Abstract
Cry1Ab toxin binding analysis was performed to determine whether resistance in laboratory-selected Ostrinia nubilalis strains is associated with target site alteration. Brush border membrane vesicles were prepared using dissected midguts from late instars of susceptible and resistant strains (Europe-R and RSTT) of O. nubilalis. Immunoblot analysis indicated that three different proteins bound to Cry1Ab toxin and were recognized by an anticadherin serum. In a comparison of resistant and susceptible strains, reduced Cry1Ab binding was apparent for all three bands corresponding to cadherin-like proteins in the Europe-R strain, while reduced binding was apparent in only one band for the RSTT strain. Real-time analysis of Cry1Ab binding to gut receptors using surface plasmon resonance suggested slight differences in affinity in both resistant strains. Additional binding analysis was conducted using 125I-labeled Cry1Ab, Cry1Ac, and Cry1Aa. Slight differences were again observed between the resistant and susceptible strains for Cry1Ab binding. However, when binding of 125I-labeled Cry1Aa was tested, a 10-fold reduction in the concentration of binding sites was observed in the Europe-R strain. Expression of the O. nubilalis cadherin gene was similar in both the resistant and susceptible strains and did not account for differences in binding. In combination, the results of the present work suggest that differences in susceptibility to Cry1A toxins in the Europe-R strain of O. nubilalis are associated with altered receptor binding, although the precise nature of this mechanism is still uncertain.
Lepidopteran insects are particularly susceptible to Cry1 toxins from Bacillus thuringiensis (Berliner) (Bt toxins), which are highly toxic after ingestion. They are solubilized and enzymatically processed by proteinases present in the insect midgut to an active protein core. Active toxins bind specifically to midgut receptors, causing the formation of lytic pores after toxin conformational change and insertion into the membrane (11, 23, 36, 38) and, ultimately, death. Although this general process is widely accepted, details on the intoxication process, such as postbinding events and receptor specificity, are still unclear (12, 48).
Despite their effectiveness for over half a century, resistance development to Bt-based insecticides has become a growing public concern, particularly with regard to transgenic crops that express Bt toxins. To prevent or at least delay resistance and sustain this new technology, deployment strategies are being developed that minimize selective pressures or reduce the selective advantage of resistance genes. Although a number of species have been successfully selected in the laboratory for resistance to Bt toxins (reviewed by Ferré and Van Rie [8]), high levels of resistance to Bt toxins and cross-resistance in the field have been reported only in Plutella xylostella (8, 43) and in glasshouse populations of Trichoplusia ni (19). In most cases, the mechanism of resistance has been proposed to involve altered toxin binding to specific receptors in the insect midgut (8, 49).
Changes in receptor conformation that interfere with toxin binding through substitution or loss of amino acids represent the most likely mechanism of resistance to Bt toxins (17). However, conclusive evidence is generally lacking to support such a mechanism. Recently, however, disruption of a cadherin superfamily gene by retrotransposon-mediated insertion has been linked to high levels of resistance to Cry1Ac in Heliothis virescens (10). The predicted amino acid sequence from the resistant gene does not possess the trans-membrane or toxin binding domains, which would prevent binding and confer high levels of resistance. In other research with this species, altered glycosylation of microvillus proteins has been associated with reduced toxin binding and pore formation as well as increased resistance to Cry1 toxins (21). In the pink bollworm, Pectinophora gossypiella, three cadherin-like alleles were associated with resistance to Cry1Ac and conferred survival to transgenic Bt cotton (34).
The European corn borer, Ostrinia nubilalis (Hübner), is a significant pest of corn throughout North America (31). Since its initial introduction in 1996, transgenic Bt corn has increased in popularity among growers for managing corn borer infestations. Resistance to B. thuringiensis subsp. kurstaki in O. nubilalis has been reported in laboratory-selected strains exposed to the Bt formulation Dipel ES (18) and to the isolated Cry1Ac (3) and Cry1Ab (5, 39) toxins. A resistance mechanism has been partially characterized only in the Dipel-selected strain, in which differences in midgut proteases were linked to resistance (26, 29).
We have selected O. nubilalis laboratory strains for resistance to Cry1Ab protoxin and observed significant levels of resistance to this toxin and cross-resistance to Cry1Ac after 7 to 10 generations (5, 39). Resistance to Cry1Ab-activated toxin in these strains was at least 100-fold (40) after 40 generations of selection. Comparisons of midgut protease activities between resistant and susceptible strains revealed no consistent differences (40). This observation, along with the narrow cross-resistance pattern, suggested that resistance in these selected strains of O. nubilalis may involve modified midgut receptors. Therefore, the objective of the present study was to determine if altered binding is associated with Cry1Ab resistance in selected O. nubilalis strains.
MATERIALS AND METHODS
O. nubilalis strains.
Populations of O. nubilalis designated Europe-S, Europe-R, and RSTT-R were established as laboratory strains. The European strain was established in 1993 from approximately 500 O. nubilalis larvae collected in the Lombardia region of northern Italy and was provided to the University of Nebraska after 20 generations of laboratory rearing. This strain was divided into two subpopulations, one exposed throughout development to Cry1Ab protoxin from the B. thuringiensis subsp. kurstaki strain HD1-9, which produces only Cry1Ab protein (Europe-R), and the other reared in the absence of protoxin (Europe-S). The RSTT-R strain resulted from a combination of individuals from the Europe-R selection line and from a selection line from insects collected in Nebraska (39). All the resistant strains were selected with Cry1Ab protoxin.
Toxin preparation.
For radioligand binding, Cry1Aa, Cry1Ab, and Cry1Ac protoxins were obtained from recombinant B. thuringiensis EG1273, EG7077, and EG11070 strains, respectively (Ecogen Inc., Langhorn, PA). Each B. thuringiensis strain was grown at 29°C for 48 h in CCY medium (42) supplemented with 10 μg/ml of tetracycline for strains EG1273 and EG7077 and 3 μg/ml of chloramphenicol for strain EG11070. Spores and crystals were collected by centrifugation at 9,700 × g at 4°C for 10 min. Pellets were washed four times with 1 M NaCl-10 mM EDTA and suspended in 10 mM KCl. Crystals were solubilized in 50 mM sodium carbonate buffer (pH 10.5) containing 10 mM dithiothreitol. Protoxins were trypsin activated (trypsin type XI from bovine pancreas; Sigma Chemical Co., St. Louis, MO) at 37°C, 2 h (1 mg trypsin per 10 mg of protoxin). Finally, activated toxins were purified by anion-exchange chromatography as described by Estela et al. (7). The protein concentration in the preparations of Bt toxins was measured by the method of Bradford (4) using the Bio-Rad protein assay (Bio-Rad Laboratories GmgH, Munchen, Germany), with bovine serum albumin (BSA) as a protein standard.
The trypsin-activated Cry1Ab toxin used for immunoblotting and real-time binding kinetics was provided by Monsanto Co. (St. Louis, MO). The Cry1Ab protoxin was treated with bovine pancreatic trypsin and purified in a 1.2-liter Q Sepharose Fast Flow column. The trypsin-resistant core protein was eluted as a single symmetrical peak by using a gradient of sodium chloride. Peak tubes were pooled and dialyzed against 50 mM carbonate/bicarbonate buffer, pH 10.25, with 50 mM sodium chloride. The activated Cry1Ab protein was recovered at 94% purity.
BBMV preparation.
Brush border membrane vesicles (BBMV) were prepared with gut tissue dissected from ice-chilled early fifth-instar larvae. The midguts were pulled gently from the larvae after both the last three abdominal segments and the head plus thorax were removed. A small glass culture tube was rolled over the length of the gut to displace the gut contents. Dissected gut tissue was transferred to a centrifuge tube containing ice-cold MET buffer (300 mM mannitol, 17 mM Tris-HCl pH 7.5, 5 mM EGTA, protease inhibitor [Complete EDTA-free protease inhibitors cocktail; Roche Applied Science, Indianapolis, IN]), vigorously vortexed, and briefly centrifuged for 5 min at 1,000 ×g to obtain the clean midguts. Guts were either frozen at −80°C or processed immediately by the differential magnesium precipitation method of Wolfersberger et al. (46). Briefly, gut tissues were homogenized on ice in a tight-fitting glass Dounce homogenizer in ice-cold MET buffer (10%, wt/vol). The homogenate was diluted with an equal volume of ice-cold 24 mM MgCl2, blended, and held on ice for 15 min before centrifugation. A low-speed centrifugation (2,500 × g for 15 min at 4°C) was used to pellet heavier cell debris, and the supernatant from the initial centrifugation was further centrifuged at 30,000 × g for 30 min at 4°C. The resulting pellet was resuspended in MET buffer and centrifuged again at 2,500 × g and then at 30,000 × g for 30 min at 4°C. The resulting pellet, which corresponded to the BBMV preparation, was resuspended in HBS-N buffer (10 mM HEPES, pH 7.4, 150 mM NaCl), flash-frozen in liquid nitrogen, and stored at −80°C until use. The protein concentration of the BBMV preparations was determined by the bicinchoninic acid method (41). Alkaline phosphatase and aminopeptidase activities were used as marker enzymes to track purification of the BBMV preparation and were 8 to 12 times higher in BBMV preparations than in the initial homogenates (data not shown).
To prepare BBMV for radioligand binding analysis, fifth-instar larvae were collected, frozen in liquid nitrogen, preserved in dry ice, and sent to the University of Valencia in Spain. Upon arrival, larvae were dissected in ice-cold MET buffer without protease inhibitors. BBMV were prepared essentially as above, and the pellet corresponding to BBMV was suspended in 0.5× MET buffer.
Immunoblotting.
Western blot assays of Cry1Ab binding to BBMV proteins were performed using a chemiluminescence Western Light kit (Tropix, Inc., Bedford, MA). Equal amounts (80 μg) of BBMV protein from each strain were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis as described by Laemmli (24), electroblotted onto a polyvinylidene difluoride (PVDF) membrane (Bio-Rad Inc., Hercules, CA) for 90 min by using a Mini Trans-Blot electrophoretic transfer cell (Bio-Rad, Hercules, CA), and blocked for 2 h at room temperature with phosphate-buffered saline (PBS; pH 8.0) containing 5% nonfat dry milk powder, 5% glycerol, 0.5% Tween 20 (freshly made). The PVDF membrane was then incubated with activated Cry1Ab (250 ng/ml) in blocking buffer overnight at 4°C and subsequently washed three times with blocking buffer. The blot was then incubated with polyclonal rabbit anti-Cry1Ab (1:2,500; provided by Monsanto Co., St. Louis, MO), washed three times, incubated with goat anti-rabbit-alkaline phosphatase (secondary antibody at 1:10,000), and washed three times with blocking buffer. The PVDF membrane was washed with assay buffer (Tropix Inc., Bedford, MA), and detection was performed with the CDP-Star chemiluminescence kit (Tropix Inc., Bedford, MA) using a fluorescence imager (Fluor-S Imager; Bio-Rad, Hercules, CA).
Western blot assays for cadherin-like proteins were performed as described above, except that a polyclonal anticadherin serum (1:3,000 at room temperature for 1 h; provided by Pioneer Hi-Bred International, Johnston, IA) developed from a 1.8-kb region of the O. nubilalis cadherin-like gene, which included the cadherin repeats 10, 11, and 12 of the protein, was used. The membrane was washed as described above, and an anti-rabbit-alkaline phosphatase serum conjugate (1:10,000 at room temperature for 1 h) was applied to the PVDF membrane. After repeated washings with assay buffer (Tropix Inc., Bedford, MA), the membrane was incubated with CDP-Star for 5 min and the image was captured as described above.
Real-time binding kinetics.
Small unilamellar vesicles (SUV) were prepared from O. nubilalis BBMV in HBS-N buffer by extrusion (30). Briefly, BBMV proteins were diluted to a concentration of 0.5 mg/ml, homogenized, and passed four times through a 400-nm polycarbonate filter (Avestin, Inc., Ottawa, Canada) in a Liposofast Basic extrusion apparatus (Avestin, Inc., Ottawa, Canada). The resulting liposome solution was subsequently passed four times through a 100-nm polycarbonate filter and finally passed 12 times through a 50-nm polycarbonate filter to give a translucent solution. Samples were aliquoted to 50 μl, frozen in liquid nitrogen, and stored at −80°C until use.
The surface plasmon resonance (SPR) detector (BIAcore 2000 system) and sensor chips (Pioneer L1) were purchased from BIAcore Inc. (Piscataway, NJ). The running and diluting buffer in all experiments was HBS-N. The methods for SPR analysis of Cry1Ab binding to O. nubilalis SUV were based on those described by Masson et al. (32, 33) and Cooper et al. (6). All the experiments were performed at 25°C, and the L1 chip was conditioned with two short pulses of detergent solution (40 mM n-octyl-β-d-glucopyranoside, 0.1 M NaOH) at 100 μl/min prior to attachment of SUV. Vesicles (0.5 mg/ml) were immobilized onto the lipophilic-modified dextran surface of the chip. Approximately 1,000 resonance units were reproducibly captured onto the chip surface after a short pulse (1 min) with NaOH (10 mM) at 30 μl/min to create more stability in the vesicle surface. This immobilization level was the minimum necessary to initiate analyte binding in these analyses. The vesicle surface was subsequently blocked with a 5-min pulse of BSA (0.1 mg/ml) and held until a stable baseline was obtained.
For real-time analysis using SPR, the activated Cry1Ab toxin was dialyzed against HBS-N (10 mM HEPES, pH 7.4, 150 mM NaCl) overnight at 4°C and then centrifuged at 16,000 × g for 10 min. The pooled supernatant was analyzed using a bicinchoninic acid assay to determine the protein concentration (41).
Binding kinetics analysis was performed by injecting activated Cry1Ab toxin at 30 μl/min for 200 s, followed by a 300-s dissociation phase. Cry1Ab (dissolved in HBS-N) concentrations varied from 100 nM up to 1,000 nM for the susceptible strain and from 500 nM up to 1,750 nM for resistant strains. At the end of each experiment, the chip surface was regenerated with two pulses of detergent solution (40 mM n-octyl-β-d-glucopyranoside, 0.1 M NaOH). A reference flow cell without BBMV immobilization was used in all experiments to avoid bulk effects (the SPR response caused by the difference in bulk refractive index between the sample and the running buffer). The experimentation was performed at least three times with two samples for each colony.
All sensorgram data were analyzed with the BIAevaluation software version 3.0 (BIAcore Inc., Piscataway NJ), by local fitting. The software directly generated parameters for a simple binding model of one analyte to one ligand. The model equation to describe the interaction can be written as dR/dt = konC(Rmax − Rt) − koffRt, where R is the response, dR/dt is the rate of toxin-receptor complex formation, C is the concentration of injected toxin, Rmax − Rt is the amount of free remaining receptor sites at time t (expressed in resonance units), kon is the association rate constant (expressed as M−1 s−1), and koff is the dissociation rate constant (expressed as s−1) (22). All models were evaluated for goodness of fit of the data to the Lingmuir (1:1) model for both association and dissociation rate constants based on a chi-square value of <10.0. Models were also verified by residual plots, which calculated the differences between the observed and the fitted curves for each data point.
125I labeling and binding assays.
Trypsin-activated Cry1Aa, Cry1Ab, and Cry1Ac were labeled with 125I by the method of chloramine-T as previously described (45). The specific radioactivities were 9.22 mCi/mg, 5.11 mCi/mg, and 110 mCi/mg for labeled Cry1Aa, Cry1Ab, and Cry1Ac, respectively. To determine the appropriate concentration of BBMV for competition assays, vesicles from each strain (0 to 0.25 mg of total vesicle protein/ml) were incubated with 81 pM [125I]Cry1Aa, 67 pM [125I]Cry1Ab, or 3 pM [125I]Cry1Ac in 100 μl of PBS (pH 7.4) containing 0.1% BSA (PBS-BSA) at room temperature for 60 min. The toxin bound to BBMV was separated from free toxin by centrifugation at 16,000×g at 4°C for 10 min. The pellet was washed twice with 500 μl of ice-cold PBS-BSA. The radioactivity in the pellet was then measured in a 1282 Compugamma CS Universal gamma counter (LKB Wallac Pharmacia, Turku, Finland) and taken as total binding. Nonspecific binding was estimated by adding a 10,000-fold excess of unlabeled toxin to the reaction mixture. Specific binding was calculated as the difference between total and nonspecific binding.
For competition assays, increasing amounts of unlabeled Cry1Aa, Cry1Ab, or Cry1Ac toxins were added to the reaction mixture containing each labeled toxin and 100 μg/ml BBMV in PBS-BSA for the experiments performed with labeled Cry1Aa or 50 μg/ml for the experiments performed with labeled Cry1Ab and Cry1Ac. The equilibrium dissociation constant (Kd) and binding site concentration (Rt) were estimated with the LIGAND software (35). Statistical tests (analysis of variance) were performed and charts were made using GraphPad Prism version 4.02 for Windows (GraphPad Software, San Diego, CA.). All binding experiments were independently performed at least twice.
Total RNA and mRNA extraction from gut tissue and Northern blotting.
Fourth-instar O. nubilalis larvae were used for dissection of gut tissue as described previously. Dissected guts were frozen in liquid nitrogen until further processing. Total RNA was isolated using the TRIzol reagent (Sigma, St. Louis, MO). Approximately 70 mg of tissue from each strain was ground in liquid nitrogen and quickly processed with TRIzol according to the manufacturer's instructions (Sigma, St. Louis, MO) to generate total RNA. Extracted RNA was diluted in Tris-HCl buffer (5 mM, pH 8.0), quantified spectrophotometrically using the absorbance ratio of >1.8 at 260/280 nm, and stored at −80°C until further use.
Fresh total RNA obtained as described above from O. nubilalis gut tissue was used for Northern blotting analysis. A cDNA probe (700 bp) was generated using primers (Fw, 5′-CCCACATTTTCATCATCGTC-3′; Rv, 5′-CATCCTCCAGTTCTATCACC-3′) designed from the O. nubilalis cadherin gene (GenBank accession number AX147207). The probe was labeled with digoxigenin (DIG) using the DIG PCR labeling kit (Roche Applied Science, Indianapolis, IN). Samples of total RNA (7.5 μg) from resistant and susceptible strains were separated by denaturing electrophoresis on a 1.2% agarose-formaldehyde gel. The RNAs were transferred overnight onto positively charged nylon membranes (Roche Applied Science, Indianapolis, IN) by capillary blotting using diethyl pyrocarbonate-treated 20× SSC transfer buffer (3.0 M NaCl, 0.3 M sodium citrate, pH 7.0) and irreversibly fixed in a baking oven at 80°C for 2 h. The blot was prehybridized for 1 h at 50°C in 5 ml of hybridization solution (DIG Easy hyb; Roche Applied Science, Indianapolis, IN). After adding the denatured probe to a fresh hybridization solution, the membrane was hybridized overnight at 50°C with continuous shaking. The blot was rinsed three times with continuous shaking in 2× SSC, 0.1% SDS at room temperature and three times with 0.5× SSC, 0.1% SDS at 68°C to eliminate nonspecific hybridizations. The membrane was washed briefly in washing buffer (maleate buffer, 0.3% Tween 20), blocked for 1 h in 0.5× blocking buffer (Roche Applied Science, Indianapolis, IN), and then incubated with anti-DIG at a 1:15,000 dilution in blocking buffer for 45 min. The membrane was washed four times (15 min each) with washing buffer, briefly washed with detection buffer (Roche Applied Science, Indianapolis, IN) for 5 min, and incubated with the chemiluminescent substrate for alkaline phosphatase (CSPD; Tropix Inc., Bedford, MA) for 5 min. The blot was exposed to chemiluminescence Biomax film (Kodak, Rochester, NY) at room temperature for 25 min. The membrane was stripped and reprobed with the O. nubilalis DIG-labeled RPS3 probe (600 bp) to standardize loadings.
RESULTS
Immunoblot assays.
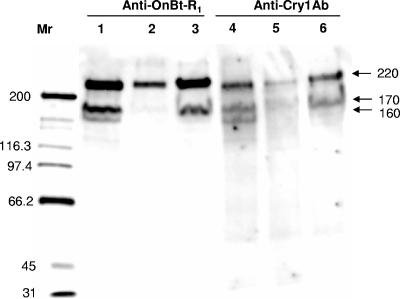
Immunoblot assays to identify potential Cry1Ab binding proteins using both anti-Cry1Ab and anti-OnBt-R1 serum are presented in Fig. 1. Signals from both Cry1Ab ligand blotting and the anticadherin Western blotting were compared among the susceptible and resistant strains. The Cry1Ab toxin bound more strongly to the 220-kDa band than the 170-kDa and 160-kDa bands (Fig. 1, lane 4) in the susceptible strain, based on signal intensities. A similar pattern was observed in the Western blot assay using the O. nubilalis anticadherin serum, with the 220-kDa band showing the strongest signal in the susceptible strain (Fig. 1, lane 1). Similar Cry1Ab binding proteins were observed in the two resistant strains; however, signal intensity was considerably reduced in the Europe-R strain compared to the susceptible strain (Fig. 1, lane 5) for all three bands. Reduction in signal intensity was also observed in this strain when the membrane was probed with the anticadherin serum (Fig. 1, lane 2). The 220-kDa protein was the only band clearly visible in the Europe-R strain for both Cry1Ab and anticadherin binding. The RSTT-R strain showed clearly reduced binding of the 160-kDa protein and a slight reduction of binding for the 220-kDa and 170-kDa proteins for both Cry1Ab and anticadherin serum. No bands were visible in the controls performed without Cry1Ab and anticadherin serum (data not shown).
FIG. 1.
Binding of anticadherin (anti-OnBt-R1) from O. nubilalis and Cry1Ab toxin to BBMV proteins (80 μg) from one susceptible and two Cry1Ab-selected strains. Lanes: 1 and 4, Europe-S; 2 and 5, Europe-R; 3 and 6, RSTT-R. Cry1Ab toxin was detected using a specific antibody against it.
Real-time binding kinetics.
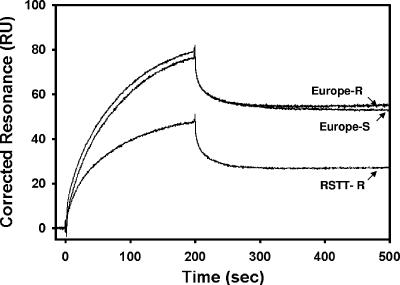
Cry1Ab binding to O. nubilalis SUV preparations as measured by SPR and differences in binding among strains are documented in the sensorgrams presented in Fig. 2. Kinetics analysis and affinity constants determined for susceptible and resistant strains are presented in Table 1. Significant differences were observed in the Cry1Ab association rates (kon) between Europe-S and both resistant strains, whereas the RSTT-R strain exhibited higher rates of Cry1Ab dissociation (koff) relative to both Europe-S and Europe-R. Overall, significant differences existed in equilibrium dissociation constants (Kd) between the Europe-S strain (35 ± 3 nM) and resistant Europe-R (61 ± 6 nM) and RSTT-R (94 ± 11 nM) strains, conferring at least a twofold lower Cry1Ab binding affinity in both resistant strains.
FIG. 2.
Representative sensorgram of Cry1Ab interacting with SUV from the O. nubilalis susceptible strain (Europe-S) and two Cry1Ab-selected strains (Europe-R and RSTT-R).
TABLE 1.
Cry1Ab kinetic parameters and equilibrium dissociation constants from Cry1Ab-susceptible and -resistant European corn borer, Ostrinia nubilalis, strains using real-time analysis (Biacore 2000)a
| Strain | kon (M−1 s−1) (103) | koff (s−1) (10−4) | Kd (nM) |
|---|---|---|---|
| Europe-S | 10.7 ± 0.3 a | 3.7 ± 0.4 a | 35 ± 3 a |
| Europe-R | 9.7 ± 0.3 b | 5.9 ± 0.5 a | 61 ± 6 b |
| RSTT-R | 10.03 ± 0.04 b | 9.5 ± 1.1 b | 94 ± 11 c |
Values are means ± standard deviations from three replicates (three batches of SUV). Means in a column that are followed by the same letter were not statistically different (least significance difference test, P < 0.05).
Binding with 125I-labeled toxins.
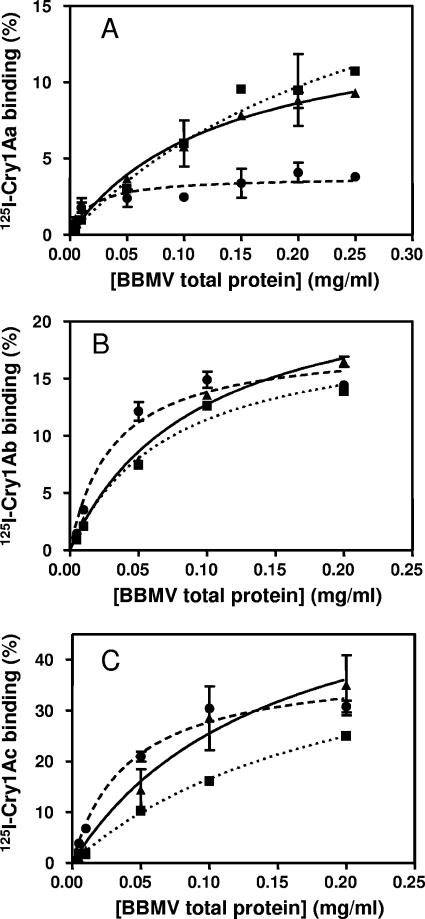
Experiments performed to determine the appropriate amount of BBMV to use in competition binding assays showed that Cry1Aa, Cry1Ab, and Cry1Ac bound specifically to BBMV from both susceptible and resistant larvae (Fig. 3). The results showed that labeled Cry1Aa reached a much lower level of maximum binding (4%) in the Europe-R strain than the Europe-S or RSTT-R strains (9%) (Fig. 3A). In contrast, assays performed with labeled Cry1Ab and Cry1Ac showed that the maximum binding of these toxins was similar among strains (Fig. 3B and C).
FIG. 3.
Specific binding of 125I-labeled toxin to increasing concentrations of BBMV from O. nubilalis. (A) Cry1Aa; (B) Cry1Ab; (C) Cry1Ac. ▴ and solid line, Europe-S; • and dashed line, Europe-R; ▪ and dotted line, RSTT-R.
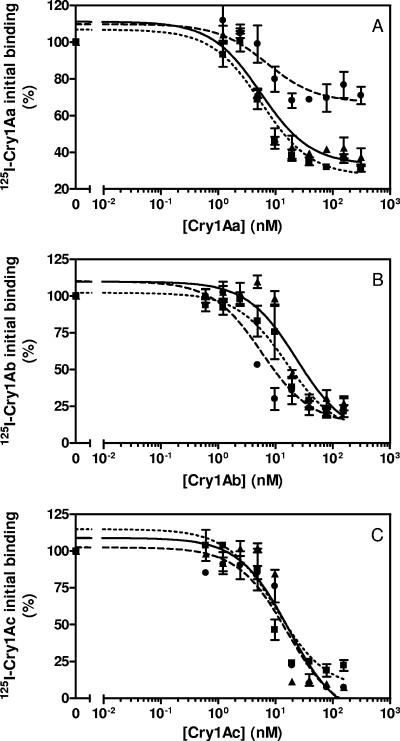
Homologous competition binding experiments (when binding of a labeled ligand is competed with the same unlabeled ligand) were performed to obtain quantitative data on binding parameters. Assays performed with labeled Cry1Aa showed a main difference in Europe-R compared to the other two strains (Fig. 4A). Quantitative analysis of the data revealed a 10-fold decrease in concentration of binding sites (Rt) (P < 0.05) (Table 2), whereas equilibrium dissociation constants (Kd) were not significantly different among strains (P > 0.05) (Fig. 4A; Table 2). The Europe-R strain also showed significant differences relative to the other two strains when tested with labeled Cry1Ab (Fig. 4B; Table 2). Kd was around 4-fold lower (i.e., higher affinity) (P < 0.05) and Rt was between 2- and 4-fold higher than in the other two strains (P < 0.05), which indicated an increase in overall affinity (Rt/Kd ratio) of at least 10-fold (Table 2). No significant changes in binding parameters were observed among strains when using labeled Cry1Ac (Fig. 4C; Table 2).
FIG. 4.
Binding of 125I-labeled toxins to BBMV from O. nubilalis strains at increasing concentrations of unlabeled competitor. (A) Cry1Aa; (B) Cry1Ab; (C) Cry1Ac. ▴ and solid line, Europe-S; • and dashed line, Europe-R; ▪ and dotted line, RSTT-R.
TABLE 2.
Binding parameters estimated from homologous competition experiments performed with labeled Cry toxins and BBMV from susceptible and resistant strains of O. nubilalisa
| Strain | Ligand | Kd (mean ± SD) (nM) | Rt (mean ± SD) (pmol/mg)b | Rt/Kd |
|---|---|---|---|---|
| Europe-S | Cry1Aa | 3.28 ± 0.02 | 1.8 ± 0.2 | 0.55 |
| Europe-R | 4 ± 3 | 0.2 ± 0.1 | 0.05 | |
| RSTT-R | 3.0 ± 0.7 | 2.3 ± 0.4 | 0.77 | |
| Europe-S | Cry1Ab | 10 ± 4 | 3.6 ± 0.9 | 0.36 |
| Europe-R | 2.37 ± 0.05 | 13 ± 3 | 5.5 | |
| RSTT-R | 11.9 ± 0.1 | 6.8 ± 0.3 | 0.57 | |
| Europe-S | Cry1Ac | 5 ± 1 | 23 ± 2 | 4.6 |
| Europe-R | 5.0 ± 0.8 | 49 ± 9 | 9.8 | |
| RSTT-R | 5 ± 3 | 30 ± 10 | 6.0 |
Values are the means and standard deviations of at least two replicates.
Rt is expressed in pmoles of binding sites per milligram of total vesicle protein.
Northern blotting.
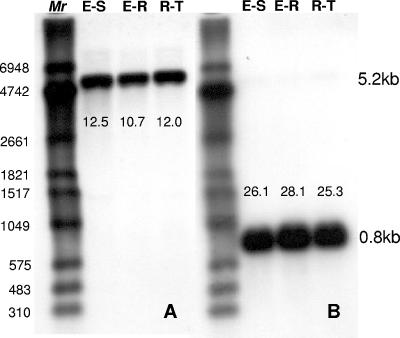
Results of Northern blotting experiments performed with a probe specific to the O. nubilalis cadherin gene are shown in Fig. 5. A single 5.2-kb mRNA band hybridized with the probe in all the strains tested (Fig. 5A). Signal intensity for the cadherin-like transcripts was similar among the selected and susceptible strains. RPS3 signal, used to standardize sample loading, was also very similar (Fig. 5B), suggesting that the cadherin gene product in all the strains was similarly expressed.
FIG. 5.
Northern analysis of OnBt-R1 expression in Cry1Ab-susceptible and -resistant O. nubilalis strains. A positive nylon membrane containing total RNA (7.5 μg/lane) was probed with DIG-labeled oligonucleotide corresponding to the OnBt-R1 gene fragment. Lanes 1, 2, and 3 represent Europe-S (E-S), Europe-R (E-R), and RSTT-R (R-T), respectively. (A) Cadherin-probe hybridization to a 5.2-kb mRNA band. (B) The membrane was stripped and reprobed with the DIG-RPS3 probe for normalization, with hybridization to a 0.8-kb mRNA band. Mr DIG-labeled RNA marker II (Roche Applied Science, Indianapolis, IN).
DISCUSSION
The key step in the specific action of Bt toxins is the binding to brush border membrane receptors (38, 45, 49). It has also been shown that alteration of binding of Bt toxins is associated with high levels of resistance (for review, see references 8, 13, 14, 21, and 37). Aminopeptidase N and cadherin-like proteins, among others, have been proposed as Bt toxin receptors in insect midguts (15, 20, 48). However, resistance to Cry1A toxins has been associated only with the cadherin receptor gene in both H. virescens (10) and P. gossypiella (34).
Bt toxin resistance in O. nubilalis has thus far only been characterized in a strain that was selected with Dipel. This strain showed a broad spectrum of resistance to Bt protoxins, being highly resistant to all Bt protoxins present in Dipel (Cry1Aa, Cry1Ab, Cry1Ac, and Cry2Aa), cross-resistant to Cry1Ba protoxin (27), and only marginally resistant to activated Cry1Ab (26). Resistance was associated with reduction of a trypsin-like protease involved in activation of the Bt protoxin form (26, 29), and no alteration in Cry1Ab and Cry1Ac binding was found in the resistant insects (28). Because the Dipel formulation contains multiple toxins, it is unclear whether such a resistance mechanism would evolve in response to selection from a single toxin.
In the present work, it was hypothesized that receptor binding differences would exist between susceptible (Europe-S) and Cry1Ab-selected (Europe-R and RSTT-R) strains of O. nubilalis. Higher resistance of the selected strains to activated Cry1Ab compared to Cry1Ab protoxin (39, 40) and a narrow spectrum of cross-resistance were observed in earlier studies (39). Additionally, there were no differences in activities of luminal gut proteases with either model substrates or with protoxin and activated Cry1Ab toxin (40). Together these results suggest that altered toxin binding to midgut receptors might be associated with resistance.
Using standard immunoblotting techniques, three proteins were found to bind Cry1Ab in the susceptible strain (Europe-S). Furthermore, the same proteins were recognized by the O. nubilalis cadherin antiserum, suggesting that they belong to the cadherin protein superfamily (9). Alternatively, the smaller bands that were recognized by the cadherin antiserum might represent degradation peptides or processed forms from the 220-kDa protein. Because the Europe-R strain exhibited a similar profile but with drastically reduced signal intensity, the results suggest that resistance in this strain is associated with a reduced concentration of the cadherin receptor. In contrast, the RSTT-R strain showed only a slight decrease in signal intensity compared with the susceptible strain with both the anticadherin antibody and with Cry1Ab.
Based on results from immunoblotting, we expected to find a marked reduction in binding of Cry1Ab to SUV from the Europe-R strain. Detailed analysis of binding kinetics using SPR indicated only slight differences among strains. The RSTT-R strain, which showed only minor differences in both the Cry1Ab ligand blot and Western blot assays using the anticadherin antibody, exhibited greater differences in Cry1Ab binding relative to the Europe-R strain. However, these minor differences appear unlikely to completely explain the resistance to Cry1Ab associated with this strain. It is worth noting that, in a previous study with a P. xylostella Cry1A-resistant strain, a study carried out with SPR did not show any major difference in binding of Cry1Ac (33), while binding analysis using 125I-labeled Cry1Ac indicated a drastic reduction of toxin binding (44).
Binding analysis with 125I-labeled Cry1Ab to BBMV from the Europe-R strain showed an increase in the overall affinity of this toxin with an almost fourfold increase in concentration of binding sites. However, when binding of 125I-labeled Cry1Aa was tested, a 10-fold reduction in the concentration of binding sites was found. No changes were observed using 125I-labeled Cry1Ac. With the RSTT-R strain, no differences with the Europe-S strain were found with either of the 125I-labeled Cry1A toxins.
Similar results to those found in the Europe-R strain have been reported for the YHD2 colony of H. virescens, in which altered Cry1Aa binding was associated with resistance to Cry1Ac and Cry1Ab, although differences in binding were detected only for Cry1Aa (25). A mutation in the cadherin receptor gene has been associated with resistance in the YHD2 strain (10). There are other examples in which resistance to Cry1A toxins has been associated with reduced binding of one toxin without measurable effects on others. In P. gossypiella strain AZP-R, resistance to Cry1A toxins was associated with lack of binding of Cry1Ab but not of Cry1Aa or Cry1Ac (13). This strain was shown to carry three mutant alleles of the cadherin gene, which was shown to be linked to resistance to Cry1A toxins (34). Other examples in which lack of binding of just one Cry1A toxin has been associated with resistance to the three Cry1A toxins have also been reported in P. xylostella (2, 14, 47). We hypothesize that the above examples reflect binding of some Cry1A toxins to BBMV proteins other than those involved in the in vivo toxic action. It is well known that Cry1Ac (but not Cry1Aa or Cry1Ab) has a high affinity for GalNAc residues and that aminopeptidase-N contains such residues (7, 16). Binding to these residues could mask a lack of binding to other “effective” receptors.
It should be noted that differences in 125I-labeled Cry1Aa binding in Europe-R were associated with differences in concentrations of binding sites rather than with affinity. This is in agreement with the results obtained with the anticadherin antibody and with Cry1Ab binding blot assays. However, the lower concentration of the cadherin protein was not supported by the results obtained on transcript abundance of the cadherin gene, suggesting that the final reduction in cadherin protein concentration must be due to posttranslational processing.
In combination, the results of the present work suggest that differences in susceptibility to Cry1A toxins in the Europe-R strain of O. nubilalis are associated with altered receptor binding, as shown by the reduced concentration of cadherin receptors, although the precise nature of this mechanism is still unclear. To our knowledge, this is the first time that an alteration in the cadherin receptor has been shown in a Bt-resistant strain at the protein level. The resistance alleles contributing to altered receptor binding in the Europe-R strain must be present in RSTT-R due to the mixed origin of the founder insects in the latter. However, this mechanism was not clearly observed in RSTT-R, suggesting that other factors may have a more important contribution to resistance in this strain. Inheritance experiments with these strains have indicated that resistance is associated with multiple loci and, therefore, suggest a complex resistance mechanism (1). Receptor isolation and further characterization will be important to clarify the differences between the strains analyzed in this study.
Acknowledgments
We are indebted to David G. Heckel for helpful suggestions on this work and to Terence Spencer for insect rearing.
Support for this project at the University of Nebraska was provided by Pioneer Hi-Bred International and a USDA Risk Avoidance and Mitigation Program grant (no. 2002-51101-01981), as well as by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (Brazil). The work at the University of Valencia was supported by an EU project (QLRT-2001-01969).
This paper is contribution no. 15002 of the Journal Series of the Nebraska Agricultural Experiment Station and contribution no. 1238 of the Department of Entomology.
REFERENCES
- 1.Alves, A. P., T. Spencer, B. E. Tabashnik, and B. D. Siegfried. 2006. Inheritance of resistance to the Cry1Ab Bacillus thuringiensis toxin in Ostrinia nubilalis (Lepidoptera:Crambidae). J. Econ. Entomol. 99:494-501. [DOI] [PubMed] [Google Scholar]
- 2.Ballester, V., F. Granero, B. E. Tabashnik, T. Malvar, and J. Ferré. 1999. Integrative model for binding of Bacillus thuringiensis toxins in susceptible and resistant larvae of the diamondback moth (Plutella xylostella). Appl. Environ. Microbiol. 65:1413-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bolin, P. C., W. D. Hutchison, and D. A. Andow. 1999. Long-term selection for resistance to Bacillus thuringiensis Cry1Ac endotoxin in a Minnesota population of European corn borer (Lepidoptera: Crambidae). J. Econ. Entomol. 92:1021-1030. [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Chaufaux, J., M. Seguin, J. J. Swanson, D. Bourguet, and B. D. Siegfried. 2001. Chronic exposure of the European corn borer (Lepidoptera: Crambidae) to CrylAb Bacillus thuringiensis toxin. J. Econ. Entomol. 94:1564-1570. [DOI] [PubMed] [Google Scholar]
- 6.Cooper, M. A., J. Carroll, E. R. Travis, D. H. Williams, and D. J. Ellar. 1998. Bacillus thuringiensis Cry1Ac toxin interaction with Manduca sexta aminopeptidase N in a model membrane environment. Biochem. J. 333:677-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Estela, A., B. Escriche, and J. Ferré. 2004. Interaction of Bacillus thuringiensis toxins with larval midgut binding sites of Helicoverpa armigera (Lepidoptera: Noctuidae). Appl. Environ. Microbiol. 70:1378-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferré, J., and J. Van Rie. 2002. Biochemistry and genetics of insect resistance to Bacillus thuringiensis. Annu. Rev. Entomol. 47:501-533. [DOI] [PubMed] [Google Scholar]
- 9.Flannagan, R. D., C. G. Yu, J. P. Mathis, T. E. Meyer, X. M. Shi, H. A. A. Siqueira, and B. D. Siegfried. 2005. Identification, cloning and expression of a Cry1Ab cadherin receptor from European corn borer, Ostrinia nubilalis (Hubner) (Lepidoptera: Crambidae). Insect Biochem. Mol. Biol. 35:33-40. [DOI] [PubMed] [Google Scholar]
- 10.Gahan, L. J., F. Gould, and D. G. Heckel. 2001. Identification of a gene associated with Bt resistance in Heliothis virescens. Science 293:857-860. [DOI] [PubMed] [Google Scholar]
- 11.Gill, S. S., E. A. Cowles, and P. V. Pietrantonio. 1992. The mode of action of Bacillus thuringiensis endotoxins. Annu. Rev. Entomol. 37:615-636. [DOI] [PubMed] [Google Scholar]
- 12.Gómez, I., J. Sánchez, R. Miranda, A. Bravo, and M. Soberón. 2002. Cadherin-like receptor binding facilitates proteolytic cleavage of helix alpha-1 in domain I and oligomer pre-pore formation of Bacillus thuringiensis Cry1Ab toxin. FEBS Lett. 513:242-246. [DOI] [PubMed] [Google Scholar]
- 13.González-Cabrera, J., B. Escriche, B. E. Tabashnik, and J. Ferré. 2003. Binding of Bacillus thuringiensis toxins in resistant and susceptible strains of pink bollworm (Pectinophora gossypiella). Insect Biochem. Mol. 33:929-935. [DOI] [PubMed] [Google Scholar]
- 14.González-Cabrera, J., S. Herrero, and J. Ferré. 2001. High genetic variability for resistance to Bacillus thuringiensis toxins in a single population of diamondback moth. Appl. Environ. Microbiol. 67:5043-5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffitts, J. S., and R. V. Aroian. 2005. Many roads to resistance: how invertebrates adapt to Bt toxins. BioEssays 27:614-624. [DOI] [PubMed] [Google Scholar]
- 16.Hayakawa, T., Y. Shitomi, K. Miyamoto, and H. Hori. 2004. GalNAc pretreatment inhibits trapping of Bacillus thuringiensis Cry1Ac on the peritrophic membrane of Bombyx mori. FEBS Lett. 576:331-335. [DOI] [PubMed] [Google Scholar]
- 17.Heckel, D. G. 1994. The complex genetic basis of resistance to Bacillus thuringiensis toxin in insects. Biocontrol. Sci. Technol. 4:405-417. [Google Scholar]
- 18.Huang, F. N., R. A. Higgins, and L. L. Buschman. 1997. Baseline susceptibility and changes in susceptibility to Bacillus thuringiensis subsp. kurstaki under selection pressure in European corn borer (Lepidoptera: Pyralidae). J. Econ. Entomol. 90:1137-1143. [Google Scholar]
- 19.Janmaat, A. F., and J. Myers. 2003. Rapid evolution and the cost of resistance to Bacillus thuringiensis in greenhouse populations of cabbage loopers, Trichoplusia ni. Phil. R. Soc. Lond. B Biol. 270:2263-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jurat-Fuentes, J. L., and M. J. Adang. 2004. Characterization of a Cry1Ac-receptor alkaline phosphatase in susceptible and resistant Heliothis virescens larvae. Eur. J. Biochem. 271:3127-3135. [DOI] [PubMed] [Google Scholar]
- 21.Jurat-Fuentes, J. L., F. L. Gould, and M. J. Adang. 2002. Altered glycosylation of 63- and 68-kilodalton microvillar proteins in Heliothis virescens correlates with reduced Cry1 toxin binding, decreased pore formation, and increased resistance to Bacillus thuringiensis Cry1 toxins. Appl. Environ. Microbiol. 68:5711-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karlsson, R., A. Michaelsson, and L. Mattsson. 1991. Kinetic analysis of monoclonal antibody-antigen interactions with a new biosensor based analytical system. J. Immunol. Methods 145:229-240. [DOI] [PubMed] [Google Scholar]
- 23.Knowles, B. H. 1994. Mechanism of action of Bacillus thuringiensis insecticidal delta-endotoxins. Adv. Insect Physiol. 24:275-308. [Google Scholar]
- 24.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 25.Lee, M. K., F. Rajamohan, F. Gould, and D. H. Dean. 1995. Resistance to Bacillus thuringiensis Cry1a delta-endotoxins in a laboratory-selected Heliothis virescens strain is related to receptor alteration. Appl. Environ. Microbiol. 61:3836-3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, H., B. Oppert, R. A. Higgins, F. Huang, L. L. Buschman, J.-R. Gao, and K. Y. Zhu. 2005. Characterization of cDNAs encoding three trypsin-like proteinases and mRNA quantitative analysis in Bt-resistant and -susceptible strains of Ostrinia nubilalis. Insect Biochem. Mol. 35:847-860. [DOI] [PubMed] [Google Scholar]
- 27.Li, H., B. Oppert, R. A. Higgins, F. Huang, L. L. Buschman, and K. Y. Zhu. 2005. Susceptibility of Dipel-resistant and -susceptible Ostrinia nubilalis (Lepidoptera: Crambidae) to individual Bacillus thuringiensis protoxins. J. Econ. Entomol. 98:1333-1340. [DOI] [PubMed] [Google Scholar]
- 28.Li, H. R., J. González-Cabrera, B. Oppert, J. Ferré, R. A. Higgins, L. L. Buschman, G. A. Radke, K. Y. Zhu, and F. N. Huang. 2004. Binding analyses of Cry1Ab and Cry1Ac with membrane vesicles from Bacillus thuringiensis-resistant and -susceptible Ostrinia nubilalis. Biochem. Biophys. Res. Commun. 323:52-57. [DOI] [PubMed] [Google Scholar]
- 29.Li, H. R., B. Oppert, R. A. Higgins, F. N. Huang, K. Y. Zhu, and L. L. Buschman. 2004. Comparative analysis of proteinase activities of Bacillus thuringiensis-resistant and -susceptible Ostrinia nubilalis (Lepidoptera: Crambidae). Insect Biochem. Mol. Biol. 34:753-762. [DOI] [PubMed] [Google Scholar]
- 30.MacDonald, R. C., R. I. MacDonald, B. P. M. Menco, K. Takeshita, N. K. Subbarao, and L.-R. Hu. 1991. Small-volume extrusion apparatus for preparation of large, unilamellar vesicles. BBA Biomembranes 1061:297-303. [DOI] [PubMed] [Google Scholar]
- 31.Mason, C. E., M. E. Rice, D. D. Calvin, J. W. Van Duyn, W. B. Showers, W. D. Hutchison, J. F. Witkowski, R. A. Higgins, D. W. Onstad, and G. P. Dively. 1996. European corn borer: ecology and management. North central regional extension publication 327. Iowa State University, Ames.
- 32.Masson, L., A. Mazza, and R. Brousseau. 1994. Stable immobilization of lipid vesicles for kinetic-studies using surface-plasmon resonance. Anal. Biochem. 218:405-412. [DOI] [PubMed] [Google Scholar]
- 33.Masson, L., A. Mazza, R. Brousseau, and B. Tabashnik. 1995. Kinetics of Bacillus thuringiensis toxin binding with brush-border membrane vesicles from susceptible and resistant larvae of Plutella xylostella. J. Biol. Chem. 270:11887-11896. [DOI] [PubMed] [Google Scholar]
- 34.Morin, S., R. W. Biggs, M. S. Sisterson, L. Shriver, C. Ellers-Kirk, D. Higginson, D. Holley, L. J. Gahan, D. G. Heckel, Y. Carriere, T. J. Dennehy, J. K. Brown, and B. E. Tabashnik. 2003. Three cadherin alleles associated with resistance to Bacillus thuringiensis in pink bollworm. Proc. Natl. Acad. Sci. USA 100:5004-5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munson, P. J., and D. Rodbard. 1980. LIGAND: a versatile computerized approach for characterization of ligand-binding systems. Anal. Biochem. 107:220-239. [DOI] [PubMed] [Google Scholar]
- 36.Rajamohan, F., M. K. Lee, and D. H. Dean. 1998. Bacillus thuringiensis insecticidal proteins: molecular mode of action. Prog. Nucleic Acid Res. 60:1-27. [DOI] [PubMed] [Google Scholar]
- 37.Sayyed, A. H., B. Raymond, M. S. Ibiza-Palacios, B. Escriche, and D. J. Wright. 2004. Genetic and biochemical characterization of field-evolved resistance to Bacillus thuringiensis toxin Cry1Ac in the diamondback moth, Plutella xylostella. Appl. Environ. Microbiol. 70:7010-7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schnepf, E., N. Crickmore, J. Van Rie, D. Lereclus, J. Baum, J. Feitelson, D. R. Zeigler, and D. H. Dean. 1998. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol. Mol. Biol. Rev. 62:775-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siqueira, H. A. A., D. Moellenbeck, T. Spencer, and B. D. Siegfried. 2004. Cross-resistance of CrylAb-selected Ostrinia nubilalis (Lepidoptera: Crambidae) to Bacillus thuringiensis delta-endotoxins. J. Econ. Entomol. 97:1049-1057. [DOI] [PubMed] [Google Scholar]
- 40.Siqueira, H. A. A., K. W. Nickerson, D. Moellenbeck, and B. D. Siegfried. 2004. Activity of gut proteinases from Cry1Ab-selected colonies of the European corn borer, Ostrinia nubilalis (Lepidoptera: Crambidae). Pest Manag. Sci. 60:1189-1196. [DOI] [PubMed] [Google Scholar]
- 41.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 42.Stewart, G. S. A. B., K. Johnstone, E. Hagelberg, and D. J. Ellar. 1981. Commitment of bacterial spores to germinate. Biochem. J. 198:101-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tabashnik, B. E., N. L. Cushing, N. Finson, and M. W. Johnson. 1990. Field development of resistance to Bacillus thuringiensis in diamondback moth (Lepidoptera, Plutellidae). J. Econ. Entomol. 83:1671-1676. [Google Scholar]
- 44.Tabashnik, B. E., N. Finson, F. R. Groeters, W. J. Moar, M. W. Johnson, K. Luo, and M. J. Adang. 1994. Reversal of resistance to Bacillus thuringiensis in Plutella xylostella. Proc. Natl. Acad. Sci. USA 91:4120-4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Van Rie, J., S. Jansens, H. Höfte, D. Degheele, and H. Mellaert. 1989. Specificity of Bacillus thuringiensis endotoxins. Importance of specific receptors on the brush border membrane of the mid-gut of target insects. Eur. J. Biochem. 186:239-247. [DOI] [PubMed] [Google Scholar]
- 46.Wolfersberger, M., P. Luethy, A. Maurer, P. Parenti, F. V. Sacchi, B. Giordana, and G. M. Hanozet. 1987. Preparation and partial characterization of amino acid transporting brush border membrane vesicles from the larval midgut of the cabbage butterfly (Pieris brassicae). Comp. Biochem. Phys. A 86:301-308. [Google Scholar]
- 47.Wright, D. J., M. Iqbal, F. Granero, and J. Ferré. 1997. A change in a single midgut receptor in the diamondback moth (Plutella xylostella) is only in part responsible for field resistance to Bacillus thuringiensis subsp. kurstaki and B. thuringiensis subsp. aizawai. Appl. Environ. Microbiol. 63:1814-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, X., M. Candas, N. B. Griko, L. Rose-Young, and L. A. Bulla. 2005. Cytotoxicity of Bacillus thuringiensis Cry1Ab toxin depends on specific binding of the toxin to the cadherin receptor BT-R1 expressed in insect cells. Cell Death Differ. 12:1407-1416. [DOI] [PubMed] [Google Scholar]
- 49.Zhuang, M. B., D. I. Oltean, I. Gómez, A. K. Pullikuth, M. Soberón, A. Bravo, and S. S. Gill. 2002. Heliothis virescens and Manduca sexta lipid rafts are involved in Cry1A toxin binding to the midgut epithelium and subsequent pore formation. J. Biol. Chem. 277:13863-13872. [DOI] [PubMed] [Google Scholar]