffer B within 30 min and then holding 95% buffer B for 10 min.
Online electrospray ionization MSn of the HPLC-separated compounds was done with a Finnigan LTQ-Fourier transform (FT) mass spectrometer (Thermo Electron Corp., Germany). Electrospray ionization parameters were adapted to the flow rate and mass range. Accurate masses (accuracy, ≤2 ppm), allowing the determination of the chemical formulas of the eluting compounds, were obtained by using the FT mass analyzer at a resolution of 100,000. Meanwhile, fragment ions were generated and analyzed in the LTQ mass analyzer. Alternatively, data-dependent fragmentation or fixed m/z fragmentation was used; the latter resulted in better sensitivity. Although the accurate FT masses in combination with MS2 experiments were sufficient to identify the acylation pattern of the compounds, MS3 experiments were done to demonstrate the presence of the second acetate group unambiguously.
RESULTS
Identification of a gene cluster for MEL biosynthesis.
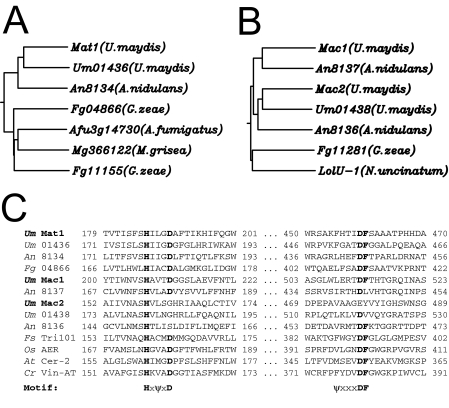
We previously identified a glycosyltransferase, Emt1, which is essential for MEL biosynthesis (10). Transcription of the emt1 gene is strongly induced under conditions of nitrogen starvation (10). To identify additional genes involved in the biosynthesis of MELs, we performed a genome-wide expression analysis using DNA microarrays. Total RNA was prepared before and after a nutritional downshift to nitrogen limitation. The RNA was used to probe Affymetrix DNA chips representing about 93% of the 6,835 genes, which are presently annotated in the U. maydis genome database MUMDB (http://mips.gsf.de/genre/proj/ustilago/). Upon nitrogen limitation, 582 genes are induced more than 2.5-fold (not shown). Remarkably, the previously identified emt1 gene showed the highest level of induction under these conditions (more than 25×). This induction level corresponds to the high expression level under conditions of nitrogen starvation as observed previously by Northern analysis (10). Among the induced genes are many whose annotations implicate a direct function in nitrogen uptake, such as import of oligopeptides, amino acids, or nucleosides (Table 1). Importantly, four genes located immediately adjacent to emt1 were identified among the genes that are induced more than 2.5-fold upon nitrogen limitation (Table 1 and Fig. 1B). These genes carry the following entry names in the U. maydis database (MUMDB): Um03116 (GenBank accession number EAK83924), Um03115 (EAK83923), Um03114 (EAK83922), and Um10636 (EAK83927). Since many fungal genes involved in secondary metabolism are arranged in clusters (36), we considered this array of coregulated genes to be a prime candidate for a MEL biosynthesis gene cluster. Database comparison revealed that Um03115 is a membrane-spanning protein of 619 amino acids which belongs to the highly conserved family of major facilitators (26). This suggests that this protein, which we termed Mmf1, is involved in the export of MEL glycolipids. The derived amino acid sequences of Um03114, Um03116, and Um10636 are related to each other and display high similarity with other fungal proteins of unknown function (Fig. 2A and B). Within the predicted sequences of these proteins we detected two short signature sequences that correspond to the Pfam domain 02458. This domain has been recently described as characteristic for the BAHD family of CoA-dependent acyltransferases, mostly derived from plants (33) (Fig. 2C). Therefore, we termed the corresponding proteins Mat1 (Um03114), Mac1 (Um03116), and Mac2 (Um10636), since they are supposed to catalyze the transfer of acetyl groups (Mat1) (see below) or acyl groups (Mac1 and Mac2) (see below) to mannosylerythritol.
TABLE 1.
Nitrogen starvation-induced genes
| Entrya | Induction (n-fold) | Predicted functionb |
|---|---|---|
| Um03117 | 25.5 | Glycosyltransferase Emt1 |
| Um11057 | 18.5 | Oligopeptide transporter |
| Um04347 | 16.5 | Oligopeptide transporter |
| Um00076 | 9.5 | Purine permease |
| Um06012 | 8.7 | General amino acid permease |
| Um03049 | 8.5 | Neutral amino acid permease |
| Um03115 | 6.9 | Major facilitator (Mmf1) |
| Um03663 | 5.9 | Uracil permease |
| Um03116 | 5.4 | Predicted protein (Mac1) |
| Um01840 | 5.2 | Nucleoside transporter |
| Um10636 | 5.0 | Predicted protein (Mac2) |
| Um03690 | 4.8 | Purine permease |
| Um00034 | 4.5 | Major facilitator |
| Um01996 | 3.9 | Multidrug resistance protein |
| Um03700 | 3.6 | Glutamate permease |
| Um02146 | 3.3 | GABA permease |
| Um01899 | 2.6 | Multidrug resistance protein |
| Um10648 | 2.5 | P-type ATPase |
| Um01062 | 2.5 | GDP-mannose transporter |
| Um04523 | 2.4 | Low-affinity ammonium transporter (Ump1) |
| Um00343 | 2.4 | General amino acid permease |
| Um03114 | 2.3 | Predicted protein (Mat1) |
| Um02159 | 2.3 | Amino acid transporter |
Entry numbers refer to the Munich Information Center for Protein Sequences U. maydis database (MUMDB).
Shown are only proteins with predicted functions related to nitrogen starvation. Proteins of the MEL cluster are indicated by bold letters. Emt1 has been described previously (10).
FIG. 2.
Mat1, Mac1, and Mac2 belong to the superfamily of acyl-CoA-dependent acyltransferases. (A) Molecular phylogeny of Mat1. Sequences were aligned using ClustalW. The following numbers are the accession numbers: for Mat1 of U. maydis, EAK83922; for Um01436 of U. maydis, EAK81770; for An8134 of A. nidulans, EAA58771; for Fg04866 of Gibberella zeae, EAA74194; for Afu3g14730 of Aspergillus fumigatus, EAL92114; for Mg366122 of Magnaporthe grisea, EAQ71714; and for Fg11155 of G. zeae, EAA75365. (B) Molecular phylogeny of Mac1 and Mac2. The following numbers are the accession numbers: for Mac1 of U. maydis, EAK83924; for An8137 of A. nidulans, EAA58774; for Mac2 of U. maydis, EAK83927; for Um01438 of U. maydis, EAK81602; for An8136 of A. nidulans, EAA58773; for Fg11281 of G. zeae, EAA78594; and for LolU-1 of Neotyphodium uncinatum, AAV68707. (C) Alignment of the short signature sequences (HXXXD and DF) that are nearly invariant in this family of potential acyltransferases (Pfam domain 02458). The HXXXD motif is believed to be part of the active center. ψ indicates a hydrophobic amino acid.
Deletion of mat1 results in the production of deacetylated MELs.
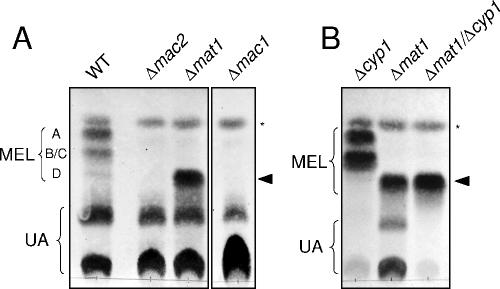
To determine the biological function of the cluster genes, deletion mutants of single genes were generated by homologous recombination in the haploid wild-type strains FB1 and MB215. Gene replacement was confirmed by PCR and Southern analysis (not shown). Mutants were then tested for glycolipid production by incubation under conditions of nitrogen limitation. Glycolipids were extracted from the culture supernatant and analyzed by TLC. Wild-type cells secrete large amounts of both ustilagic acid (UA) and MELs (Fig. 3A). MELs secreted by wild-type cells are separated into three spots with different mobilities. The band which displays the highest mobility corresponds to fully acetylated MEL A, while the monoacetylated derivatives MEL B and MEL C migrate significantly more slowly (21, 32). Wild-type cells produce only minor amounts of the completely deacetylated MEL D, which is visible in TLC as a faint band with low mobility (Fig. 3A). In contrast, mat1 mutant cells secreted large amounts of MELs that migrated as a single prominent spot with the same mobility as the deacetylated MEL D (Fig. 3A). The production of ustilagic acid is not affected by the mat1 deletion.
FIG. 3.
Analysis of glycolipid production by TLC. Ethyl acetate extracts of extracellular glycolipids of wild-type and mutant strains were separated by TLC. Wild-type strains produce both MEL and UA. MELs are separated according to their degree of acetylation: fully acetylated MEL A displays the highest mobility, while the partially acetylated forms MEL B and MEL C and the deacetylated form MEL D migrate significantly more slowly. The asterisk denotes an as yet unknown glycolipid, which belongs neither to the MEL class nor to the UA class. (A) Deletion of either mac1 or mac2 results in complete loss of MEL production, while UA secretion is not affected. The Δmat1 mutant strain secretes deacetylated MEL D, as indicated by the filled arrowhead. (B) Glycolipid production of strain SH21Δmat1Δcyp1, which is deficient for ustilagic acid production and secretes only deacetylated MEL D (filled arrowhead). This double mutant strain was derived from a cross between FB1Δmat1 and MB215Δcyp1.
Mass spectrometry of wild-type and mutant MEL derivatives.
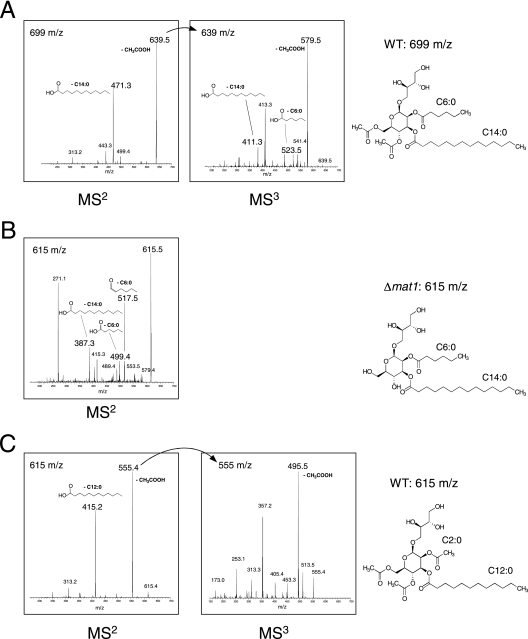
To determine the molecular structures of MELs produced by wild-type and mat1-deficient strains, we used LC/MSn. In general, structural analysis of MELs is hampered by the fact that wild-type cells of U. maydis produce a complex mixture of many different MEL derivatives. These differ not only in the length of the acyl chains at positions R-2 and R-3 but also in the degree of acetylation at positions R-4 and R-6 (Fig. 1A) (21, 32). To facilitate mass spectrometric identification of mutant MEL derivatives, we constructed a strain which is defective in ustilagic acid production and thus produces only MEL glycolipids. To this aim, strain FB1Δmat1 was crossed with MB215Δcyp1, which carries a deletion of the cyp1 gene encoding a P450 monooxygenase essential for ustilagic acid synthesis (10). We isolated a double mutant from among the progeny (SH21Δmat1Δcyp1) which secretes solely the mutant MEL glycolipids (Fig. 3B). Culture supernatants of wild-type strain MB215 and strain SH21Δmat1Δcyp1 were extracted with ethyl acetate and subjected to LC/MSn. In wild-type extracts, we identified a glycolipid compound m/z 699 [M+Na]+ which could not be detected in extracts of the mat1 mutant strain (Fig. 4A). Instead, the mutant secretes a novel glycolipid species m/z 615 [M+Na]+ (Fig. 4B). Fragmentation analysis of these compounds revealed that both glycolipids contain C6:0 and C14:0 fatty acids at positions R-2 and R-3, respectively. However, the wild-type MEL (m/z = 699) carries acetyl groups at both R-4 and R-6, while the corresponding mutant glycolipid (m/z = 615) is completely deacetylated (Fig. 4B). This indicates that mat1 mutants produce the corresponding MEL variant (C6:0; C14:0) only in its deacetylated form.
FIG. 4.
Mass spectrometry of wild-type and mutant MEL derivatives. Supernatants of wild-type and mutant strains were extracted with ethyl acetate and subjected to LC/MS analysis. (A) Wild-type cells secrete a compound with m/z 699 [M+Na]+. Tandem (MS2) and triple (MS3) mass fragmentation revealed that this MEL variant is completely acetylated at both C-4 and C-6. At position C-2 it carries a C6:0 fatty acid, and at position C-3 it carries a C14:0 fatty acid. (B) Δmat1 mutant cells secrete a completely deacetylated derivative of this glycolipid with m/z 615. (C) A fully acetylated glycolipid with a molecular weight of m/z 615 is found in wild-type strains. Fragmentation mass analysis (MS2 and MS3) revealed that it differs from the wild-type MEL (m/z 615) by the lengths of the esterified fatty acids at positions C-2 and C-3, which are C2:0 and C12:0, respectively.
Next we asked whether we could also detect this deacetylated MEL in the wild-type extract. Although a glycolipid with m/z = 615 [M+Na]+ can be detected in wild-type extracts, this substance has a different retention time in LC from the mutant glycolipid with m/z 615 (not shown). Mass spectrometry revealed that this glycolipid is fully acetylated at R-4 and R-6 but carries C2:0 and C12:0 acyl groups at the R-2 and R-3 positions of the mannosylerythritol moiety (Fig. 4C). The observation that the fully acetylated wild-type glycolipid and the mutant deacetylated glycolipid display the same total mass (m/z = 615) can be explained by the fact that in the mutant the loss of the two acetyl groups (−84) is just compensated by the presence of six additional CH2 groups (+84). This result was confirmed by Fourier transform mass spectrometry, which allows prediction of the molecular formula by precise mass determination. While the exact mass of the wild-type glycolipid was determined as m/z = 615.2984, the mat1 mutant secreted a MEL with m/z 615.3724 (data not shown). These values correspond to the molecular formulas C28H48O13Na (calculated mass = 615.2987) for the wild-type MEL and C30H56O11Na (calculated mass = 615.3715) for the mutant MEL. This indicates that both hydroxyl groups at R-4 and R-6 of the mannosylerythritol are unesterified in the MEL derivative (m/z = 615) produced by the mat1 deletion mutant. By the same method, molecular formulas of additional MEL derivatives secreted by the mat1 mutant were estimated. In all cases, the mutant MELs contained only 11 oxygen atoms (data not shown), indicating the presence of two free hydroxyl groups. Taken together, these data clearly demonstrate that Mat1 is required for acetylation of the mannosylerythritol lipids.
Heterologously expressed Mat1 acetyltransferase is able to acetylate MELs in vitro at both the 4′-OH and 6′-OH groups.
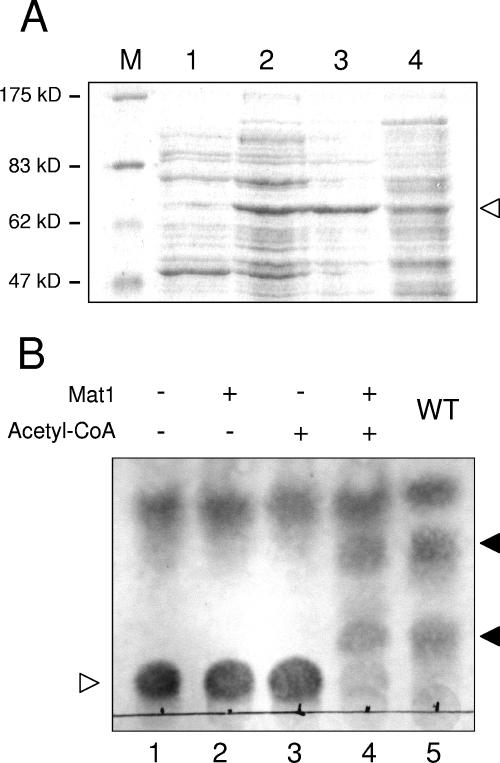
To determine whether Mat1 directly catalyzes the acetylation of MELs, the mat1 open reading frame was amplified by PCR and cloned into a bacterial expression vector (see Materials and Methods). Mat1 overexpression was induced by addition of IPTG (Fig. 5A), and cell extracts were tested for acetyltransferase activity in vitro. We used deacetylated MEL D, which was purified from culture supernatants of the SH21Δmat1Δcyp1 double mutant strain, as the substrate in the enzyme assay. This strain conveniently secretes only the deacetylated MEL, since mutation of cyp1 prevents synthesis of cellobiose lipids and the absence of mat1 abolishes acetylation of MELs. This deacetylated substrate was incubated with a crude extract from Mat1-overexpressing E. coli cells. The enzyme reaction was performed in the presence and absence of acetyl-CoA (see Materials and Methods). Only if both Mat1-containing cell extract and acetyl-CoA were added were faster-migrating forms that correspond to the mono- and diacetylated forms detected in thin-layer chromatography (Fig. 5B). Acetylation of MEL D was also confirmed by mass spectrometry (data not shown). This demonstrates that Mat1 acts in vitro as an acetyl-CoA-dependent MEL acetyltransferase. Tandem mass spectrometry confirmed that the reaction products were acetylated at both the C-4 and C-6 positions (not shown). This indicates that the U. maydis MEL acetyltransferase Mat1 displays relaxed regioselectivity and is able to acetylate mannosylerythritol lipids at two different hydroxyl groups.
FIG. 5.
In vitro acetyltransferase activity of Mat1. (A) Mat1 was overexpressed in E. coli, and cell extracts were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The arrowhead indicates the Mat1 protein, which is only partially soluble. M, size marker; lane 1, crude extract from uninduced cells; lane 2, crude extract from induced cells; lane 3, pellet from induced cells; lane 4, supernatant from induced cells. (B) TLC analysis of Mat1 in vitro acetyltransferase activity. Deacetylated MEL D (lane 1) was incubated with Mat1-containing cell extract in the absence of acetyl-CoA (lane 2), with acetyl-CoA and mock cell extract (lane 3), and with both Mat1 and acetyl-CoA (lane 4). For comparison, acetylated MELs (MEL A, MEL B, and MEL C) extracted from a wild-type strain are shown (lane 5). The empty arrowhead indicates deacetylated MEL D, and the filled arrowheads indicate the spots corresponding to partially and fully acetylated MEL variants.
The putative acyltransferases Mac1 and Mac2 are essential for MEL production.
The proteins encoded by mac1 and mac2 also belong to the BAHD superfamily of acyltransferases, which suggests that these enzymes might be responsible for the transfer of the acyl groups to mannosylerythritol. While mat1 mutants were still able to produce a significant amount of secreted MEL, deletion of either mac1 or mac2 resulted in complete loss of MEL production (Fig. 3A). Therefore, we propose that Mac1 and Mac2 catalyze the transfer of short- and medium-chain fatty acids to the C-2 and C-3 positions of mannosylerythritol. Since we could not detect any partially acylated mannosylerythritol lipids in either the mac1 mutant or the mac2 mutant, we were not able to conclude which enzyme is responsible for acylation of either position. The complete lack of MEL production by mac1 and mac2 single mutants indicates that acylation at both positions appears to be prerequisite for glycolipid secretion. In addition, the mac1 and mac2 genes cannot replace each other, although they share a high degree of sequence similarity (23% amino acid identity). This supports the notion that the acyltransferases Mac1 and Mac2 display high regioselectivity and acylate specifically either the 2′-OH or the 3′-OH group of the mannosylerythritol substrate.
Genetic synteny of the MEL cluster in Aspergillus nidulans and U. maydis.
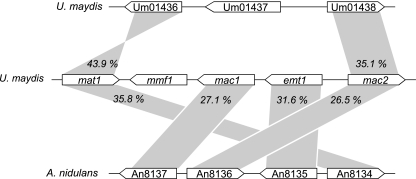
Database comparisons revealed that four of the five MEL cluster genes have close homologs in the ascomycete Aspergillus nidulans that are also arranged in a large cluster (Fig. 6). Most remarkably, there are no homologs of this gene cluster in the closely related species Aspergillus fumigatus and Aspergillus oryzae, whose genomic sequences have been recently released. The genetic synteny shared between U. maydis and A. nidulans points to a common evolutionary origin of the MEL cluster genes, maybe by horizontal gene transfer. It could also suggest that A. nidulans synthesizes glycolipids similar or identical to mannosylerythritol lipids; however, it is not known whether or under which conditions the A. nidulans genes are expressed. Since the A. nidulans cluster lacks a potential export protein, it is feasible that the putative glycolipids might not be secreted. Limited synteny is also observed between the MEL gene cluster and another region within the U. maydis genome. Mat1 and Mac2 are highly similar to the predicted polypeptides Um01436 and Um01438, respectively. These genes are located within a novel iron uptake cluster that contains a nonribosomal peptide synthase (Um01434) involved in the synthesis of an as yet uncharacterized ferrichrome siderophore and a ferric reductase (Um01439) which is required for the uptake of siderophore-bound iron (R. Kahmann, personal communication). This could indicate that this cluster might specify a siderophore which is additionally modified by acetylation and/or acylation.
FIG. 6.
Extended synteny between the U. maydis MEL biosynthesis cluster and a corresponding gene cluster of unknown function in A. nidulans. Additional synteny is observed between mat1 and mac2 and the U. maydis genes Um01436 and Um01438. Numbers indicate identity at the amino acid sequence level.
DISCUSSION
In this work, we report the identification of the first biosynthesis gene cluster for a fungal extracellular glycolipid. The U. maydis MEL cluster specifies five proteins, one glycosyltransferase, three acyltransferases, and one export protein of the major facilitator family. All enzymes are most probably involved in the biosynthesis and export of the commercially important biosurfactant mannosylerythritol. By mutation analysis, we demonstrated that the putative acyltransferases Mac1 and Mac2 are both essential for MEL biosynthesis. In addition, the acetyltransferase Mat1 catalyzes the acetylation of mannosylerythritol lipids in U. maydis.
While fungal genes of the primary metabolism are rarely clustered, gene clusters have been observed preferentially for genes involved in the production of secondary metabolites (11, 34). This might reflect their recent introduction from a different species by lateral gene transfer. It has been proposed that the organization of secondary metabolites in gene clusters would confer an advantage for such spreading among different species (36). This “selfish cluster” hypothesis argues that only cluster organization guarantees the transfer of complete biosynthesis pathways upon horizontal gene transfer. The observed synteny of the MEL biosynthesis gene cluster with a similar cluster in the quite distantly related fungus A. nidulans strongly favors the idea of lateral gene transfer between these species or some of their recent progenitors.
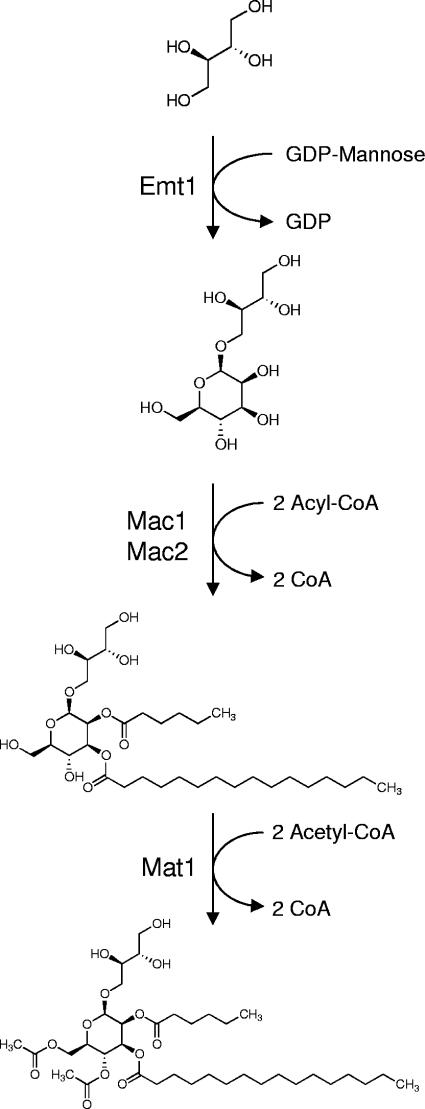
Mutational and biochemical analysis of the MEL biosynthesis cluster allows us to propose a biosynthesis pathway leading to MEL formation (Fig. 7). The first step would be the generation of mannosylerythritol by mannosylation of erythritol, which is most probably catalyzed by the previously described glycosyltransferase Emt1 (10). The intermediate mannosylerythritol has been isolated in significant amounts from MEL-producing cells (1). We assume that this disaccharide is subsequently acylated with fatty acids of various lengths by the putative acyltransferases Mac1 and Mac2 at positions C-2 and C-3. This acylation reaction appears to be essential for secretion because deletion of either mac1 or mac2 abolished MEL production completely. Currently, we cannot specify in which order these two enzymes act on their substrates or which acyltransferase is responsible for acylation of C-2 and C-3. In general, MELs secreted by U. maydis carry a short fatty acid (C2 to C8) at position C-2 and a medium or long fatty acid (C10 to C18) at C-3 (21, 32). This implies that Mac1 and Mac2 differ not only in regioselectivity but also in their preference for the length of the acyl-CoA cofactor. Using mass spectrometry, we identified MEL derivatives which carried an acetyl group at C-2. This implies that at least one of the acyltransferases, presumably the one which catalyzes the transfer of the short-chain fatty acid, also accepts acetyl-CoA as a donor. For the MEL-producing fungus Candida antarctica, it has been previously shown that medium-length fatty acids are derived from longer fatty acids by partial peroxisomal β-oxidation (20). Inspection of the genome sequence revealed that U. maydis contains both mitochondrial and peroxisomal proteins for fatty acid degradation. Thus, we assume that a similar peroxisomal chain-shortening pathway also exists in U. maydis.
FIG. 7.
Proposed biosynthetic route for mannosylerythritol lipids. The first step of MEL biosynthesis is condensation of mannosyl and erythritol catalyzed by the glycosyltransferase Emt1. Mac1 and Mac2 are proposed to transfer short- and medium-chain fatty acids to positions R-2 and R-3. The last step, acetylation of deacetylated MEL at positions R-4 and R-6, is catalyzed by a single enzyme, Mat1.
The final step of MEL biosynthesis is the acetylation of the fully acylated mannosylerythritol lipids. Acetylation is not essential for glycolipid secretion, since strains deficient for mat1 secrete large amounts of deacetylated MEL D (Fig. 3). Also, wild-type strains secrete MEL D, albeit in very small amounts (Fig. 3A) (32). Here we showed that acetylation is catalyzed by the acetyl-CoA-dependent acetyltransferase Mat1. Mat1 displays a relaxed regioselectivity and is able to transfer acetyl groups to both positions C-4 and C-6 of the mannosyl moiety. Mat1 overexpressed in E. coli can perform this reaction in vitro, which indicates that no further enzymes are involved in this reaction. Wild-type cells secrete a large fraction of MELs which are acetylated at only one position. This could indicate that the second acetylation reaction is significantly slower than the first one. In this scenario, partially acetylated glycolipids are secreted before the second acetylation happens.
Database comparisons revealed that Mat1, Mac1, and Mac2 are closely related to each other and constitute a novel family of fungal acyl/acetyltransferases. We could identify two short amino acid sequence motifs which were first described for the plant deacetylvindoline acetyltransferase and which catalyze the last step of vindoline biosynthesis (33). A number of related plant enzymes which are involved in the production of diverse secondary metabolites have been identified (7). The first representative of this superfamily to be crystallized was vinorine synthase (25). The high level of sequence conservation suggested that the nearly invariable motif HXXXD is part of the active center. Interestingly, Mac2 and some of the related fungal enzymes lack the aspartate residue of this motif, indicating that it might be dispensable for catalysis (Fig. 2C). The crystal structure of vinorine synthase supports this notion, since it demonstrates that in the active center the side chain of Asp164 points away from His160 and the active site. Therefore, Asp164 is unlikely to participate directly in catalysis but is rather important for maintaining the geometry of the active site (25). Remarkably, the acetyltransferase Mat1 can acetylate the mannosyl group at two distinct positions, at least in vitro. Similar relaxed regioselectivity has also been observed in other fungal acetyltransferases, e.g., taxadiene acetyltransferase (3). Mac1 and Mac2 display some similarity to the LolU-1 protein, which has been identified in the fungal endophyte Neotyphodium uncinatum as part of a cluster involved in the production of loline alkaloids (31). This protein has been proposed to act as a transcriptional regulator of loline biosynthesis, but its similarity with fungal acetyltransferases indicates a more direct role for this enzyme, most probably in acetylation of loline to acetylloline.
Secretion of MELs is supposed to be catalyzed by the major facilitator Mmf1, which is part of the gene cluster. We recently generated a knockout mutant for mmf1. This mutant is unable to produce extracellular MELs (M. Caliaro and M. Bölker, unpublished data), indicating that the major facilitator Mmf1 is essential for secretion. Mmf1 appears to display only limited specificity for its substrates, since deacetylated MELs are secreted as efficiently as the acetylated wild-type form. In addition, the observed spectrum of MELs carrying acyl groups of different lengths supports the notion that the putative exporter does not distinguish between these derivatives. Such a broad specificity is typical for members of the large family of major facilitators, which are often involved in multidrug resistance (4). The U. maydis Mmf1 protein displays the highest level of sequence similarity to Mfs1-1 from Coprinus cinereus, whose gene is located within the mating type-determining region of this basidiomycetous fungus (8). This could indicate a potential role of this exporter in the function of the mating type locus, e.g., for secretion of glycolipids which enhance diffusion of the hydrophobic lipopeptide pheromones, as has been suggested for U. maydis (10). Expression of the MEL biosynthesis gene cluster is highly induced under conditions of nitrogen starvation. Comparison of the promoter regions of the cluster genes revealed no obvious conserved sequence motifs which could be involved in the coregulation of these genes. However, in the promoter regions of the newly identified genes we identified several GATA sequences, as has also been described for the putative promoter of the emt1 gene (10). This supports a role for a potential GATA factor homologous to the general nitrogen regulator AreA from A. nidulans in the regulation of this gene cluster. Such a protein is present in the U. maydis genome (Um04252).
The identification of the MEL biosynthesis gene cluster is expected to allow interesting applications. We have already demonstrated that strains deleted for mat1 produce only the fully deacetylated mannosylerythritol lipid MEL D. Thus, our mutants represent the first example of metabolic engineering of extracellular glycolipids. We can also envisage that specific production of fully acetylated MEL A can be triggered by overexpression of Mat1 acetyltransferase. This could be of commercial value because MEL A has some interesting properties not shared by the other less acetylated variants. Only MEL A dramatically increases gene transfection efficiency of liposomes (14), while the partially acetylated derivatives MEL B and MEL C do not. In addition, MEL A forms large vesicles, called coacervates, whereas MEL B is unable to form such structures (13). Since U. maydis can easily be genetically modified, it is an ideal organism to design novel glycolipid biosurfactants with specific and novel properties by metabolic engineering.
Acknowledgments
This work was supported by the special grant program SFB395 from the Deutsche Forschungsgemeinschaft (DFG).
We thank Attila Pinter for his technical help during the construction of deletion mutants and Björn Sandrock for critical reading of the manuscript.
REFERENCES
- 1.Boothroyd, B., J. A. Thorn, and R. H. Haskins. 1956. Biochemistry of the ustilaginales. XII. Characterization of extracellular glycolipids produced by Ustilago sp. Can. J. Biochem. Physiol. 34:10-14. [PubMed] [Google Scholar]
- 2.Cameotra, S. S., and R. S. Makkar. 2004. Recent applications of biosurfactants as biological and immunological molecules. Curr. Opin. Microbiol. 7:262-266. [DOI] [PubMed] [Google Scholar]
- 3.Chau, M., K. Walker, R. Long, and R. Croteau. 2004. Regioselectivity of taxoid-O-acetyltransferases: heterologous expression and characterization of a new taxadien-5alpha-ol-O-acetyltransferase. Arch. Biochem. Biophys. 430:237-246. [DOI] [PubMed] [Google Scholar]
- 4.Del Sorbo, G., H. Schoonbeek, and M. A. De Waard. 2000. Fungal transporters involved in efflux of natural toxic compounds and fungicides. Fungal Genet. Biol. 30:1-15. [DOI] [PubMed] [Google Scholar]
- 5.Deml, G., T. Anke, F. Oberwinkler, B. M. Giannetti, and W. Steglich. 1980. Schizonellin A and B, new glycolipids from Schizonella melanogramma. Phytochemistry 19:83-87. [Google Scholar]
- 6.Fluharty, A. L., and J. S. O'Brien. 1969. A mannose- and erythritol-containing glycolipid from Ustilago maydis. Biochemistry 8:2627-2632. [DOI] [PubMed] [Google Scholar]
- 7.Grothe, T., R. Lenz, and T. M. Kutchan. 2001. Molecular characterization of the salutaridinol 7-O-acetyltransferase involved in morphine biosynthesis in opium poppy Papaver somniferum. J. Biol. Chem. 276:30717-30723. [DOI] [PubMed] [Google Scholar]
- 8.Halsall, J. R., M. J. Milner, and L. A. Casselton. 2000. Three subfamilies of pheromone and receptor genes generate multiple B mating specificities in the mushroom Coprinus cinereus. Genetics 154:1115-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haskins, R. H., and J. A. Thorn. 1951. Biochemistry of the ustilaginales. VII. Antibiotic activity of ustilagic acid. Can. J. Bot. 29:585-592. [Google Scholar]
- 10.Hewald, S., K. Josephs, and M. Bölker. 2005. Genetic analysis of biosurfactant production in Ustilago maydis. Appl. Environ. Microbiol. 71:3033-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hohn, T. M., S. P. McCormick, and A. E. Desjardins. 1993. Evidence for a gene cluster involving trichothecene-pathway biosynthetic genes in Fusarium sporotrichioides. Curr. Genet. 24:291-295. [DOI] [PubMed] [Google Scholar]
- 12.Im, J. H., T. Nakane, H. Yanagishita, T. Ikegami, and D. Kitamoto. 2001. Mannosylerythritol lipid, a yeast extracellular glycolipid, shows high binding affinity towards human immunoglobulin G. BMC Biotechnol. 1:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imura, T., H. Yanagishita, and D. Kitamoto. 2004. Coacervate formation from natural glycolipid: one acetyl group on the headgroup triggers coacervate-to-vesicle transition. J. Am. Chem. Soc. 126:10804-10805. [DOI] [PubMed] [Google Scholar]
- 14.Inoh, Y., D. Kitamoto, N. Hirashima, and M. Nakanishi. 2001. Biosurfactants of MEL-A increase gene transfection mediated by cationic liposomes. Biochem. Biophys. Res. Commun. 289:57-61. [DOI] [PubMed] [Google Scholar]
- 15.Kakugawa, K., M. Tamai, K. Imamura, K. Miyamoto, S. Miyoshi, Y. Morinaga, O. Suzuki, and T. Miyakawa. 2002. Isolation of yeast Kurtzmanomyces sp. I-11, novel producer of mannosylerythritol lipid. Biosci. Biotechnol. Biochem. 66:188-191. [DOI] [PubMed] [Google Scholar]
- 16.Kämper, J. 2004. A PCR-based system for highly efficient generation of gene replacement mutants in Ustilago maydis. Mol. Genet. Genomics 271:103-110. [DOI] [PubMed] [Google Scholar]
- 17.Kimura, M., I. Kaneko, M. Komiyama, A. Takatsuki, H. Koshino, K. Yoneyama, and I. Yamaguchi. 1998. Trichothecene 3-O-acetyltransferase protects both the producing organism and transformed yeast from related mycotoxins. Cloning and characterization of Tri101. J. Biol. Chem. 273:1654-1661. [DOI] [PubMed] [Google Scholar]
- 18.Kitamoto, D., S. Akiba, C. Hioki, and T. Tabuchi. 1990. Extracellular accumulation of mannosylerythritol lipids by a strain of Candida antarctica. Agric. Biol. Chem. 54:31-36. [Google Scholar]
- 19.Kitamoto, D., H. Isoda, and T. Nakahara. 2002. Functions and potential applications of glycolipid biosurfactants—from energy-saving materials to gene delivery carriers. J. Biosci. Bioeng. 94:187-201. [DOI] [PubMed] [Google Scholar]
- 20.Kitamoto, D., H. Yanagishita, K. Haraya, and K. Kitamoto. 1998. Contribution of a chain-shortening pathway to the biosynthesis of the fatty acids of mannosylerythritol lipid (biosurfactant) in the yeast Candida antarctica: effect of β-oxidation inhibitors on biosurfactant synthesis. Biotechnol. Lett. 20:813-818. [Google Scholar]
- 21.Kurz, M., C. Eder, D. Isert, Z. Li, E. F. Paulus, M. Schiell, L. Toti, L. Vertesy, J. Wink, and G. Seibert. 2003. Ustilipids, acylated beta-D-mannopyranosyl D-erythritols from Ustilago maydis and Geotrichum candidum. J. Antibiot. (Tokyo) 56:91-101. [DOI] [PubMed] [Google Scholar]
- 22.Lemieux, R. U. 1953. Biochemistry of the ustilaginales. VIII. The structures and configurations of the ustilic acids. Can. J. Chem. 31:396-417. [Google Scholar]
- 23.Lemieux, R. U., J. A. Thorn, C. Brice, and R. H. Haskins. 1951. Biochemistry of the ustilaginales. II. Isolation and partial characterization of ustilagic acid. Can. J. Chem. 29:409-414. [DOI] [PubMed] [Google Scholar]
- 24.Li, C., and W. H. Wong. 2001. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA 98:31-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma, A. D., L. F. Brass, and C. S. Abrams. 1997. Pleckstrin associates with plasma membranes and induces the formation of membrane projections: requirements for phosphorylation and the NH2-terminal PH domain. J. Cell Biol. 136:1071-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pao, S. S., I. T. Paulsen, and M. H. Saier, Jr. 1998. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62:1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rau, U., L. A. Nguyen, S. Schulz, V. Wray, M. Nimtz, H. Roeper, H. Koch, and S. Lang. 2005. Formation and analysis of mannosylerythritol lipids secreted by Pseudozyma aphidis. Appl. Microbiol. Biotechnol. 66:551-559. (First published 10 July 2004; doi: 10.1007/s00253-004-1672-9.) [DOI] [PubMed] [Google Scholar]
- 28.Ron, E. Z., and E. Rosenberg. 2001. Natural roles of biosurfactants. Environ. Microbiol. 3:229-236. [DOI] [PubMed] [Google Scholar]
- 29.Schmitt, M. E., T. A. Brown, and B. L. Trumpower. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18:3091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schulz, B., F. Banuett, M. Dahl, R. Schlesinger, W. Schäfer, T. Martin, I. Herskowitz, and R. Kahmann. 1990. The b alleles of U. maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain-related motif. Cell 60:295-306. [DOI] [PubMed] [Google Scholar]
- 31.Spiering, M. J., C. D. Moon, H. H. Wilkinson, and C. L. Schardl. 2005. Gene clusters for insecticidal loline alkaloids in the grass-endophytic fungus Neotyphodium uncinatum. Genetics 169:1403-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spoeckner, S., V. Wray, M. Nimtz, and S. Lang. 1999. Glycolipids of the smut fungus Ustilago maydis from cultivation on renewable resources. Appl. Microbiol. Biotechnol. 51:33-39. [Google Scholar]
- 33.St-Pierre, B., P. Laflamme, A. M. Alarco, and V. De Luca. 1998. The terminal O-acetyltransferase involved in vindoline biosynthesis defines a new class of proteins responsible for coenzyme A-dependent acyl transfer. Plant J. 14:703-713. [DOI] [PubMed] [Google Scholar]
- 34.Tudzynski, B., and K. Hölter. 1998. Gibberellin biosynthetic pathway in Gibberella fujikuroi: evidence for a gene cluster. Fungal Genet. Biol. 25:157-170. [DOI] [PubMed] [Google Scholar]
- 35.Wakamatsu, Y., X. Zhao, C. Jin, N. Day, M. Shibahara, N. Nomura, T. Nakahara, T. Murata, and K. K. Yokoyama. 2001. Mannosylerythritol lipid induces characteristics of neuronal differentiation in PC12 cells through an ERK-related signal cascade. Eur. J. Biochem. 268:374-383. [DOI] [PubMed] [Google Scholar]
- 36.Walton, J. D. 2000. Horizontal gene transfer and the evolution of secondary metabolite gene clusters in fungi: an hypothesis. Fungal Genet. Biol. 30:167-171. [DOI] [PubMed] [Google Scholar]