Abstract
Low signal intensity due to poor probe hybridization efficiency is one of the major drawbacks of rRNA-targeted in situ hybridization. There are two major factors affecting the hybridization efficiency: probe accessibility and affinity to the targeted rRNA molecules. In this study, we demonstrate remarkable improvement in in situ hybridization efficiency by applying locked-nucleic-acid (LNA)-incorporated oligodeoxynucleotide probes (LNA/DNA probes) without compromising specificity. Fluorescently labeled LNA/DNA probes with two to four LNA substitutions exhibited strong fluorescence intensities equal to or greater than that of probe Eub338, although these probes did not show bright signals when they were synthesized as DNA probes; for example, the fluorescence intensity of probe Eco468 increased by 22-fold after three LNA bases were substituted for DNA bases. Dissociation profiles of the probes revealed that the dissociation temperature was directly related to the number of LNA substitutions and the fluorescence intensity. These results suggest that the introduction of LNA residues in DNA probes will be a useful approach for effectively enhancing probe hybridization efficiency.
The rRNA-targeted fluorescence in situ hybridization (FISH) technique has become a routine molecular tool for analyses of the microbial community (1, 3) since it was first applied by DeLong and coworkers in 1989 (8). However, the technique has limitations that need to be addressed before it can be applied to the detection of a wide range of diverse microbes (3). The major limitation is low fluorescence intensity due to various factors, such as low rRNA content of cells, impermeability of cell walls, and poor probe hybridization efficiency owing to secondary and tertiary structures of the rRNA and effects of ribosomal proteins (3).
Low probe hybridization efficiency is especially problematic. Fuchs et al. mentioned that half of newly designed probes failed to produce detectable signals during in situ hybridization (11). To overcome this problem, Amann and coworkers systematically investigated the accessibility to target sites with hundreds of 16S and 23S rRNA-targeted probes and finally concluded that the hybridization efficiency of probes depended on the accessibility associated with regional variations in the three-dimensional structure of the ribosome, resulting in different hybridization efficiencies within rRNA molecules (5, 10, 11). However, a further study on accessibility using a native ribosome model revealed that the hindrance of target sites by ribosomal proteins was negligible for a paraformaldehyde-fixed sample (4). In addition, rRNA base pairing was more important than rRNA tertiary structure for probe hybridization (4), suggesting that the denaturing of rRNA secondary structure is a crucial factor for probe hybridization. The probes used for the accessibility analyses were designed with theoretical dissociation temperatures (Tds), which were calculated using the formula developed by Suggs et al. (37), of between 48 and 60°C. Hybridization was performed at 46°C. Nonetheless, there were still problems hybridizing with some target sites, especially at stem regions of predicted rRNA secondary structures (5, 10, 11). The theoretical Td-based probe design was not sufficient to guarantee hybridization in situ (42).
Recently, Yilmaz and Noguera proposed a comprehensive model, in which the concept of the affinity of a probe to its folded nucleic acid target was introduced as a complementary idea to that of accessibility (42). In that model, accessibility was regarded as the ability of the probe to access the target site, regardless of whether hybridization will occur. The affinity was defined as the overall Gibbs free energy change (ΔG°overall), which was estimated using a model consisting of the DNA-RNA (probe-rRNA hybrid), DNA-DNA (probe self-folding), and RNA-RNA (rRNA self-folding) interactions, and it was considered to be a significant predictor of hybridization efficiency (42). Based on the affinity concept, the probes were elongated to satisfy the ΔG°overall value for efficient hybridization. Consequently, over 90% of rRNA-targeted probes yielded moderate to high brightness after hybridization reached equilibrium (43). However, elongating probes in order to increase affinity may restrict the flexibility of probe design and decrease mismatch discrimination ability.
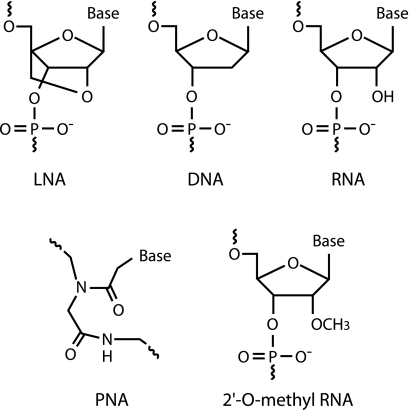
Recently, locked nucleic acid (LNA) has been developed as a novel RNA derivative nucleotide analog, in which the ribose ring is constrained by a methylene linkage between the 2′ oxygen and the 4′ carbon (Fig. 1), resulting in the locked C3′-endo conformation (18, 27). The introduction of LNA residues in oligonucleotides stabilizes the duplex by either preorganization or increased base stacking (increasing melting temperature [Tm] 1 to 8°C against DNA and 2 to 10°C against RNA by one LNA substitution in an oligonucleotide) (6, 18, 23, 26, 29-31, 40). In addition, LNA is able to hybridize with DNA or RNA according to Watson-Crick base-pairing rules with higher selectivity (13, 14, 18, 24). Due to such advantages, LNA or LNA/DNA mixmers (hereinafter called LNA/DNA) have been used for single-nucleotide polymorphism analysis, real-time PCR, microarray, and other applications (13, 14, 20, 39). In FISH analysis, LNA-incorporated oligodeoxynucleotide probes (LNA/DNA probes) have been successfully applied to human chromosomes, micro-RNAs in zebrafish, mRNA in yeast cells, and so on (16, 25, 34, 35, 38, 41). These studies demonstrated that LNA/DNA probes could bring about high binding affinities with target sequences, resulting in sensitive detection.
FIG. 1.
Structures of LNA, DNA, RNA, PNA, and 2′-O-methyl RNA. The ribose ring of LNA is constrained by a methylene linkage between the 2′ oxygen and the 4′ carbon.
From this background of knowledge, we were convinced that the use of such high-affinity LNA/DNA probes could be one solution for overcoming the problem of low hybridization efficiency of probes with rRNA in whole cells.
MATERIALS AND METHODS
Bacterial culture and fixation.
Escherichia coli TOPO10 (Invitrogen) was cultivated in LB medium at 37°C. Comamonas testosteroni (NBRC14951) was grown in medium 802 containing polypepton (10 g/liter), yeast extract (2 g/liter), and MgSO4 · 7H2O (1 g/liter) at 30°C. Anaerolinea thermophila UNI-1, isolated in our laboratory, was cultivated in medium containing sucrose (6.8 g/liter) and yeast extract (1 g/liter) at 55°C as previously described elsewhere (33). The cells were harvested during exponential growth phase, washed once in phosphate-buffered saline (PBS; 130 mM NaCl, 10.8 mM Na2HPO4, 4.2 mM NaH2PO4 [pH 7.2]), and fixed with 3% paraformaldehyde for 4 h at 4°C. After fixation, the cells were washed twice in PBS and stored in ethanol-PBS solution at −20°C until further processing.
Oligonucleotide probes.
Oligonucleotide probes used in this study are listed in Table 1. To evaluate the improvement of hybridization efficiency, we selected five dim probe sequences (Eco227, Eco468, Eco621, Eco836, and Eco1068) and two bright probe sequences (Eco252 and Eco681) according to the results of Behrens et al. (5). LNA/DNA probes used in this study had one to four LNA substitutions. DNA and LNA/DNA probes labeled with Cy3 at the 5′ end were purchased from Proligo (Kyoto, Japan) and ThermoElectron (Ulm, Germany), respectively. For nomenclature of the probes, LNA/DNA probes were prefixed with the letter “L,” and the number of the LNA substitution was indicated after the E. coli position by a hyphen followed by a numeral. LNAs were written in uppercase letters, and DNAs were written in lowercase letters, as shown in Table 1.
TABLE 1.
Oligonucleotide probes used in this study
| Probe nameb | Sequence (5′-3′)c | Relative intensityd ± SD | FAde (% FA) | Reference |
|---|---|---|---|---|
| Eco468 | aac gtc aat gag caa agg t | 0.05 ± 0.03 | 5 | 11 |
| LEco468-1 | aac gtc aat gag caa aGg t | 0.52 ± 0.07 | 13 | This study |
| LEco468-2 | aac gtc aat gaG caa aGg t | 0.87 ± 0.04 | 24 | This study |
| LEco468-2p | aac gtc Aat gag cAa agg t | 0.83 ± 0.05 | 24 | This study |
| LEco468-3 | aac gtc aat gAG caa aGg t | 1.08 ± 0.02 | 35 | This study |
| Eco621 | aga tgc agt tcc cag gtt | 0.13 ± 0.02 | 18 | 11 |
| LEco621-1 | aga tgc agt tcc caG gtt | 0.76 ± 0.07 | 30 | This study |
| LEco621-1p | aga tgc agt Tcc cag gtt | 0.82 ± 0.05 | 27 | This study |
| LEco621-2 | aga tgc agt tcC caG gtt | 1.12 ± 0.02 | 53 | This study |
| LEco621-3 | aga tgc agt tcC caG Gtt | 1.03 ± 0.03 | 62 | This study |
| Eco1068 | aca ttt cac aac acg agc | 0.44 ± 0.05 | 6 | 11 |
| LEco1068-1 | aca ttt cac aac acg aGc | 0.57 ± 0.07 | 12 | This study |
| LEco1068-2 | aca ttt cac aac acg AGc | 0.74 ± 0.11 | 17 | This study |
| LEco1068-2p1 | aca ttt cac aac aCg aGc | 0.87 ± 0.06 | 24 | This study |
| LEco1068-2p2 | aca ttt Cac aac acg aGc | 0.80 ± 0.03 | 23 | This study |
| LEco1068-3 | aca ttt cac aac aCg AGc | 0.95 ± 0.05 | 29 | This study |
| LEco1068-3p | aca ttt Cac aac aCg aGc | 0.97 ± 0.06 | 32 | This study |
| LEco1068-4 | aca ttt Cac aac aCg AGc | 1.00 ± 0.05 | 42 | This study |
| Eub338 | gct gcc tcc cgt agg agt | 1.00 ± 0.05 | 46 | 2 |
| LEub338-1 | gct gcc tcc cgt aGg agt | 1.03 ± 0.02 | 63 | This study |
| LEub338-2 | gCt gcc tcc cgt aGg agt | 1.07 ± 0.02 | 82 | This study |
| LEub338-2v3 | gct gcc Tcc cgt agg Agt | 1.04 ± 0.02f | 75 | This study |
| Eco227 | tcc cat ctg ggc aca tc | 0.79 ± 0.07 | 16 | 11 |
| Eco252 | agc cgt tac ccc acc ta | 0.98 ± 0.02 | 19 | 11 |
| Eco681 | cat ttc acc gct aca cct | 1.05 ± 0.03 | 19 | 11 |
| Eco836 | cca cgc ctc aag ggc ac | 0.52 ± 0.06 | 13 | 11 |
| GAM42a | gcc ttc cca cat cgt tt | 0.99 ± 0.04 | 22 | 22 |
| LGAM42a-1 | gcc ttc cca cAt cgt tt | 1.01 ± 0.04 | 32 | This study |
| LGAM42a-2 | gcc ttC cca cAt cgt tt | 1.07 ± 0.04 | 49 | This study |
| BET42a | gcc ttc cca ctt cgt tt | NDa | ND | 22 |
| LBET42a-1 | gcc ttc cca cTt cgt tt | ND | ND | This study |
| LBET42a-2 | gcc ttC cca cTt cgt tt | ND | ND | This study |
ND, not determined.
LNA/DNA probes have “L” at the beginning of the names and the number of LNA is indicated after the E. coli position.
Bases written in uppercase letters are LNAs, and those in lowercase letters are DNAs. Underlined letters indicate mismatch sequences (Eub338 for A. thermophila and GAM42a for C. testosteroni).
Except where indicated otherwise, relative intensity indicates the brightness of the probe at 0% formamide relative to the fluorescence intensity of Eub338 at 0% formamide.
Results show the formamide concentration that yields the 50% probe dissociation.
Result shows the brightness of the probe at 55% formamide relative to the fluorescence intensity of Eub338 at 0% formamide.
FISH.
In situ hybridization was performed as described elsewhere (33). The sample was mixed with PBS containing 0.001% sodium dodecyl sulfate and heated at 60°C for 1 min. Then prewarmed low-melting-point agarose was added at a concentration of 0.1% and reheated at 60°C for 1 min (12). Eight microliters of the mixture was applied to a 10-well glass slide (Matsunami, Osaka, Japan) and air dried at 60°C for 10 min. Dehydration was performed through ethanol series (50, 80, and 95% for 5, 1, and 1 min, respectively). Hybridization was carried out in buffer (20 mM Tris-HCl [pH 7.2], 0.01% sodium dodecyl sulfate, 900 mM sodium chloride, 0 to 80% formamide) with 0.5 μM of labeled probe for 3 to 4 h at 46°C in the dark. To remove excess probe, the slide was immersed in the same buffer for 15 min at 48°C.
Microscopic evaluation and quantitative analysis of fluorescence intensity.
The samples were covered with SlowFade antifade reagent (Invitrogen) and evaluated on a BX50F microscope (Olympus, Tokyo, Japan) equipped with a 100-W high-pressure mercury burner. For image acquisition, a DP70 color charge-coupled-device camera (Olympus) was used and the exposure time was fixed for all preparations. Fluorescence intensities were quantified by using Quantity One image analysis software (Bio-Rad, Hercules, CA). All experiments were performed in triplicate.
RESULTS
In situ hybridization with DNA probes.
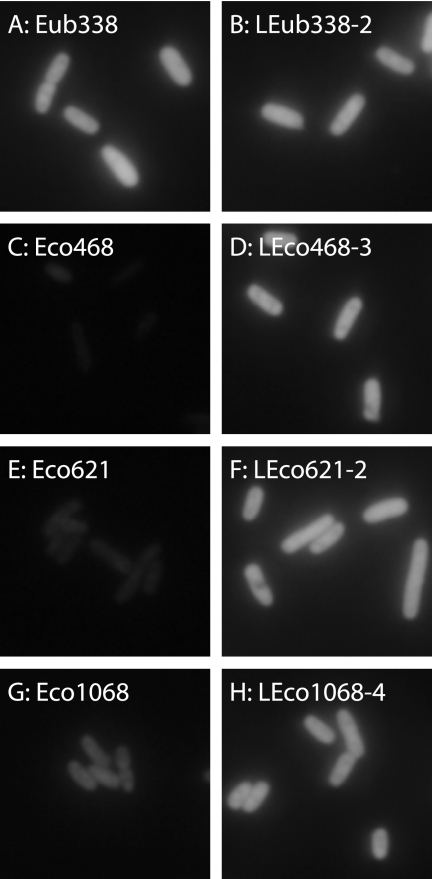
First of all, we quantified the fluorescence intensities of DNA probes (Table 1). In this study, the fluorescence intensity was used as an indicator of probe hybridization efficiency. Furthermore, the fluorescence intensity of each probe was expressed as a relative intensity, based on defining the fluorescence intensity of Eub338 at 0% formamide as 1.00. The relative intensities of DNA probes and epifluorescence micrographs of Eub338, Eco468, Eco621, and Eco1068 at 0% formamide are shown in Table 1 and Fig. 2, respectively. Eco227 (class IV), Eco468 (class VI), Eco621 (class VI), Eco836 (class V), and Eco1068 (class V) were reported as dim probes, and Eco252 and Eco681 were classified in the brightest group (class I) by Behrens et al. (5). In this study, Eco468 and Eco621 showed weak signals (relative intensities, 0.05 ± 0.03 and 0.13 ± 0.02, respectively) as reported by Behrens et al. (5), while Eco227, Eco836, and Eco1068 gave relatively strong signals (relative intensities, 0.79 ± 0.07, 0.52 ± 0.06, and 0.44 ± 0.05, respectively). On the other hand, Eco252 and Eco681 (relative intensities, 0.98 ± 0.02 and 1.05 ± 0.03, respectively) employed as bright probes yielded approximately the same intensities as did Eub338, suggesting that the Eub338 probe produced quite strong signals under the experimental conditions of this study. Based on these findings, we selected three probes, Eco468, Eco621, and Eco1068, which gave weaker signals (low hybridization efficiencies), for further investigation.
FIG. 2.
Epifluorescence micrographs of DNA and LNA/DNA probe-conferred signals at 0% formamide in hybridization buffer. The same exposure time (250 ms) was used for all probes.
In situ hybridization with LNA/DNA probes.
Relative intensities of LNA/DNA probes at 0% formamide are listed in Table 1, and some examples of epifluorescence photomicrographs are shown in Fig. 2. One-LNA-substituted probes, designated LEco468-1, LEco621-1 and LEco1068-1, showed higher fluorescence intensities than those of the respective DNA probes. The increments of LEco468-1 and LEco621-1 were significant (changes in relative intensity [Δrelative intensity] were 0.47 and 0.63, respectively), whereas that of LEco1068-1 was trivial (Δrelative intensity, 0.13). Two-LNA-substituted probes (LEco468-2, LEco621-2, and LEco1068-2) were applied, and stronger signals were obtained relative to the single-substitution LNA/DNA probes. The fluorescence intensity of LEco621-2 surpassed that of Eub338 (Table 1). By substituting three bases, LEco468-3 yielded stronger signals than that of Eub338 (Table 1) and LEco1068-3 was close to Eub338. Finally, LEco1068-4 (four substitutions) yielded the same fluorescence intensity as did Eub338 (Table 1). Ultimately, after the substitution of two to four bases in the probes, the relative intensities of Eco468, Eco621, and Eco1068 increased by 22-, 8.6-, and 2.3-fold, respectively.
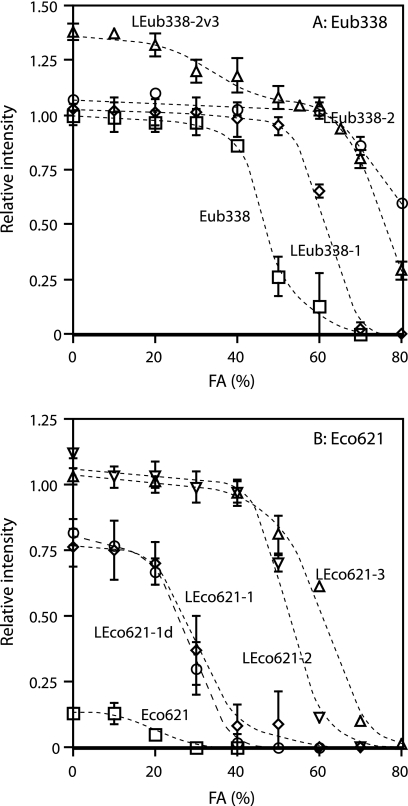
Next we investigated the positional effects of LNA substitutions. It is known that substitutions with isolated LNA residues in central positions have a greater effect on Tm than substitutions clustered at termini (7, 19). The following probes were compared: LEco468-2 versus LEco468-2p, LEco621-1 versus LEco621-1p, LEco1068-2 versus LEco1068-2p1 versus LEco1068-2p2, LEco1068-3 versus LEco1068-3p, and LEub338-2 versus LEub338-2v3 (Table 1). It was found that probes having LNA substitutions at different positions yielded different intensities, but the effect was less than that of an additional LNA substitution (Table 1). Interestingly, under low-stringency hybridization conditions, nonspecific hybridization was observed for LEub338-2v3 but not LEub338-2 (Fig. 3). This suggests that specificity might be influenced by the positions of the LNA substitutions. LEub338-2v3 was able to hybridize specifically under high-stringency hybridization conditions (see below).
FIG. 3.
Dissociation curves of Eub338 and Eco621 sequences. Formamide (FA) concentration in hybridization buffer was increased instead of elevating hybridization temperature. (A) Eub338 sequence. Eub338, squares; LEub338-1, diamonds; LEub338-2, circles; and LEub338-2v3, triangles. (B) Eco621 sequence. Eco621, squares; LEco621-1, diamonds; LEco621-1d, circles; LEco621-2, inverted triangles; and LEco621-3, triangles.
Effects of LNA substitutions on probe affinity.
Next, we investigated whether probe affinities were enhanced by substituting with LNA. Since affinity directly correlates with Tm, changes in affinity can be estimated by examining the Tm shift. Nevertheless, hybridization at high and various temperatures is not recommended because of the adverse effects of high temperatures on the samples and the technical difficulty of setting up such an experiment. Therefore, in this study, we changed the formamide concentration in the hybridization buffer and determined the formamide concentration that yields 50% probe dissociation (FAd; the FAd is equivalent to the Td of the probe). Determined values of FAd for all probes are listed in Table 1, and dissociation curves of the two representative sequences (Eub338 and Eco621) are shown in Fig. 3. The augmentation of the FAd of each probe was observed with an increasing number of LNA substitutions. The increment was 5 to 20% for any single substitution. These results confirmed that the affinities of probes were improved by LNA substitutions, resulting in enhanced fluorescence. Although the FAd increased with substituted LNA number in Eub338, little enhancement in intensity was observed (Table 1 and Fig. 3). In addition, the series of GAM42a and LEco621-2 and LEco621-3 showed behavior similar to that of Eub338 (Table 1).
Effects of LNA substitutions on probe specificity.
Since substitutions of LNA bases for DNA bases brought about significant improvements in in situ hybridization efficiency, the effects of LNA substitutions on probe specificity were investigated. First, to examine the discrimination ability of LNA/DNA probes against single-mismatch sequences, the GAM42a probe, which is specific for the Gammaproteobacteria (E. coli) and has a single mismatch relative to Betaproteobacteria (C. testosteroni), was employed (22). As previously indicated, without the competitor probe BET42a, it was difficult for the GAM42a probe to differentiate E. coli from C. testosteroni under hybridization conditions that did not result in a reduction of fluorescence intensity (22). The LNA/DNA probes LGAM42a-1 and LGAM42a-2 have LNA substitutions at the mismatch position (Table 1). Likewise, we could not distinguish between E. coli and C. testosteroni with the LNA/DNA probes without a loss of signal intensity. Hybridization with the competitor probes LBET42a-1 for LGAM42a-1 and LBET42a-2 for LGAM42a-2 successfully differentiated E. coli from C. testosteroni without compromising signal intensity.
Next, the Eub338 probe, which is specific for most of the domain Bacteria but has two mismatches relative to A. thermophila, a member of the Chloroflexi subphylum group I (33), was employed. As was previously described elsewhere (33), Eub338 did not hybridize to A. thermophila. Likewise, the LNA/DNA probes LEub338-1, LEub338-2, and LEub338-2v3 resulted in no signals from A. thermophila under the optimized hybridization conditions for each probe.
DISCUSSION
Improvement of in situ hybridization efficiency.
The results presented here demonstrate that the introduction of LNA nucleotides into DNA probes is useful for improving the efficiency of hybridization with rRNA targets. Three probe sequences, Eco468, Eco621, and Eco1068, evaluated in this study showed dramatic signal enhancements after substituting two to four LNA bases for DNA bases. The difference was attributable to increased probe affinity for the target sequence, which was further verified by elevation of the FAd.
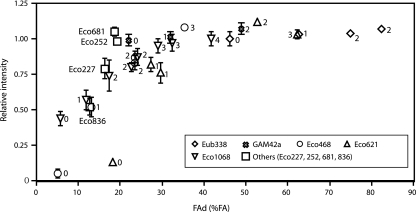
One significant finding in this study is that the number of LNA substitutions in the oligonucleotide is the most important factor for improving hybridization efficiency. In each tested probe sequence, the number of LNA nucleotides correlated with the fluorescence intensity before the signal reached a plateau (Fig. 4). In addition, the number of LNA nucleotides correlated with the FAd (Table 1). From these results, we observed that the fluorescence intensity correlated with the FAd and reached a plateau when the FAd was higher than approximately 35% (Fig. 4). In other words, the hybridization efficiency of probes having a FAd of over 35% formamide must be high. This observation is in agreement with the study of Yilmaz and Noguera (42), in which elongated probes were found to improve hybridization efficiency but generally increase FAd. On the other hand, it was difficult to predict the hybridization efficiency of LNA/DNA probes according to the concept of ΔG°overall proposed by Yilmaz and Noguera (42) because the thermodynamic values of LNA substitutions are available to date for only the LNA/DNA-DNA hybrid (23) (data not shown). If additional information, such as the thermodynamic effects of LNA substitutions on LNA/DNA-RNA hybrid, were available, the theoretical prediction of hybridization efficiency using thermodynamic parameters might be possible, which could be helpful for determining the number of LNA substitutions in oligonucleotides.
FIG. 4.
Relationship between FAd and relative intensities of the probes at 0% formamide (FA) used in this study. The number of LNA nucleotides is indicated for each probe. Diamonds, Eub338 sequence; crosses, GAM42a sequence; circles, Eco468 sequence; triangles, Eco621 sequence; inverted triangles, Eco1068 sequence; and squares, Eco227, Eco252, Eco681, and Eco836.
Besides the LNA used in this study, attempts have been made to improve hybridization efficiency by using artificial nucleotide analogs, which have higher binding affinity than DNA. Peptide nucleic acid (PNA) (Fig. 1) is well known for its applications to the FISH technique (28, 36). Because of the uncharged backbone of PNA, PNA probes show higher affinities than do DNA probes, with increased sequence selectivity (9). However, PNA probe synthesis is limited in terms of probe length and sequence. Therefore, it is sometimes difficult to design PNA probes with sufficient specificities (28). On the other hand, although the number of substitutable LNA nucleotides in a probe is limited, there are no other restrictions for designing LNA/DNA probes. Hence, LNA/DNA probes offer added design flexibility relative to PNA probes.
The nucleotide analog 2′-O-methyl RNA (Fig. 1) can also be used for probe synthesis. 2′-O-methyl RNA probes show improved hybridization efficiency for extracted rRNA (21). However, LNA has a greater effect on Tm than does 2′-O-methyl RNA (15, 19), indicating an advantage of LNA substitutions for significant improvement in probe affinity.
In addition to utilizing artificial nucleotide analogs, Yilmaz and Noguera reported an improvement of hybridization efficiency by probe elongation (42). However, LNA/DNA probes have several advantages over elongated probes. One advantage is the added flexibility of probe design. The elongation may restrict the available sequences for which a probe could be designed. Another advantage of LNA/DNA probes is accessibility. Yilmaz and Noguera showed that elongated probes hybridized insufficiently after short hybridization times, possibly due to limited accessibility to target sites (42). On the other hand, hybridization for 3 h was sufficient without formamide for the LNA/DNA probes used in this study.
Specificity of LNA/DNA probes.
The specificity of oligonucleotide probes was investigated by using the GAM42a and Eub338 probes. In this study, LNA substitutions were carried out at the mismatch positions (Table 1). Practically, LNA/DNA probes can be readily used for in situ hybridization as hybridization with competitor probes allowed easy discrimination for single mismatches without a loss of signal intensity. Furthermore, two mismatches can easily be discriminated without competitor probes.
The effects of LNA substitutions on mismatch discrimination have been investigated. The sequence selectivity of the LNA/DNA oligonucleotides is, if anything, improved compared to that of the corresponding DNA oligonucleotides (18, 24). Furthermore, LNA oligonucleotides show increased ΔTms between the fully complementary sequence and the one-mismatch sequence compared to those of the DNA oligonucleotides (13, 14, 24). Thus, substituting multiple LNA residues might result in higher specificities. However, LNA/DNA probes in which LNA and DNA nucleotides alternate showed strong nonspecific binding due to greatly increased affinity and it was difficult to determine the FAd and the optimum hybridization conditions for these probes (data not shown). The number of LNA substitutions needs to be controlled in order to maximize both the hybridization efficiency and the specificity.
Conclusion and future perspectives.
From these results, we can recommend an approach to probe design that will enhance hybridization efficiency. The oligonucleotide should have a dissociation point higher than 35% formamide, which can normally be achieved by one to four LNA substitutions in a probe of typical length (i.e., 18 to 25 bases). The positions of the LNA substitutions within the oligonucleotide and the continuity of LNA substitutions (7, 19) may be important in some cases. Additional aspects of LNA/DNA probe design, such as the effects of LNA substitutions on probe secondary structure, will require further study. It is, however, recommended that LNA substitutions at self-complementary regions should be avoided due to strong base pairing in LNA-LNA hybrids (17, 32).
Our results also suggest that LNA/DNA probes will be applicable for the identification of microorganisms in natural environments. LNA/DNA probes targeting variable regions of 16S rRNA, where hybridization efficiency is often low (11), would be designable without compromising specificity. Furthermore, the introduction of LNA residues in oligonucleotides probably improves hybridization efficiency for not only rRNA but also mRNA and DNA. LNA/DNA probes would be useful for the detection of mRNA and genes on chromosomes with oligonucleotide probes.
Acknowledgments
We thank Masahiro Okawara at Nagaoka University of Technology for his technical assistance. Yuji Sekiguchi at the National Institute of Advanced Industrial Science and Technology and Madan Tandukar at Nagaoka University of Technology are acknowledged for reading the manuscript.
This study was supported financially by research grants from the New Energy and Industrial Technology Development Organization (NEDO), the Ministry of Education, Culture, Sports, Science and Technology, Japan, and the Japan Society for the Promotion of Science.
REFERENCES
- 1.Amann, R. I., B. M. Fuchs, and S. Behrens. 2001. The identification of microorganisms by fluorescence in situ hybridisation. Curr. Opin. Biotechnol. 12:231-236. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behrens, S., B. M. Fuchs, F. Mueller, and R. Amann. 2003. Is the in situ accessibility of the 16S rRNA of Escherichia coli for Cy3-labeled oligonucleotide probes predicted by a three-dimensional structure model of the 30S ribosomal subunit? Appl. Environ. Microbiol. 69:4935-4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behrens, S., C. Rühland, J. Inácio, H. Huber, Á. Fonseca, I. Spencer-Martins, B. M. Fuchs, and R. Amann. 2003. In situ accessibility of small-subunit rRNA of members of the domains Bacteria, Archaea, and Eucarya to Cy3-labeled oligonucleotide probe. Appl. Environ. Microbiol. 69:1748-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bondensgaard, K., M. Petersen, S. K. Singh, V. K. Rajwanshi, R. Kumar, J. Wengel, and J. P. Jacobsen. 2000. Structural studies of LNA:RNA duplexes by NMR: conformations and implications for RNase H activity. Chemistry 6:2687-2695. [DOI] [PubMed] [Google Scholar]
- 7.Braasch, D. A., S. Jensen, Y. Liu, K. Kaur, K. Arar, M. A. White, and D. R. Corey. 2003. RNA interference in mammalian cells by chemically-modified RNA. Biochemistry 42:7967-7975. [DOI] [PubMed] [Google Scholar]
- 8.DeLong, E. F., G. S. Wickham, and N. R. Pace. 1989. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science 243:1360-1363. [DOI] [PubMed] [Google Scholar]
- 9.Egholm, M., O. Buchardt, L. Christensen, C. Behrens, S. M. Freier, D. A. Driver, R. H. Berg, S. K. Kim, B. Norden, and P. E. Nielsen. 1993. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen-bonding rules. Nature 365:566-568. [DOI] [PubMed] [Google Scholar]
- 10.Fuchs, B. M., K. Syutsubo, W. Ludwig, and R. Amann. 2001. In situ accessibility of Escherichia coli 23S rRNA to fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 67:961-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuchs, B. M., G. Wallner, W. Beisker, I. Schwippl, W. Ludwig, and R. Amann. 1998. Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 64:4973-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishii, K., M. Mußmann, B. J. MacGregor, and R. Amann. 2004. An improved fluorescence in situ hybridization protocol for the identification of bacteria and archaea in marine sediments. FEMS Microbiol. Ecol. 50:203-212. [DOI] [PubMed] [Google Scholar]
- 13.Jacobsen, N., J. Bentzen, M. Meldgaard, M. H. Jakobsen, M. Fenger, S. Kauppinen, and J. Skouv. 2002. LNA-enhanced detection of single nucleotide polymorphisms in the apolipoprotein E. Nucleic Acids Res. 30:e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacobsen, N., M. Fenger, J. Bentzen, S. L. Rasmussen, M. H. Jakobsen, J. Fenstholt, and J. Skouv. 2002. Genotyping of the apolipoprotein B R3500Q mutation using immobilized locked nucleic acid capture probes. Clin. Chem. 48:657-660. [PubMed] [Google Scholar]
- 15.Kierzek, E., A. Ciesielska, K. Pasternak, D. H. Mathews, D. H. Turner, and R. Kierzek. 2005. The influence of locked nucleic acid residues on the thermodynamic properties of 2′-O-methyl RNA/RNA heteroduplexes. Nucleic Acids Res. 33:5082-5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kloosterman, W. P., E. Wienholds, E. de Bruijn, S. Kauppinen, and R. H. Plasterk. 2006. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat. Methods 3:27-29. [DOI] [PubMed] [Google Scholar]
- 17.Koshkin, A. A., P. Nielsen, M. Meldgaard, V. K. Rajwanshi, S. K. Singh, and J. Wengel. 1998. LNA (locked nucleic acid): an RNA mimic forming exceedingly stable LNA:LNA duplexes. J. Am. Chem. Soc. 120:13252-13253. [Google Scholar]
- 18.Koshkin, A. A., S. K. Singh, P. Nielsen, V. K. Rajwanshi, R. Kumar, M. Meldgaard, C. E. Olsen, and J. Wengel. 1998. LNA (locked nucleic acids): synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedron 54:3607-3630. [Google Scholar]
- 19.Kurreck, J., E. Wyszko, C. Gillen, and V. A. Erdmann. 2002. Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res. 30:1911-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Latorra, D., K. Campbell, A. Wolter, and J. M. Hurley. 2003. Enhanced allele-specific PCR discrimination in SNP genotyping using 3′ locked nucleic acid (LNA) primers. Hum. Mutat. 22:79-85. [DOI] [PubMed] [Google Scholar]
- 21.Majlessi, M., N. C. Nelson, and M. M. Becker. 1998. Advantages of 2′-O-methyl oligoribonucleotide probes for detecting RNA targets. Nucleic Acids Res. 26:2224-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manz, W., R. Amann, W. Ludwig, M. Wagner, and K.-H. Schleifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 23.McTigue, P. M., R. J. Peterson, and J. D. Kahn. 2004. Sequence-dependent thermodynamic parameters for locked nucleic acid (LNA)-DNA duplex formation. Biochemistry 43:5388-5405. [DOI] [PubMed] [Google Scholar]
- 24.Mouritzen, P., A. T. Nielsen, H. M. Pfundheller, Y. Choleva, L. Kongsbak, and S. Møller. 2003. Single nucleotide polymorphism genotyping using locked nucleic acid (LNA). Expert Rev. Mol. Diagn. 3:27-38. [DOI] [PubMed] [Google Scholar]
- 25.Nelson, P. T., D. A. Baldwin, W. P. Kloosterman, S. Kauppinen, R. H. Plasterk, and Z. Mourelatos. 2006. RAKE and LNA-ISH reveal microRNA expression and localization in archival human brain. RNA 12:187-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Obika, S., D. Nanbu, Y. Hari, J.-I. Andoh, K.-I. Morio, T. Doi, and T. Imanishi. 1998. Stability and structural features of the duplexes containing nucleoside analogues with a fixed N-type conformation, 2′-O,4′-C-methyleneribonucleosides. Tetrahedron Lett. 39:5401-5404. [Google Scholar]
- 27.Obika, S., D. Nanbu, Y. Hari, K.-I. Morio, Y. In, T. Ishida, and T. Imanishi. 1997. Synthesis of 2′-O,4′-C-methyleneuridine and -cytidine. Novel bicyclic nucleosides having a fixed C3-endo sugar puckering. Tetrahedron Lett. 38:8735-8738. [Google Scholar]
- 28.Perry-O'Keefe, H., S. Rigby, K. Oliveira, D. Sørensen, H. Stender, J. Coull, and J. J. Hyldig-Nielsen. 2001. Identification of indicator microorganisms using a standardized PNA FISH method. J. Microbiol. Methods 47:281-292. [DOI] [PubMed] [Google Scholar]
- 29.Petersen, M., K. Bondensgaard, J. Wengel, and J. P. Jacobsen. 2002. Locked nucleic acid (LNA) recognition of RNA: NMR solution structures of LNA:RNA hybrids. J. Am. Chem. Soc. 124:5974-5982. [DOI] [PubMed] [Google Scholar]
- 30.Petersen, M., C. B. Nielsen, K. E. Nielsen, G. A. Jensen, K. Bondensgaard, S. K. Singh, V. K. Rajwanshi, A. A. Koshkin, B. M. Dahl, J. Wengel, and J. P. Jacobsen. 2000. The conformations of locked nucleic acids (LNA). J. Mol. Recognit. 13:44-53. [DOI] [PubMed] [Google Scholar]
- 31.Petersen, M., and J. Wengel. 2003. LNA: a versatile tool for therapeutics and genomics. Trends Biotechnol. 21:74-81. [DOI] [PubMed] [Google Scholar]
- 32.Schmidt, K. S., S. Borkowski, J. Kurreck, A. W. Stephens, R. Bald, M. Hecht, M. Friebe, L. Dinkelborg, and V. A. Erdmann. 2004. Application of locked nucleic acids to improve aptamer in vivo stability and targeting function. Nucleic Acids Res. 32:5757-5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sekiguchi, Y., H. Takahashi, Y. Kamagata, A. Ohashi, and H. Harada. 2001. In situ detection, isolation, and physiological properties of a thin filamentous microorganism abundant in methanogenic granular sludges: a novel isolate affiliated with a clone cluster, the green non-sulfur bacteria, subdivision I. Appl. Environ. Microbiol. 67:5740-5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Silahtaroglu, A., H. Pfundheller, A. Koshkin, N. Tommerup, and S. Kauppinen. 2004. LNA-modified oligonucleotides are highly efficient as FISH probes. Cytogenet. Genome Res. 107:32-37. [DOI] [PubMed] [Google Scholar]
- 35.Silahtaroglu, A. N., N. Tommerup, and H. Vissing. 2003. FISHing with locked nucleic acids (LNA): evaluation of different LNA/DNA mixmers. Mol. Cell. Probes 17:165-169. [DOI] [PubMed] [Google Scholar]
- 36.Stender, H., K. Lund, K. H. Petersen, O. F. Rasmussen, P. Hongmanee, H. Miörner, and S. E. Godtfredsen. 1999. Fluorescence in situ hybridization assay using peptide nucleic acid probes for differentiation between tuberculous and nontuberculous mycobacterium species in smears of mycobacterium cultures. J. Clin. Microbiol. 37:2760-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suggs, S. V., T. Hirose, T. Miyake, E. H. Kawashima, and M. J. Johnson. 1981. Use of synthetic oligodeoxyribonucleotides for the isolation of specific cloned DNA sequences, p. 683-963. In D. Brown and C. F. Fox (ed.), Developmental biology using purified genes. Academic Press, Inc., New York, N.Y.
- 38.Thomsen, R., P. S. Nielsen, and T. H. Jensen. 2005. Dramatically improved RNA in situ hybridization signals using LNA-modified probes. RNA 11:1745-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tolstrup, N., P. S. Nielsen, J. G. Kolberg, A. M. Frankel, H. Vissing, and S. Kauppinen. 2003. OligoDesign: optimal design of LNA (locked nucleic acid) oligonucleotide capture probes for gene expression profiling. Nucleic Acids Res. 31:3758-3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vester, B., and J. Wengel. 2004. LNA (locked nucleic acid): high-affinity targeting of complementary RNA and DNA. Biochemistry 43:13233-13241. [DOI] [PubMed] [Google Scholar]
- 41.Wienholds, E., W. P. Kloosterman, E. Miska, E. Alvarez-Saavedra, E. Berezikov, E. de Bruijn, H. R. Horvitz, S. Kauppinen, and R. H. Plasterk. 2005. MicroRNA expression in zebrafish embryonic development. Science 309:310-311. [DOI] [PubMed] [Google Scholar]
- 42.Yilmaz, L. S., and D. R. Noguera. 2004. Mechanistic approach to the problem of hybridization efficiency in fluorescent in situ hybridization. Appl. Environ. Microbiol. 70:7126-7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yilmaz, L. S., H. E. Ökten, and D. R. Noguera. 2006. Making all parts of the 16S rRNA of Escherichia coli accessible in situ to single DNA oligonucleotides. Appl. Environ. Microbiol. 72:733-744. [DOI] [PMC free article] [PubMed] [Google Scholar]